Abstract
Introduction
Two-dimensional strain analysis is a powerful analysis modality, however, clinical utilization has been limited by variability between different analysis systems and operators. We compared strain in adults and children using vendor-specific and vendor-independent software to evaluate variability.
Methods
One hundred and ten subjects (50/110 pediatric, 80/110 normal left ventricular function) had echocardiograms with a General Electric ultrasound scanner between September 2010 and January 2012. Left ventricular longitudinal strain was derived with EchoPAC (General Electric, v10.8.1), a vendor-specific software, and Velocity Vector Imaging (Siemens, v3.5), which is vendor-independent. Three independent readers analyzed all the echocardiograms yielding 330 datasets.
Results
Mean left ventricular global longitudinal Lagrangian strain was −18.1 ± SD 4.4% for EchoPAC and −15.3 ± SD 4.1% for Velocity Vector Imaging. Velocity Vector Imaging yielded lower absolute global longitudinal Lagrangian strain by mean 2.9 (±SD 2.7, p < 0.0001), and lower regional longitudinal strain. These differences persisted in normal subjects versus those with cardiomyopathy. Longitudinal strain differences were slightly higher in the pediatric cohort. There was no significant difference in inter-observer longitudinal strain and a small difference in intra-observer strain between analysis systems. On repeat measurements, a significant change in global longitudinal Lagrangian strain occurred after the difference exceeded 3–5 strain points for EchoPAC and Velocity Vector Imaging, respectively.
Conclusion
Velocity Vector Imaging produces lower left ventricular longitudinal strain values versus EchoPAC for the same echo images. Both systems have similar inter-observer variability, Velocity Vector Imaging slightly higher intra-observer variability. A statistically significant change in global longitudinal Lagrangian strain occurs with changes >3–5 strain points on repeat measurements. Strain values between the systems are not interchangeable.
Keywords: Two-dimensional strain echocardiography, left ventricular function, cardiac deformation, Digital Imaging and Communications in Medicine, vendor-independent software
Introduction
Two-dimensional strain echocardiography (2DSE) is a robust technique to analyze cardiac deformation and function. 2DSE calculates Lagrangian strain by tracking stable acoustic backscatter from 2D echocardiographic images through the cardiac cycle, and has been validated via in vitro and in vivo models and against tagged MRI.1–5 While 2DSE has shown promising clinical utility in several settings, widespread utilization is hampered by variability between manufacturer-specific software with different tracking algorithms. Investigation of this topic is necessary to meet the clinical demands of the future, as discussed in “A Suggested Roadmap for Cardiovascular Ultrasound Research for the Future”.6 One proposed technique to overcome vendor-dependency is to perform 2DSE analysis on images in the Digital Imaging and Communications in Medicine (DICOM) format with vendor independent software. DICOM images are derived from the native polar scan-line (raw) data and contain data in Cartesian coordinates. Syngo Velocity Vector Imaging (VVI, Siemens Medical Solutions USA, Inc., Mountain View, CA) has been validated for 2DSE analysis of DICOM data.1,5,7 However, the proprietary nature of 2D strain software makes it difficult to a priori define the level of agreement between different analysis systems. There are limited data comparing vendor-specific and vendor-independent analysis systems in a broad cohort that includes adult and pediatric patients. Less is known about variability in regional strain. Therefore, we compared LV longitudinal strain in 60 adult and 50 pediatric patients to assess agreement and variability of polar versus a DICOM-based analysis system to measure LV global longitudinal strain (GLS) and LV regional longitudinal strain (RLS).
Methods
Study design and patient population
Retrospective analysis was performed on echocardiograms obtained between September 2010 and January 2012 at the Cleveland Clinic Divisions of Cardiovascular Medicine and Pediatric Cardiology. All echocardiograms had been obtained for routine clinical evaluations using a standardized 2D strain acquisition protocol. The Institutional Review Board approved the study protocol. A total of 60 adult and 50 pediatric (<18 years of age) consecutive echocardiograms that satisfied the following criteria were analyzed.
Inclusion and exclusion criteria
All patients above the age of 12 months with four-chambered hearts and an echocardiogram suitable for 2D strain analysis were included. Only one echocardiogram was selected per subject. Suitability of the echocardiogram included: adequate visualization of the entire left ventricle including a clearly defined endocardial border, ECG tracing accompanying the 2D clip, minimum clip length of three cardiac cycles, and frame rate optimized to patient’s heart rate within a range of 60–90 frames/second. Subjects were excluded if they had complex congenital heart disease or if they had a history of cardiac surgery.
Sample size determination
We based sample size determination on the number of patients needed to show the difference in the width of the limits of agreement (LOA; as defined by Bland–Altman analysis8) between pediatric and adult subjects studied. Our sample size was calculated in order to have a power of 80% (assuming a two-tailed alpha of 0.05) to detect if one group had a 50% larger width of LOA that the other. Using Statistica 8.0 software (StatSoft Inc., Tulsa, OK) we obtained a sample size of 50 subjects per group.
