Abstract
Alcohol exposure is a major reason of morbidity and mortality all over the world, with much of detrimental consequences attributing to alcoholic liver disease (ALD). With the continued ethanol consumption, alcoholic fatty liver disease (AFLD, the earliest and reversible form of ALD) can further develop to more serious forms of alcoholic liver damage, including alcoholic steatohepatitis, fibrosis/cirrhosis, and even eventually progress to hepatocellular carcinoma and liver failure. Furthermore, cell trauma, inflammation, oxidative stress, regeneration, and bacterial translocation are crucial promoters of ethanol-mediated liver lesions. AFLD is characterized by excessive fat deposition in liver induced by excessive drinking, which is related closely to the raised synthesis of fatty acids and triglyceride, reduction of mitochondrial fatty acid β-oxidation, and the aggregation of very-low-density lipoprotein (VLDL). Although little is known about the cellular and molecular mechanisms of AFLD, it seems to be correlated to diverse signal channels. Massive studies have suggested that liver steatosis is closely associated with the inhibition of silent information regulator 1 (SIRT1) and the augment of lipin1 β/α ratio mediated by ethanol. Recently, serine/arginine-rich splicing factor 10 (SFRS10), a specific molecule functioning in alternative splicing of lipin 1 (LPIN1) pre-mRNAs, has emerged as the central connection between SIRT1 and lipin1 signaling. It seems a new signaling axis, SIRT1–SFRS10–LPIN1 axis, acting in the pathogenesis of AFLD exists. This article aims to further explore the interactions among the above three molecules and their influences on the development of AFLD.
Impact statement
ALD is a major health burden in industrialized countries as well as China. AFLD, the earliest and reversible form of ALD, can progress to hepatitis, fibrosis/cirrhosis, even hepatoma. While the mechanisms, by which ethanol consumption leads to AFLD, are complicated and multiple, and remain incompletely understood. SIRT1, SFRS10, and LIPIN1 had been separately reported to participate in lipid metabolism and the pathogenesis of AFLD. Noteworthy, we found the connection among them via searching articles in PubMed and we had elaborated the connection in detail in this minireview. It seems a new signaling axis, SIRT1–SFRS10–LIPIN1 axis, acting in the pathogenesis of AFLD exists. Further study aimed at SIRT1–SFRS10–LIPIN1 signaling system will possibly offer a more effective therapeutic target for AFLD.
Keywords: Ethanol, silent information regulator 1, serine/arginine-rich splicing factor 10, lipin1, alcoholic fatty liver disease
Introduction
Alcoholic liver disease (ALD), a liver disease originating from heavy chronic, acute or chronic-binge ingestion of ethanol, has progressed to huge health challenge for humanity worldwide.1 Generally speaking, not all drinkers are suffering from ALD, which is largely dependent on host susceptibility. Plenty of factors have been observed as the correlation factors for the progression and severity of ALD, such as the absolute amount of ethanol intake, sex, race, obesity and metabolic syndrome, genetic factors, hepatitis virus infection, and lifestyle factors (such as smoking).1–3 Alcoholic fatty liver disease (AFLD), the initial reaction of the liver to ethanol abuse, happens to greater than 90% of heavy drinkers.2 In reality, AFLD is asymptomatic and even reversible after cessation of ethanol exposure in the patients without other risk factors.4 AFLD derives from the hepatic adipopexis, which is closely linked to ethanol-mediated lipodystrophy including enhanced lipogenesis and diminished lipolysis.5 Despite little known about the molecular mechanisms of the ethanol-mediated hepatic lipid accumulation, the following pathogenic factors may conduce to the liver steatosis:2,6,7 (1) The inhibition of SIRT1 mediated by ethanol evokes a series of cascade, impacting the pathways of lipogenesis, fatty acid β-oxidation, and the intake/secretion of lipoprotein. (2) Augmented hepatic afflux of free fatty acids from adipose tissue and of chylomicrons from the intestinal mucosa. (3) Ethanol-induced impairments of adenosine monophosphate activated kinase (AMPK) signaling foster lipogenesis and halt lipid catabolism by affecting the activity of peroxisome proliferating-activated receptor (PPAR) α or sterol regulatory element binding protein 1 (SREBP-1) c. (4) Acetaldehyde, one of metabolites of ethanol, does harm to mitochondria and microtubules, subsequently leading to the abatement of NADH oxidation and the gather of VLDL.