Data acquisition
All echocardiograms were acquired on a General Electric (GE) Vivid 7 or Vivid E9 (General Electric Medical Systems, Horten, Norway) ultrasound platform using an S4, S5 or S8 curved-array transducer. Echo images were acquired in the apical 4-, 2- and long-axis (three chamber) views. The target frame rates were set to approximate the patient’s heart rate, within a range of 60–90 frames per second (fps). The clips, which contained a minimum of three beats, were saved in polar (raw) and DICOM formats. DICOM transformation was done at acoustic (native) frame rates without compression (i.e. default compression to 30 fps was disabled). Prior to analysis, a single cardiac cycle from each of the clips was selected to be used across observers. For both analysis systems, this was usually the R-R interval selected by the software system with a minority of cases where the R-R was manually adjusted to optimize tracking. All analyses were done independently by three trained investigators (SA, KN, AB). For intra-observer analysis strain measurements were repeated on 20 randomly selected studies (10 adult, 10 pediatric) by all three observers after several months to minimize measurement bias.
GE EchoPAC strain analysis
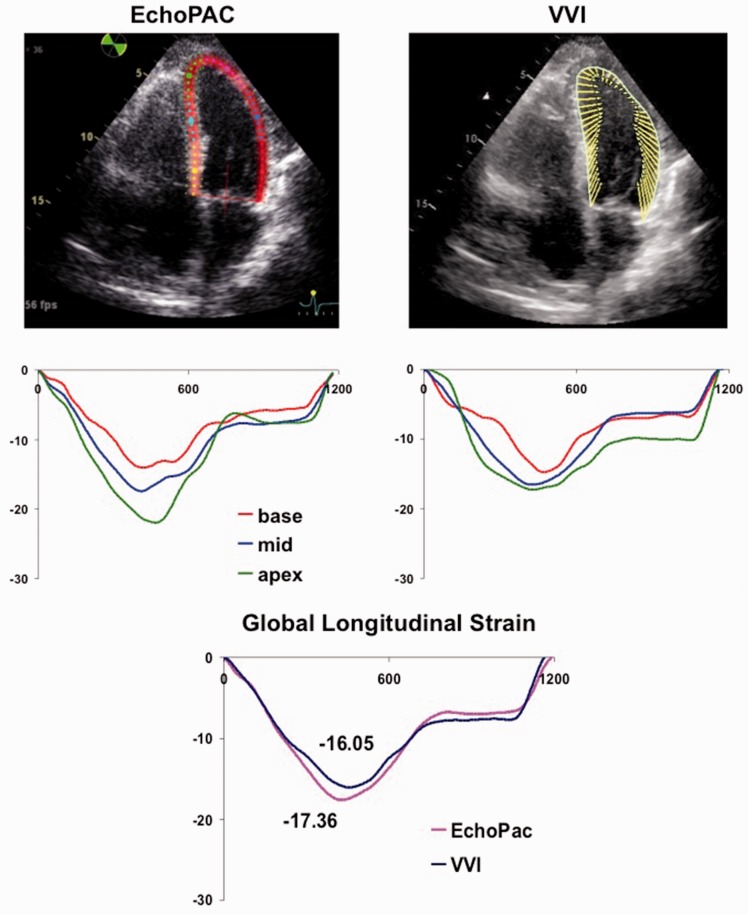
Polar raw data were analyzed using GE EchoPAC PC analysis software (GE, v10.8.1) as described.9 A region of interest (ROI) that extended over the entire LV wall thickness was constructed by initial manual placement of 3–6 points close to the endocardial perimeter, with its width and position adjusted to attain optimal tracking (Figure 1). Only a single, reference cardiac cycle was analyzed in each clip; this R-R was changed from the default in a small number of cases to optimize tracking. Following analysis of all three LV apical views, a “bulls-eye” was generated with regional strain measurements, consistent with standardized myocardial segmentation and nomenclature.10 A total of 18 regional segments were produced comparable to VVI measurements as below.
Figure 1.
Representative regional strain curves for the same patient analyzed with EchoPAC (left) and VVI (right). Global strain curve for the same patient from EchoPAC (pink line) and VVI (blue line) shown at bottom of Figure 1.
Syngo VVI strain analysis
DICOM images at native acoustic frame rates were analyzed using syngo VVI software (Siemens, v3.5) as described previously.5,11 ROI for all VVI tracings was made with a minimum of six points placed over the endocardial LV surface. While the software has the ability to perform epicardial tracking, we experienced superior tissue tracking using the endocardial-tracking feature alone. Therefore, all VVI analyses for echocardiograms were performed with the “Epi” feature disabled, with ROI represented as a thin subendocardial line (Figure 1). For the majority of studies, strain analysis was performed over three consecutive cardiac cycles for VVI studies, which was the default for the VVI software. This included the R-R interval used for EchoPAC analysis above. For a small number of studies, R-R interval was adjusted to exclude 1 or 2 cardiac cycles to optimize tracking, with the same cardiac cycle used by all observers.
For all analyses, the maximum negative strain value over the period of cardiac cycle (nadir of the strain curve) was recorded for peak longitudinal strain. Peak GLS was calculated as the average of 18 RLSs.