To date, several crucial molecules have been verified as direct or indirect targets of ethanol in rodents or humans, including SIRT1, AMPK, SREBP-1, nuclear factor kappa B, PPARγ co-activator-1α (PGC-1α), SFRS10, lipin1 and so on.6 Among the molecules above mentioned, SIRT1 seems to be the upstream modulator of this signaling network, while lipin-1 has been shown as one of vital downstream molecules causing the onset and progress of AFLD.6 More interestingly, SFRS10, a vital splicing factor acting in alternative splicing of LPIN1 pre-mRNAs, has appeared as the pivotal hinge between SIRT1 and lipin-1 signaling.8
Recently, expanding attentions have been paid to SIRT1–SFRS10–LPIN1 axis in the advance of AFLD. Considerable experiments have witnessed the visible increase of lipin1 β/α ratio mediated by ethanol in the liver of AFLD model or obese humans, which is largely generated via the impairments of SIRT1–SFRS10–LPIN1 axis.6,9,10 The nutritional or pharmacological modulations in any step of SIRT1–SFRS10–LPIN1 signaling axis might be a potential treatment target of human AFLD.
Ethanol downregulates hepatic SIRT1
SIRT1, a pivotal modulator of intrahepatic lipid metabolism and inflammatory responses via its deacetylation activity in the body, is a nicotinamide adenine dinucleotide (NAD+)-dependent class III protein deacetylase.11 SIRT1 could act on the acetylation status of a great deal of targets, subsequently modulating numerous lipid metabolism pathways including lipogenesis, fatty acid β-oxidation, as well as lipoprotein intake and secretion in liver.6 SIRT1 is predominately located in the nucleus and is activated by high NAD+ levels.11 Ethanol exposure may downregulate SIRT1 via several mechanisms as follows:6 the reduced ratio of NAD+ /NADH mediated by ethanol, reactive oxygen species (ROS),5 an increased level of SIRT1 inhibitors: miR-21712 or miR-34a, disruption of adiponectin signaling induced by ethanol,13 an elevated lipocalin 2 (Lcn2).14
In the liver, ethanol oxidizes to acetaldehyde via the catalysis of alcohol dehydrogenase (ADH), microsomal ethanol oxidation system and catalase, then oxidizing to acetic acid by acetaldehyde dehydrogenase 2 (ALDH2).15 The redox state has been changed in the procedure of ethanol metabolism, which reduces the ratio of NAD+ /NADH as the result of the conversion from NAD+ to NADH. Then NAD+-dependent deacetylated activity of SIRTl attenuates, as a result of the reduced concentration of NAD+ induced by ethanol.11
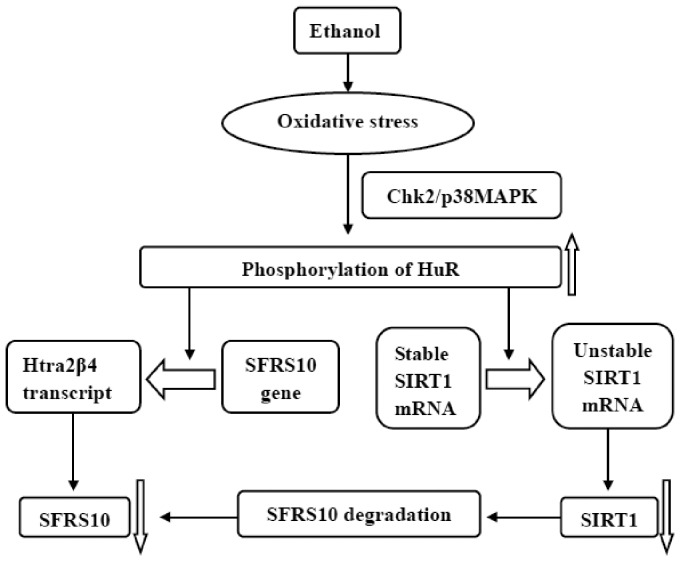
Moreover, ROS are released in the process of ethanol metabolism in liver, particularly via the cytochrome P450 family 2, subfamily E, polypeptide 1 microsomal ethanol oxidizing pathway.1 ROS mediate SIRT1 exported from nucleus to cytoplasm and disturb SIRT1 activity via its nucleocytoplasmic shuttling.6 Nevertheless, ethanol-mediated transport of SIRT1 was widely impeded by pre-incubation with N-acetylcysteine (an antioxidant).6 Besides, oxidative stress impairs SIRT1 expression via phosphorylating human antigen R (HuR), a widely expressed mRNA-binding protein in mammalian genome, belonging to the family of embryonic lethal abnormal vision protein.16,17 HuR has been confirmed to upregulate SIRT1 expression by binding and stabilizing SIRT1 mRNA.17 Generally, HuR binds with 3′-untranslated region (UTR) of SIRT1 mRNA to increase SIRT1 mRNA stability and protein levels.18,19 While phosphorylation of HuR at residue serine 100 by checkpoint kinase 2 (Chk2) promoted the complex of [HuR–SIRT1mRNA] to dissociate, concomitantly damaging the SIRT1 mRNA stability and decreasing protein levels in human diploid fibroblasts after exposure to hydrogen peroxide (H2O2)19 (Figure 1). Additionally, ethanol exposure enhances gut permeability and bacteria/endotoxin translocation, thereafter increasing lipopolysaccharide (LPS) coming into the portal circulation.5 LPS, acetaldehyde, and acetate have been confirmed to promote the production of ROS in cultured hepatic cells,5 subsequently inhibiting SIRT1 activity and disrupting SIRT1 signaling.20
Figure 1.
Ethanol downregulates SIRT1 and SFRS10 protein levels by phosphorylation of HuR. Ethanol induces phosphorylation of HuR via oxidative stress, and then impairs SIRT1 and SFRS10 expression
In cultured mouse AML-12 hepatocytes and in the livers of chronically ethanol-fed mice, miR-217 (an endogenous inhibitor of SIRT1) is upregulated to impair SIRT1, via binding to the 3′UTR of SIRT1 mRNAs and causing mRNA cleavage or suppressing its translation.12 Besides, it has also been confirmed that both miR-217 overexpression and alcohol consumption inhibit SIRT1 deacetylase activity in AML-12 cells.12 Utilizing primary kupffer cells (KCs) or cultured RAW 264.7 macrophages, Yin et al. had demonstrated that ethanol obviously increased miR-217 abundance and notably magnified LPS-induced miR-217 expression.20 Simultaneously, the study showed that SIRT1 mRNA and protein, human transformer-2-beta gene mRNA and lipin1α mRNA expression were both markedly lessened, whereas total lipin1 and lipin1β mRNA were enhanced in the livers of mice injected with adenovirus-miR-217 compared with controls.20 Taken together, ethanol administration might worsen steatosis and inflammation in hepatocytes, via disruption of miR-217–SIRT1–SFRS10–lipin1 axis.20 HuR and miR-217 may exert synergistic effects in signal channels of SIRT1 impairment, although no direct proofs confirm that ethanol increases the number of phosphorylated HuR degrading SIRT1 mRNA. Moreover, miR-34a (a negative regulator of SIRT1) is markedly promoted in ethanol-consumed hepatobiliary cell lines and in the livers of mice administrated with ethanol.21
In cultured hepatic cells, cultured macrophages, KCs, even in rat H4IIEC3 cells, adiponectin has been shown to obviously upregulate SIRT1 protein levels in a dose-dependent manner.6 Conversely, knocking down both AdipoRs 1 and 2 thoroughly eliminated the augment of SIRT1 by adiponectin.6 Ethanol can affect the expression and secretion of adiponectin in adipose tissue.13 Moreover, hypoadiponectinemia and aberrant hepatic adiponectin signaling are involved in liver steatosis in several AFLD animal models.22 Meanwhile, Lcn2, a new discovered molecule functioning in the pathogenesis of AFLD, has been shown at an increased level in liver induced by ethanol or adenovirus-miR-217.6,14,20 Intriguingly, overexpression of Lcn2 mediated by adenovirus in the hepatocytes or mouse liver exacerbated the proceeding of AFLD, which is linked to the inhibition of SIRT1 protein.6,14 Additionally, ethanol may inhibit SIRT1 activity via interfering in its post-translational modifications, including acetylation, phosphorylation as well as sumoylation.6 Altogether, ethanol downregulates hepatic SIRT1 in multifarious manners.