Statistical analysis
Continuous variables were expressed as mean ± standard deviation (SD). Relationship between polar and DICOM measurements was assessed with Pearson correlation, and agreement (bias) between techniques was evaluated with Bland–Altman analysis. t-tests were applied to assess differences between groups. All inter-technique (polar vs. DICOM) comparisons were made based on the average of three strain measurements by three observers. Variability in measurements between the two analysis systems was tested with two-way analysis of variance (ANOVA) with calculation of intra- and interobserver standard error of measurement (SEMintra, SEMinter). SEMintra expresses the random error by a typical observer, and SEMinter is a measurement of the mean variation between different observers.12 The differences in SEM between the two analysis systems were tested with a t-test specific to this scenario,13 calculated as:
where var(SEM2) = 2SEM4/v(SEM)
SEM calculations were performed assuming a random effects model.14 Thereafter, the SEM was used to calculate the minimum change that needs to occur in successive measurements in order to have a statistically significant change in that measurement, termed the minimum discernable difference (MDD). The MDD in two successive measurements made by the same (MDDintra) and different observers (MDDinter) for a 95% confidence interval (CI) was calculated as: √2 × 1.96 × SEM.14
Statistical analyses were performed on SPSS (release 11.0, SPSS Inc., Chicago, IL, USA), JMP Pro (release 9.0.0, SAS Institute Inc., Cary, NC, USA) and Microsoft Excel (release 14.2.4, Microsoft Corporation, Redmond, WA, USA). p < .05 was considered statistically significant. Although all statistical analyses were conducted on the signed GLS data (which are generally negative), for ease of discussion we will adopt the general convention of reporting the absolute value (magnitude) of the GLS measurements and their differences.
Results
Global strain
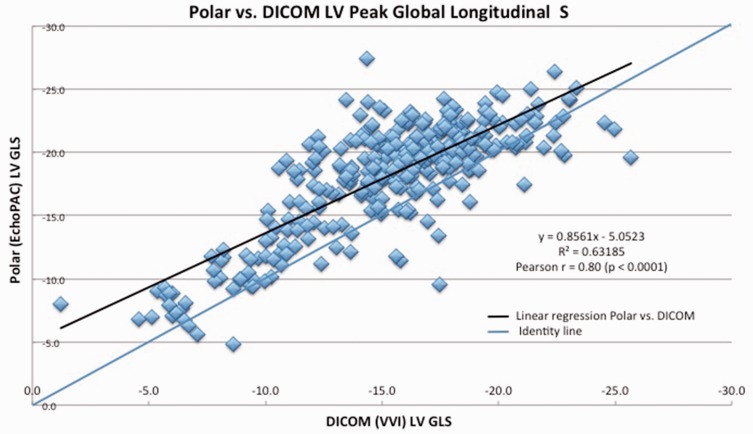
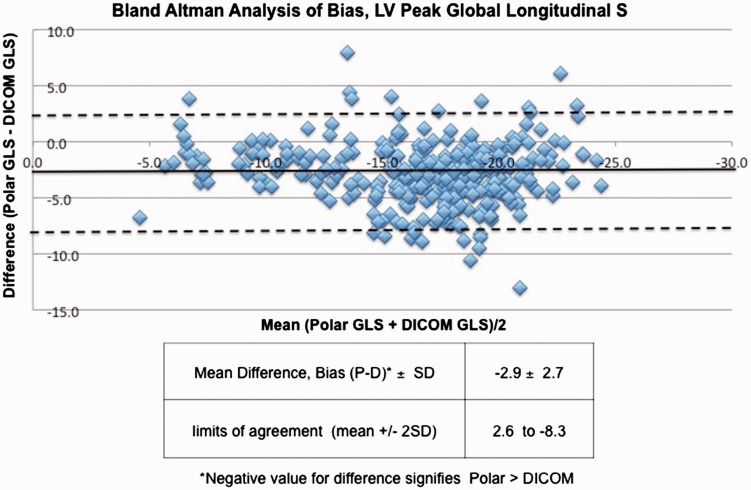
Patient characteristics are shown in Table 1. In the whole population of 110 subjects, the LV Peak GLS by EchoPAC (polar) was −18.1 ± SD 4.4% and −15.3 ± SD 4.1% with VVI (DICOM). Pearson analysis of polar and DICOM GLS yielded r = 0.8 (p < 0.0001) (Figure 2). The mean difference (EchoPAC GLS – VVI GLS) was 2.9 ± SD 2.7 % (p < 0.0001) (Figure 3), with EchoPAC producing higher absolute GLS (higher negative strain) for the entire cohort. This trend persisted when comparing EchoPAC vs VVI GLS for subjects with normal LV function versus those with cardiomyopathy (Table 2). The difference between EchoPAC and VVI was higher among pediatric subjects on Student’s t-test (p = 0.02) (Table 3).
Table 1.
Patient characteristics.