SFRS10 and AFLD
A splicing factor: SFRS10
Alternative splicing of pre-mRNAs significantly generates the proteome diversity and organismal complexity.23 Serine-arginine rich (SR) proteins, a well-conserved class of RNA-binding proteins, act as important regulators of constitutive mRNA splicing.24 SFRS10, previously named RA301 and human transformer-2β (htra2β), belongs to the family of SR-like proteins and exerts an important role in alternative splicing of pre-mRNAs.25 It has been demonstrated that SFRS10 adjusts the alternative splicing of the calcitonin/calcitonin gene-related peptide, the survival motor neuron 1 protein, the tau protein and lipin 1.6,23
SFRS10 gene contains 10 exons and 9 introns and forms at least five diverse transcripts (htra2β1–5)25 and two protein isoforms (full length Tra2β protein and Tra2β-3) in human through selective splicing.26 Interestingly, the functionally active and full length Tra2β (namely SFRS10) is encoded by the main isoform tra2β1 including all exons except for exon 2.25 SFRS10 contains an RNA recognition motif (RRM), which lies between N-terminal serine-arginine dipeptide rich (RS)1 domain and C-terminal RS2 domain.27 Besides, there exist two motifs termed ribonucleoprotein 1 (RNP1) and RNP2 in the RRM of SFRS10.28 Both the RRM region and RS1 are demanded for SFRS10 splicing activity.27 RRM recognizes and combines with specific RNA sequence to determine splicing specificity and ensure pre-mRNA substrates entering into the splicing pathway,29 while the RS regions and its phosphorylation status modulate RNA interactions:23 The phosphorylation of RS1 region decreases RNA binding to RRM, whereas the unphosphorylated RS2 region facilitates the combination.23 Nevertheless, the shorter protein Tra2β-3 includes a effective RRM sequence but excludes the domain of RS1, and it may serve as a forceful splicing inhibitor resulting from the competitive combination with the same RNA goals.26 SFRS10 resides both in nucleus and cytoplasm, and the nuclear localization signals (NLSs), which present in RS1 domain, play a vital role in SFRS10 nucleocytoplasmic shuttling and subcellular distribution:29 NLSs generally foster SFRS10 localizing to the nuclear, while its phosphorylation status at serine residues guide SFRS10 to cytoplasm.23
Interestingly, it seems that there exists an autoregulation in the expression level of the full length Tra2β protein, which is associated with itself negative feedback loop:26,27,30 for instance, high SFRS10 expression levels could combine with four AGAA rich exonic splicing enhancers existent in exon 2 and promote the retention of exon 2, resulting in the formation of tra2β4 transcript that does not encode protein and ultimately resulting in the decrease of SFRS10 level.26,27,30 On the contrary, SFRS10 at low concentration will fail to recognize its exon 2 and lead to the exon 2 skipping, eventually inducing the production of the functional full-length protein.26,27,30 It is the autoregulation of SFRS10 that can explain why SFRS10 expression levels just mildly cut down in the liver of heterozygous mice.10
Oxidative stress impairs SFRS10 expression via phosphorylating HuR
In acute stage (within 4–6 h) after exposure to arsenite (an oxidant), a gastric cancer cell line (AGS) preferentially formed tra2β 4 transcript, which switched off SFRS10 synthesis and transported SFRS10 to cytoplasm.25 Interestingly, AGS cells induced tra2β 1 mRNA expression and SFRS10 reaccumulation in nucleus after 6 h.25 Similar results were also observed in another gastric cancer cell line (KATO III), colon cancer cell lines (T84 and HCT116), HeLa cells, and rat gastric mucosa.25 Analogously, H2O2 also triggered tra2β 4 mRNA expression in AGS cells and other cells.25 HuR functions in nucleus and mediates stress responses by stabilizing and/or promoting the expression of target mRNAs, while SFRS10 includes AU-rich elements in exon 2, in which exists a HuR-binding motif.16 HuR may participate in splicing regulation of SFRS10 pre-mRNA after exposure to oxidative stress (Figure 1). Akaike et al. have demonstrated that both Chk2- and mitogen-activated protein kinase p38 (p38MAPK)-dependent phosphorylation of HuR induce SFRS10 exon 2 retention and interact with the 39-nucleotide proximal domain of exon 2, reducing the expression of SFRS10 in HCT116 cells exposed to sodium arsenite.16 Chk2- and p38MAPK-mediated phosphorylation of nuclear HuR at residue serine 88 and/or tyrosine 118 urges the association between HuR and exon 2a of SFRS10 pre-mRNA in colon cancer cells exposure to oxidants, and promotes the formation of TRA2β 4 transcript that does not translate.16 Oxidative stress may promote the retention of SFRS10 exon 2 to reduce SFRS10 expression via phosphorylating HuR.
Nevertheless, SFRS10 has been confirmed to be induced in cultured astrocytes during hypoxia followed by reoxygenation,31 middle carotid artery occlusion, silicosis, arteriosclerosis, nerve injury,32 and breast cancer.28 Interestingly, the increase of SFRS10 in cultured vascular smooth muscle cells (VSMCs), were restrained by diphenyl iodonium (a NADPH oxidase inhibitor) or PD98059 (MAPK kinase inhibitor), which has been speculated in connection with the production of superoxide anion (O2−) as well as the activation of the MAPK cascade.33 Speculatively, ROS produced in the hepatic ethanol metabolism may induce the generation of SFRS10, which is conflicting with the experimental results above. While we may owe this paradox to the different time periods of oxidative stress.