| Patient characteristics | All n = 110 | Adult n = 60 | Pediatric n = 50 |
|---|---|---|---|
| Male: female | 69:41 | 34:26 | 35:15 |
| Median age, years (range) | 32 (2–87) | 56 (19–87) | 11 (2–18) |
| Mean Heart Rate, beats/min ± SD | 76 ± 17 | 71 ± 11 | 80 ± 19 |
| Mean Frame Rate, frame/sec ± SD | 73 ± 12 | 68 ± 11 | 75 ± 12 |
| LV dimensions |
Mean ± SD (range) |
Mean ± SD (range) |
Mean ± SD (range) |
| LV End Diastolic volume, cm3/m2 | 93.7 ± 48.8 (27.1–378) | 99 ± 49.9 (49–378) | 81.1 ± 36.6 (27.1–203) |
| LV End Diastolic volume, Peds Z score | 0.0 ± 2.3 (−3.4 to 7.6) | ||
| LV End Systolic volume, cc/m2 | 44.4 ± 39.8 (11.4–327) | 47.6 ± 42.2 (18–327) | 32.1 ± 15.2 (11.4–67.4) |
| LV End Systolic volume, Peds Z score | −0.2 ± 3.2 (−4.4 to 12.5) | ||
| LV Mass, grams | 220.9 ± 119.4 (42–623) | 237.1 ± 120.4 (135–623) | 166.9 ± 86.1 (42–468) |
| LV Mass, Peds Z score | −0.2 ± 1.3 (−3.86 to 3.55) | ||
| LV Ejection Fraction, % | 55.9 ± 13.5 (14–79.6) | 55.3 ± 14.3 (14–71) | 60.4 ± 10 (28.5–79.6) |
| LV Shortening Fraction, % | 33.5 ± 10.2 (5.3–64.7) | 32.6 + 11.3 (5.3–64.7) | 35.5 ± 8.1 (21.7–62.7) |
| LV pathology |
n (%) |
n (%) |
n (%) |
| Normal | 80 (73%) | 35 (58%) | 45 (90%) |
| Hypertrophic CM | 12 (11%) | 11 (18%) | 1(2%) |
| Dilated CM | 7 (6%) | 4 (6.7%) | 3 (6%) |
| Ischemic CM | 2 (2%) | 2 (3.3%) | 0 |
| Idiopathic CM | 1 (1%) | 1 (1.7%) | 0 |
| Non Ischemic CM | 4 (4%) | 4 (6.7%) | 0 |
| Restrictive CM | 1 (1%) | 1 (1.7%) | 0 |
| Low-normal systolic function | 3 (3%) | 2 (3.3%) | 1 (2%) |
Figure 2.
Pearson correlation between polar and DICOM GLS, linear regression analysis.
Figure 3.
Bland–Altman’s analysis of peak GLS between polar (EchoPAC) and DICOM (VVI) strain analysis.
Table 2.
Polar vs. DICOM global longitudinal strain, normal vs cardiomyopathy.
| Reader 1 | Normal n = 84 | Cardiomyopathy n = 27 | p |
|---|---|---|---|
| EchoPAC GLS | −20.1 | −13.9 | <0.0001 |
| VVI GLS | −16.1 | −10.9 | <0.0001 |
| p | <0.0001 | 0.03 |
Table 3.
Polar vs. DICOM global longitudinal strain, adult and pediatric patients.
| Mean polar GLS (±SD) | Mean DICOM GLS (±SD) | Differencea (Polar − DICOM) | Adults vs Ped (Polar-DICOM) | |
|---|---|---|---|---|
| Adult (n = 60) | −16.8 (±4.8) | −14.2 (±4.2) | −2.5 (±2.6) | t = 2.35, DF 324, p = 0.02 |
| Pediatric (n = 50) | −19.8 (±3.1) | −16.6 (±3.4) | −3.2 (±2.8) |
Negative value for difference signifies polar > DICOM.
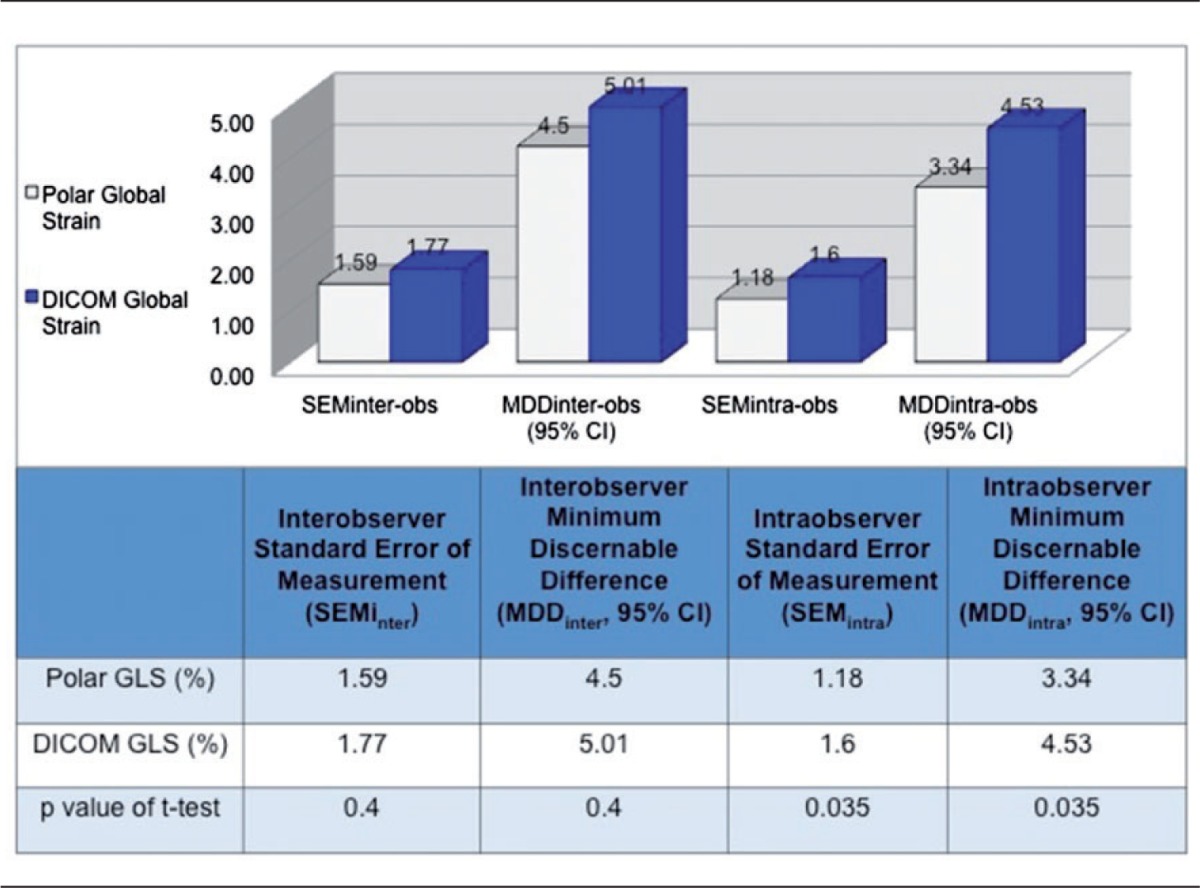