Impairment of SFRS10 expression takes part in the steatosis
SFRS10 protein functions as a pivotal regulator of alternative splicing of multiple genes, which has been observed in the lysate from rat liver with a specific antibody.34 It has inferred that SFRS10 may act as a core in the pathogenesis of AFLD, which is involved in the augment of hepatic lipogenic genes expression, VLDL secretion, and hypertriglyceridemia.6,10
Decreased expression of SFRS10 has been witnessed by Western blot in liver of high-fat diet fed (HFD-fed) mice.10 Pihlajamaki et al.10 constructed the model of SFRS10 knockdown in HepG2 cells by transfection of SFRS10-specific siRNA, in which triglyceride (TG) accumulation has raised by 1.4-fold and the mRNA levels of major lipogenic genes have increased approximately 1.5- to 2-fold, such as SREBP-1c, fatty acid synthase (FAS), acetyl-CoA carboxylase (ACC) 1, and diacylglycerol O-acyltransferase 2 (DGAT2). Interestingly, the similar results have been testified again in C2C12 myotubes.10 Conversely, experimental overexpression of SFRS10 meaningfully lessened the transcription of several lipogenic genes in Hepa1c cells,10 such as FAS, 1-acylglycerol-3-phosphate-O-acyltransferase 2, and DGAT2. In addition, they also witnessed a remarkable elevated level of the TG-enriched VLDL fraction in plasma.10 Moreover, they have also confirmed that this pattern reliably counted on raised hepatic VLDL secretion in heterozygous mice via the application of tyloxapol.10 These data have manifested that depressed SFRS10 protein expression indeed takes part in the steatosis.
Lipin1 and AFLD
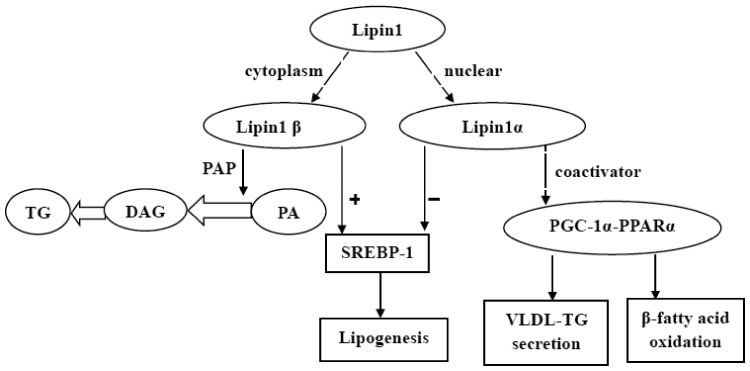
Lipin1, a mammalian Mg2+-dependent phosphatidic acid phosphohydrolase (PAP) generated by the gene of LPIN1, possesses bidirectional functions for modulating lipid metabolism in the body1 (Figure 2). For one thing, lipin1 acts as PAP-1 protein participating in glycerol phosphate pathway and facilitating TG synthesis, by dephosphorylating phosphatidic acid to produce diacylglycerol (DAG).1,8 For another, lipin1 serves as a transcriptional co-activator associating with PGC-1α, PPARα, or SREBP-1 to regulate the lipid metabolism.8,35,36 LPIN1 gene can encode three diverse isoforms, namely lipin-1 α, β, and γ, by alternative splicing of pre-mRNA in human.1,37 Lipin-1α and lipin-1β have been found principally settling in liver and skeletal muscle, while lipin-1γ primarily in brain tissue regulating lipid metabolism.37 Lipin1β, chiefly in the cytoplasm, transforms phosphatidate to DAG fostering TG synthesis and excessive hepatic fat accumulation by its PAP enzyme activity.8 Whereas lipin1α, mainly in the nucleus, acts as transcriptional co-activator and associates with PGC-1α, PPARα and SREBP-1, stimulating fatty acid oxidation, and suppressing lipid synthesis.8 Therefore, we speculate that it is lipin-1α and β which mainly participate in lipid metabolism in liver.
Figure 2.
Lipin1 functions in lipid metabolism. Lipin1β, chiefly in cytoplasm, transforms phosphatidate (PA) to DAG fostering TG synthesis by its PAP activity. Whereas lipin1α, mainly in nucleus, acts as transcriptional co-activator associating with PGC-1α and PPARα, stimulating β-fatty acid oxidation and VLDL-TG secretion. Lipin1α and lipin1β both participate in lipid synthesis by regulating SREBP-1 signaling
Lipin1α directly interplays with and coactivates PPARα and PGC-1α, and increases the expression of genes encoding enzymes associated closely with β-fatty acid oxidation,1 covering mitochondrial long-chain acyl-CoA dehydrogenase and mitochondrial medium-chain acyl-CoA dehydrogenase, acyl-CoA synthetase, acyl-CoA oxidase, carnitine palmitoyl transferase I, and fatty acid binding protein.38 Furthermore, lipin1α restrains SREBP-1 expression and activity, resulting in weakened hepatic lipogenesis by downregulating lipid and cholesterol biosynthetic enzymes expression,1,35,36 including FAS, mitochondrial glycerol-3-phosphate acyltransferase1, ACC, stearoyl-CoA desaturase, ATP citrate lyase, and malic enzyme.1,39 In hepatocytes, ethanol may interdict lipin1 nuclear entry and upregulate SREBP-1 activity via triggering mammalian target of rapamycin complex 1-phosphorylated lipin1 signaling.1,36,40 Moreover, lipin-1–PAP activity plays important role on SREBP-1 activation and the exacerbation of hepatic lipid accumulation.1,8 Paradoxically, chronic ethanol consumption to lipin1 LKO mice, which can abrogate the elevation of ethanol-induced intrahepatic PAP activity, has showed a significantly augmented lipid accumulation in liver, suggesting that lipin1 PAP activity might prevent from AFLD.41 Lipin1 PAP activity may exert a protective role on AFLD via mediating autophagy, urging lipid storage, normalizing phosphatidic acid levels, regulating fatty acid catabolism or restraining inflammation.1,42,43
Under the physiology condition, it is the lipoprotein of VLDL which carries TG migration to plasma from liver.44 Perturbations of VLDL assembly and secretion affect lipid homeostasis of liver and plasma lipoprotein profile.45 Hu et al. have showed that the rates of VLDL–TG secretion was noticeably relieved in the livers of WT mice fed with an ethanol-containing diet for four-weeks, while was strikingly elevated in lipin1 LKO mice fed a control diet.41 More importantly, they found ethanol significantly blocked the elevation of VLDL–TG secretion in lipin-1LKO mice.41 As mentioned above, lipin1 truly involve in the regulation of VLDL–TG secretion and intrahepatic lipid homeostasis. The functions of lipin1 in VLDL-TG secretion rely on nuclear lipin1-PGC-1α signaling45 rather than its PAP activity:46 PGC-1α overexpression significantly provoked VLDL-TG secretion and the efficiently secretion of newly synthesized TG in HepG2 cells.45 Collectively, lipin1α promotes VLDL–TG secretion by coactivating PGC-1α. Ethanol may induce the onset and progress of AFLD via destruction of lipin1 nuclear entry, disrupted activity for fatty acid catabolism and VLDL–TG secretion45 by hepatic abnormal PGC-1α/PPARα signaling.1,41
Lipin1 is a nucleocytoplasmic shuttling protein, and its activities are modulated by protein post-translational modifications, as well as its nucleocytoplasmic localization1 which is associated with 14-3-3 proteins and NLS.6,8 Furthermore, post-translational modifications of lipin1 largely control its nucleocytoplasmic localization such as sumoylation, acetylation or phosphorylation of lipin-1.1,8 Lipin1α is localized in both the cytoplasm and the nucleus of hepatocytes,6 but its transcriptional co-activator activity works mainly in nucleus.8 Both lipin1α and β are bedecked with sumoylation at two same sumoylation sites (K566/596).1 Sumoylation fosters lipin1α nuclear reservation and its transcriptional coactivator behaviors,47,48 whereas lipin1β activity is not dramatically changed by sumoylation.48 Hu et al. have revealed that chronic alcohol ingestion induced the diminished sumoylation and elevated acetylation levels of hepatic lipin1 in mouse livers, concomitantly the upregulation of lipin1 in cytoplasm and downregulation in nucleus.47 The capacity of ethanol to regulate lipin1 subcellular localization may be induced by impairment of SIRT1-mediated acetylation/deacetylation–sumoylation switch.1,9 Additionally, the form of serine phosphorylation of lipin1 enhances the nuclear export and its migration to the endoplasmic reticulum membrane, where lipin1 PAP activity converts phosphatidate to DAG,1 whereas serine dephosphorylation facilitates its cytoplasmic distribution.49,50
Hepatic lipin1 LKO caused obvious elevations of ROS and numerous proinflammatory cytokines mRNA levels, such as IL-1β, tumor necrosis factor-alpha (TNF-α), serum amyloid A-1, and LCN-2 in mice fed with control diet.41 Besides, lipin-1α directly associates with nuclear factor of activated T cells c4 (NFATc4) to inhibit NFATc4 transcriptional activity in adipose cells, which, in turn, lessens the activation of TNF-α and IL-6 and suppresses inflammation.20,41 It seems to suggest that lipin1 owns anti-inflammatory and antioxidant activities. Ablation of lipin1 in mice adipocyte augments the condition of AFLD probably by remarkably lowering adipocyte adiponectin gene transcription, serum total or high molecular weight multimeric form adiponectin protein levels and hepatic adiponectin receptor 1 and 2.1,51,52 Unexpectedly, myeloid cell-specific lipin1 deletion in mice alleviated alcoholic hepatitis (AH) and liver damage, but mildly accentuated ethanol-mediated steatosis.53 Altogether, lipin1 in different tissues may exert different features.