Table 4 shows inter- and intraobserver variabilities, quantified by standard error of the mean (SEMinter, SEMintra) and MDD, for polar (EchoPAC) and DICOM (VVI) global strain. For polar GLS the SEMinter was 1.59 strain points (1.59%) and SEMintra was 1.18%. For DICOM analysis GLS SEMinter was 1.77% and SEMintra was 1.60%. This yielded an interobserver MDD (MDDinter; the smallest change between two successive measurements necessary to claim statistically significant difference is present) of 4.50% for polar measurements and 5.01% for DICOM measurements. On repeat reads of 20 studies, the MDDintra was 3.34% for polar and 4.53% for DICOM global strain, respectively. By t-tests, the intra-observer variability was slightly higher with VVI-derived strain when compared to the EchoPAC analysis (p = 0.035), while the differences in inter-observer variability between the methods was not statistically significant (Table 4).
Table 4.
Standard errors of measurement (SEM) and minimum discernible difference (MDD) for inter- and intra-observer polar and DICOM strain analysis.
Regional strain
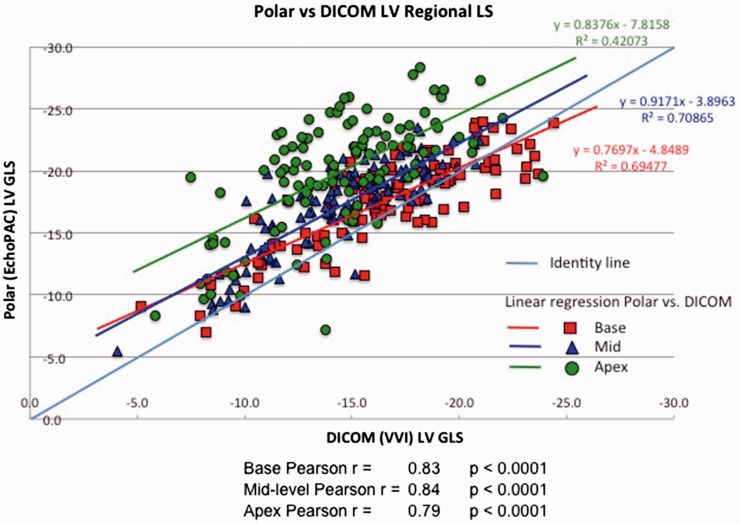
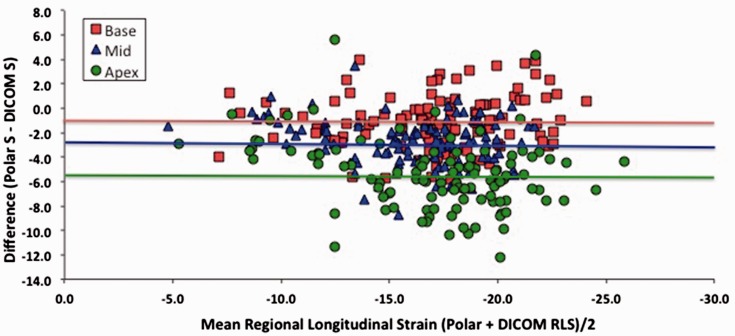
Figure 4 shows results of regional strain comparisons between polar and DICOM analysis after stratifying LV segments as basal, mid or apical. Basal and mid-level regional strains correlated well between analysis systems, r = 0.83 and r = 0.84, respectively, with slightly lower correlation at the apex; r = 0.79, p < 0.0001. As with global strain, EchoPAC analysis of polar images consistently produced higher regional peak absolute strain values than DICOM for corresponding segments (p < 0.001 for all comparisons) (Figure 5 and Table 5). Furthermore, the difference increased in base to apex direction, being −1.1% at base, −2.6% at the mid level and −5.5% at the apex (p < 0.0001 for trend).
Figure 4.
Pearson’s correlation between polar and DICOM RLS, linear regression analysis.
Figure 5.
Bland–Altman’s analysis of regional strain differences between polar and DICOM analysis (horizontal line indicates mean difference for that level).
Table 5.
Regional strain values and differences between polar and DICOM analysis.
| DICOM (VVI) | Polar (EchoPAC) | Differencea (Polar – DICOM) | |
|---|---|---|---|
| Mean basal strain (±SD), 95% CI | −16.5 (±3.9), −8.7 to −24.4 | −17.6 (±3.6), −10.4 to −24.8 | −1.1 (p < 0.0001) |
| Mean mid-level strain (±SD), 95% CI | −14.8 (±3.3), −8.2 to −21.3 | −17.4 (±3.5), −10.3 to −24.5 | −2.6 (p < 0.0001) |
| Mean apical strain (±SD), 95% CI | −14.3 (±3.8), −6.7 to −21.9 | −19.8 (±4.6), −10.6 to −29 | −5.5 (p < 0.0001) |
Negative value for difference signifies polar > DICOM.