In the physical conditions, the ratio of lipin1β to lipin1α is appropriate, while the balance between lipin-1α and β will be destroyed by ethanol. Besides, ethanol also contributes to lipin1 expression, cytoplasm localization as well as PAP function.1 Ethanol-mediated disorder of lipin1 may depend on four main mechanisms as following:8 ethanol upregulates LPIN1 promoter activity and lipin1 expression, in which ethanol metabolism, AMPK-SREBP-1 signaling, enhanced endogenous GC levels and acetylated histone H3-Lysine 9 play essential roles,1,47 ethanol facilitates lipin1 expression in cytoplasm and enhances its PAP function,8,47 abated lipin1 nuclear entry resulting from its hyperacetylation and hyposumoylation induced by ethanol,47 markedly increased proportion of lipin-1β to α regulated by ethanol, through SIRT1–SFRS10–LPIN1 axis in the liver. Additionally, pretreatment with either cyanamide (an ALDH2 depressor) or 4-methylpyrazole (an ADH depressor) in AML-12 hepatocytes, substantially interdicted the effects of ethanol have on lipin-1.41,47 Ethanol’s metabolite of acetate also involved in ethanol-lipin-1 signaling in hepatocytes.41,47 It is credible to speculate that ethanol metabolism exerts essential roles in its effects on lipin-1. Collectively, ethanol exposure can dysregulate hepatic lipin-1 expression and function in different ways conducing to the fat accumulation and development of AFLD.
Ethanol causes AFLD via SIRT1–SFRS10–LPIN1 axis
You et al. have demonstrated that the occurrence and development of AFLD is linked closely to the disarrangement of the signaling network mediated by ethanol-induced SIRT1 impairment.6 Inhibition of SIRT1 disarranges the activities of diverse target molecules, including AMPK, SREBP-1, PPARα, and PGC-1α, forkhead transcription factor O1,54 histone H3, β-catenin, SFRS10, and lipin1.6 A meaningful augment of the ratio of LPIN1 β/α has been seen in liver of HFD-fed mice as well as obese humans, in parallel with decreased SFRS10 protein levels.10 SFRS10 may emerge as a connection hub between SIRT1 and lipin1 in the occurrence and development of steatosis.
Ethanol induces an elevated lipin1 β/α ratio parallel to the inhibition of SIRT1 and SFRS10
Compared hepatocyte-specific SIRT1 knockout (Sirt1LKO) mice with wild-type (WT) mice both fed with a chow diet, the knockout of hepatic SIRT1 not only facilitated the mRNA levels of total LPIN1 and LPIN1β but also the LPIN1β/α ratio.9 Nevertheless, resveratrol, a known SIRT1 excitant, interdicted the polyubiquitination and expression of lipin-1,55,56 and relieved liver steatosis and liver injury.6,56 Interestingly, the study with Sirt1LKO mice also revealed the sharp reduction of both mRNA and protein expression levels of SFRS10, as a result of the impairment of SIRT1.9 The similar results were pronounced in cultured AML-12 hepatocytes, too.9 Going a step further, the above conclusions were verified in comparison of WT mice fed with ethanol or SIRT1 LKO mice fed with control diets and WT controls.9 Of note, SIRT1 absence in liver induced by ethanol significantly enhanced both the upregulation of LPIN1β/α and SFRS10 inhibition, comparing ethanol-fed SIRT1 LKO mice with all other groups.9 At the same time, the quantity of lipin1 in cytoplasm increased obviously, while the number in nuclear dramatically decreased in ethanol-fed WT mice or Sirt1LKO mice.9 More significantly, the nucleocytoplasmic shuttling of lipin1 was reinforced evidently in the Sirt1LKO mice fed with ethanol than all other groups.9
In another study, they established a cellular alcoholic steatosis model using AML-12 hepatocytes.9 Transfecting AML-12 cells with SIRT1wt or SFRS10wt could reverse the adipopexis and prevent the disproportion of Lpin1β/α ratio induced by ethanol widely.9 While these protective results mediated by SIRT1/SFRS10 disappeared when overexpression of SIRT1H363Y, SIRT1siRNA, or SFRS10siRNA which restrained the levels of SIRT1 or SFRS10.9 A further study has been carried out to appraise the relation between knockdown SFRS10 and endogenous LPIN1.10 Results showed that SFRS10 knockdown upregulate the ratio of LPIN1β to LPIN1α in HepG2 cells.10 Notably, SFRS10 knockdown enhances the expression