Discussion
2DSE is a semi-automated function analysis method with many potential applications.15–17 However, this technology has not seen widespread clinical utilization, with variability in measurements being a significant limitation.18 An important source of variation may be differences in post-processing algorithms among commercially available software.19,20 Even less is known about regional variability among strain analysis programs. Consequently, we investigated the agreement and reproducibility of vendor-specific (EchoPAC) and vendor-neutral (VVI) analysis software in a well-powered, mixed cohort of adult and pediatric patients with normal and diseased LVs. Additionally, we determined the minimum threshold of variation needed to show a statistically significant difference between measurements. We found vendor-independent VVI (DICOM-derived) produced lower LV GLS values compared to vendor-specific (polar-derived) strain from EchoPAC. This bias remained consistent for regional strain measurements, with minimal difference in basal segments and the greatest difference noted at the apex. The findings from this study expand upon reports of variability in the echo strain literature. Biaggi et al.21 investigated 47 healthy subjects with EchoPAC and VVI and also found significantly lower GLS magnitude from VVI analysis. Similar findings were reported when EchoPAC and VVI were compared to tagged harmonic phase (HARP) magnetic resonance imaging (MRI), considered to be the reference standard for non-invasive assessment of function.11 This study in 30 adult patients found lower absolute GLS values by both VVI and EchoPAC compared to HARP, with VVI giving significantly lower values compared to EchoPAC and EchoPAC having better correlation and agreement with HARP MRI compared to VVI. More recently, a large study20 of 817 Japanese subjects was conducted using ultrasounds platforms and analysis software from three different vendors. The investigation found low inter-vendor agreement and concluded 2D strain data is not interchangeable within analysis systems. Data from pediatric cohorts is more sparse; similar findings were reported in a study of 34 children using different strain analysis systems.22
Most papers in the literature have reported variability in global strain, however, data on regional strain are more limited. Sensitive evaluation of regional dysfunction is an appealing promise of 2DSE, with important clinical implications. For example, a recent study showed the discriminative ability of 2DSE to differentiate cardiac amyloidosis from other causes of LV hypertrophy.23 Using EchoPAC, the researchers found a relative “apical sparing” pattern (comparing apical LS values to mid-wall and basal LV LS) that was an accurate and reproducible finding to differentiate the disease states. Our investigation found discrepancies in strain between the analysis systems with increase in the difference from the base to the apex, complicating the interpretation of regional strain for studies analyzed with different software. One possible explanation for the discrepancy may be sub-optimal tracking at the apex by VVI causing an underestimation of strain values. In our study, VVI produced lower strain values at the apex than basal and mid-wall strains, a finding in contrast to tagged MRI data that show strain is highest at the apex, and raising the possibility of sub-optimal tracking by VVI at the apex.24 These findings suggest comparisons of regional strain between analysis systems should be interpreted with caution, and should likely be limited to consistent measurements with the same analysis software.
The source of differences in polar and DICOM-derived strain is likely multi-factorial, and related to the properties of the individual software systems. Some investigators have noted the impact of DICOM compression on strain variability,25 however, for our investigation this compression was disabled to minimize variability. Another source of variation may be differences in the ROI for strain calculations. EchoPAC analysis incorporates the full thickness of the myocardium, whereas VVI’s tracking is primarily based on following reference points along the endocardial border.11 EchoPAC measures the change in distance in speckles in the ROI, whereas VVI tracks the change in a curvilinear outline of the endocardium using the mitral annulus and apex as references. However, this does not explain why VVI consistently produced lower LS magnitude, as one would expect higher longitudinal values along the endocardial surface given the inherent contractile properties of myocardium. Also, the EchoPAC algorithm defaults to analysis of 1 R-R interval, while VVI incorporates a three-beat algorithm. Thus, beat-to-beat variability in strain is a potential source of difference between these systems. A combination of these differences may explain the different results produced by these software systems, resulting in values that are not interchangeable it seems, even for the same echo study. A recent study came to a similar conclusion after analyzing strain from different ultrasound systems with four strain software packages. The researchers found lowest difference in GLS when using the same analysis software for different ultrasound systems19 and concluded post-processing played a greater role in strain discordance than image acquisition.
While there is ample data regarding variability in strain between different strain analysis systems, less is known about the significance of this variability when the software is judged independently. Therefore we applied standard error of measurement (SEM) analysis, a novel method of evaluating variability in measurements.12–14 SEM is a measure of the distribution of individual measurements around their three-mean value, thus enabling the construction of CIs of a measurement. In our study, we found the variability of both systems are similar when evaluated independently, with the polar-based method having a slight edge for lower intraobserver variability. Analysis of MDD values, another unique analysis for this investigation, showed a change of 3 to 5 strain points may occur between inter/intra observer measurements before a statistically significant change in the strain value has taken place. This finding is important to consider when using 2DSE for clinical assessment of ventricular function. 2DSE may potentially uncover subclinical disease at an earlier stage than current conventional functional analysis methods and enable sensitive longitudinal evaluation of function.15,17,23,26,27 The results of our study suggest discretion should be exercised before interpreting changes between studies as “significant.” For example, a decrease in GLS of two strain points between serial echos (on the same strain analysis systems) may not signify a significant drop in function between echo studies.
Collaboration between clinicians, researchers and industry promises standardization of strain echocardiography, as presented in the consensus statement from the European Association of Cardiovascular Imaging and the American Society of Echocardiography.18 A follow-up investigation indicates incorporation of these recommendations may lead to a decrease in strain variability,28 and more investigations are needed in large and heterogeneous populations to further evaluate.
Study limitations
Strain analysis was conducted between 2010 and 2012, thus interpretation of this study’s findings applies to this timeframe. However, several subsequent studies have shown similar variability among strain analysis systems, which continues to be a key issue in the contemporary era.29,30 Circumferential and radial strain measurements were not investigated for comparison between polar and DICOM-based analysis. Review of the literature during the study design phase indicted longitudinal strain to be the most reliable of the echo strain parameters, therefore, we chose to focus on this parameter. Another limitation is the sole use of VVI as the vendor-neutral DICOM-based strain analysis software when several analysis systems were potentially available for analysis. This study potentially may have benefited from inclusion of other vendor-neutral systems, but VVI was chosen because of good validation of the technology.5 Other studies, discussed above, have investigated inter-vendor strain variability on several analysis systems. However, to our knowledge our investigation is unique for SEM and MDD analysis that adds greater insight to the known variability in multi-vendor strain analysis. Finally, inclusion of a pediatric cohort and detailed analysis of regional strain are unique features of this investigation.
Conclusions
In a large and heterogeneous population of adult and pediatric patients, analysis of DICOM images with VVI produces lower absolute GLS values compared to EchoPAC analysis of polar images from the same echocardiogram. These differences are consistent across adult and pediatric cohorts and on regional strain analysis. Differences in strain values increase in a base to apex pattern, with largest differences at the apex. A change of 3 to 5 strain points may occur before a statistically significant difference is seen on subsequent measurements. Both analysis systems have similar inter-observer variability when evaluated independently, with EchoPAC having slightly lower intra-observer differences. Strain values from EchoPAC and VVI analysis are not interchangeable.
Acknowledgements
We thank Neil Greenberg, PhD, Cleveland Clinic Division of Cardiovascular Medicine, Heart and Vascular Institute, Cleveland Clinic, and Lisa Beachler, Division of Pediatric Cardiology, Cleveland Clinic.
Declaration of conflicting interests
The author(s) declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Ethical approval
The Institutional Review Board at Cleveland Clinic, approved the study protocol.
Guarantor
SA is the guarantor of this paper.
Contributorship
SA, KN, PG, ZP, FE, JDT contributed to the design of the study; SA, KN, AB, PG were involved in data collection; SA and ZP performed the data analysis; SA wrote the first draft; all authors contributed to and approved the final version.
References
- 1.Korinek J, Wang J, Sengupta PP, et al. Two-dimensional strain-A Doppler-independent ultrasound method for quantitation of regional deformation: Validation in vitro and in vivo. J Am Soc Echocardiogr 2005; 18(12): 1247–1253. [DOI] [PubMed] [Google Scholar]
- 2.Notomi Y, Lysyansky P, Setser RM, et al. Measurement of ventricular torsion by two-dimensional ultrasound speckle tracking imaging. J Am Coll Cardiol 2005; 45(12): 2034–2041. [DOI] [PubMed] [Google Scholar]
- 3.Edvardsen T, Gerber BL, Garot J, et al. Quantitative assessment of intrinsic regional myocardial deformation by Doppler strain rate echocardiography in humans: Validation against three-dimensional tagged magnetic resonance imaging. Circulation 2002; 106(1): 50–56. [DOI] [PubMed] [Google Scholar]
- 4.Roes SD, Mollema SA, Lamb HJ, et al. Validation of echocardiographic two-dimensional speckle tracking longitudinal strain imaging for viability assessment in patients with chronic ischemic left ventricular dysfunction and comparison with contrast-enhanced magnetic resonance imaging. AJC 2009; 104(3): 312–317. [DOI] [PubMed] [Google Scholar]
- 5.Pirat B, Khoury DS, Hartley CJ, et al. A novel feature-tracking echocardiographic method for the quantitation of regional myocardial function. J Am Coll Cardiol 2008; 51(6): 651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaul S, Miller JG, Grayburn PA, et al. A suggested roadmap for cardiovascular ultrasound research for the future. J Am Soc Echocardiogr 2011; 24(4): 455–464. [DOI] [PubMed] [Google Scholar]
- 7.Amundsen BH, Helle-Valle T, Edvardsen T, et al. Noninvasive myocardial strain measurement by speckle tracking echocardiography. J Am Coll Cardiol 2006; 47(4): 789–793. [DOI] [PubMed] [Google Scholar]
- 8.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. The Lancet 1986; 1(8476): 307–310. [PubMed] [Google Scholar]
- 9.GE Medical Systems. In: Systems GM (ed.) Technical publications EchoPAC PC user manual. 1st ed. Horten, Norway: General Electric Co., 2008, pp.1–627.
- 10.Cerqueira M. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart: A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. J Nucl Cardiol 2002; 9(2): 240–245. [DOI] [PubMed] [Google Scholar]
- 11.Bansal M, Cho G-Y, Chan J, et al. Feasibility and accuracy of different techniques of two-dimensional speckle based strain and validation with harmonic phase magnetic resonance imaging. J Am Soc Echocardiogr 2008; 21(12): 1318–1325. [DOI] [PubMed] [Google Scholar]
- 12.Yuan XP, Bach D, Skanes A, et al. Assessment of intra- and interobserver variability of pulmonary vein measurements from CT angiography. Acad Radiol 2004; 11(11): 1211–1218. [DOI] [PubMed] [Google Scholar]
- 13.Tong S, Cardinal HN, McLoughlin RF, et al. Intra- and inter-observer variability and reliability of prostate volume measurement via two-dimensional and three-dimensional ultrasound imaging. Ultrasound Med Biol 1998; 24(5): 673–681. [DOI] [PubMed] [Google Scholar]
- 14.Eliasziw M, Young SL, Woodbury MG, et al. Statistical methodology for the concurrent assessment of interrater and intrarater reliability: Using goniometric measurements as an example. Phys Ther 1994; 74(8): 777–788. [DOI] [PubMed] [Google Scholar]
- 15.Geyer H, Caracciolo G, Abe H, et al. Assessment of myocardial mechanics using speckle tracking echocardiography: Fundamentals and clinical applications. J Am Soc Echocardiogr 2010; 23(4): 351–369. [DOI] [PubMed] [Google Scholar]
- 16.Blessberger H, Binder T. Two dimensional speckle tracking echocardiography: Basic principles. Heart 2010; 96(9): 716–722. [DOI] [PubMed] [Google Scholar]
- 17.Hare JL, Brown JK, Leano R, et al. Use of myocardial deformation imaging to detect preclinical myocardial dysfunction before conventional measures in patients undergoing breast cancer treatment with trastuzumab. Am Heart J 2009; 158(2): 294–301. [DOI] [PubMed] [Google Scholar]
- 18.Voigt J-U, Pedrizzetti G, Lysyansky P, et al. Definitions for a common standard for 2D speckle tracking echocardiography: Consensus document of the EACVI/ASE/Industry Task Force to standardize deformation imaging. J Am Soc Echocardiogr 2015; 28(2): 183–193. [DOI] [PubMed] [Google Scholar]
- 19.Negishi K, Lucas S, Negishi T, et al. What is the primary source of discordance in strain measurement bet. Ultrasound Med Biol 2013; 39(4): 714–720. [DOI] [PubMed] [Google Scholar]
- 20.Takigiku K, Takeuchi M, Izumi C, et al. Normal range of left ventricular 2-dimensional strain. Circ J 2012; 76(11): 2623–2632. [DOI] [PubMed] [Google Scholar]
- 21.Biaggi P, Carasso S, Garceau P, et al. Comparison of two different speckle tracking software systems: Does the method matter? Echocardiography 2011; 28(5): 539–547. [DOI] [PubMed] [Google Scholar]
- 22.Koopman LP, Slorach C, Hui W, et al. Comparison between different speckle tracking and color tissue Doppler techniques to measure global and regional myocardial deformation in children. J Am Soc Echocardiogr 2010; 23(9): 919–928. [DOI] [PubMed] [Google Scholar]
- 23.Phelan D, Collier P, Thavendiranathan P, et al. Relative apical sparing of longitudinal strain using two-dimensional speckle-tracking echocardiography is both sensitive and specific for the diagnosis of cardiac amyloidosis. Heart 2012; 98(19): 1442–1448. [DOI] [PubMed] [Google Scholar]
- 24.Moore CC, Lugo-Olivieri CH, McVeigh ER, et al. Three-dimensional systolic strain patterns in the normal human left ventricle: Characterization with tagged MR imaging. Radiology 2000; 214(2): 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Risum N, Ali S, Olsen NT, et al. Variability of global left ventricular deformation analysis using vendor dependent and independent two-dimensional speckle-tracking software in adults. J Am Soc Echocardiogr 2012; 25(11): 1195–1203. [DOI] [PubMed] [Google Scholar]
- 26.Buss SJ, Emami M, Mereles D, et al. Longitudinal left ventricular function for prediction of survival in systemic light-chain amyloidosis: Incremental value compared with clinical and biochemical markers. J Am Coll Cardiol 2012; 60(12): 1077–1078, www.sciencedirect.com/science/article/pii/S073510971202089X. [DOI] [PubMed] [Google Scholar]
- 27.Migrino RQ, Aggarwal D, Konorev E, et al. Early detection of doxorubicin cardiomyopathy using two-dimensional strain echocardiography. Ultrasound Med Biol 2008; 34(2): 208–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Marwick TH, Fukuda N, et al. Improvement in strain concordance between two major vendors after the strain standardization initiative. J Am Soc Echocardiogr 2015; 28(6): 642–647. [DOI] [PubMed] [Google Scholar]
- 29.Nagata Y, Takeuchi M, Mizukoshi K, et al. Intervendor variability of two-dimensional strain using vendor-specific and vendor-independent software. J Am Soc Echocardiogr 2015; 28(6): 630–641. [DOI] [PubMed] [Google Scholar]
- 30.Costa SP, Beaver TA, Rollor JL, et al. Quantification of the variability associated with repeat measurements of left ventricular two-dimensional global longitudinal strain in a real-world setting. J Am Soc Echocardiogr 2014; 27(1): 50–54. [DOI] [PubMed] [Google Scholar]