of LPIN1β in parallel with the moderate downregulation of LPIN1α, without altering the total expression of LPIN1.10 Moreover, the similar results responding to SFRS10 knockdown have been observed in C2C12 myotubes.10 These conclusions were confirmed again in liver of SFRS10 heterozygous mice, HFD-fed mice as well as obese humans.10 On the contrary, overexpression of SFRS10 in Hepa1c cells downregulated LPIN1β/α ratio in comparison with GFP control.10 In HepG2 cells cotransfected with SFRS10-specific siRNA, the LPIN1β knockdown not only prevented the SFRS10 knockdown-mediated upregulation of the genes of fatty acid synthesis and TG synthesis, but also impeded the raised lipogenesis, the accumulation of TG and lysophosphatidic acid(an intermediate product in the TG synthesis pathway) induced by SFRS10 knockdown.10 Collectively, these research have indicated that the downregulation of SFRS10 triggers the expression of LPIN1β and its functions activating the overexpression of lipogenic genes and lipogenesis.10 Accordingly, we may draw a conclusion that the disorder of lipin1 signaling induced by ethanol derive from the inhibition of SIRT1–SFRS10. Further, a team measured the mRNA levels of SIRT1, SFRS10, and Lpin1β/α from the AH patients’ liver samples and normal controls, and confirmed that the destruction of the SIRT1–SFRS10–LPIN1 axis advances and exacerbates human AH.9 It seems to support the presupposition that SIRT1–SFRS10–LPIN1 axis exerts a momentous position in the pathogenesis of AFLD.
Impairment of hepatic SIRT1 fostered the mRNA levels of total lipin-1,9 while SFRS10 knockdown enhanced the ratio of LPIN1β/α without altering the total expression of LPIN1.10 It seems to suggest the existence of another connectors between the SIRT1 and lipin1, such as AMPK, SREBP-1 and histone H3. The increased expression of lipin1 gene mediated by ethanol has been confirmed in association with the repression of AMPK as well as the activated SREBP-1.47 Moreover, ethanol consumption dramatically heightened the correlations of acetylated lysine 9 of histone H3 with the promoter region of LPIN1 containing the sterol regulatory element in vitro and in vivo.47
SIRT1–SFRS10–LPIN1 axis and AFLD
The mechanisms of SIRT1 impairment damaging SFRS10 expression and activity are still unknown. SIRT1 may augment SFRS10 protein stability and expression levels in hepatocytes via regulating the acetylation status of SFRS10, and then protecting SFRS10 from proteosome degradation.9 Additionally, ethanol exposure may impair SIRT1–SFRS10 signaling by oxidative stress and phosphorylated HuR, further aggravating the onset and severity of AFLD via increasing lipin-1β expression. While the mechanism of reduction of SFRS10 derived from impaired SIRT1 is still required for further inquiry.
Pre-mRNA splicing usually happens in a spliceosome complex, whose assembly needs a series of cascade steps and the involvement of multitude factors, such as the small nuclear ribonucleoproteins (snRNPs, including U1, U2, U4-U6), heterogeneous ribonucleoproteins (hnRNPs), and SR proteins or SR like proteins (such as SFRS10).34,57,58 Interestingly, SR proteins or SR like proteins and hnRNPs lean towards competing with each other in identification of RNA sequence elements (enhancers or silencers).58 The former ordinarily collects components of the spliceosome towards the exon, inducing exon inclusion and further driving splicing, while the latter usually restrains the formation of spliceosome, guiding complete or partial exon exclusion and further inhibiting splicing.59,60 SFRS10 has been testified to specifically combine with RNA, which contains AGAA/GGAA-rich sequences, through the NGAA sequence present in its RRM.61,62 Interestingly, there includes a sequence GGAA in the alternatively spliced exon 6 of LPIN1.10 One study has shown that high SFRS10 protein levels induce the skipping of LPIN1 exon 6 in the plasmid transfected by a minigene construct, while SFRS10 knockdown mediated by siRNA fosters the inclusion of exon 6.10 Lipin-1β, whose generation relies on inclusion of exon 6 of LPIN1, functions as PAP-1 fostering expression of lipogenic genes and hepatic fat accumulation.63 Whereas the site of SFRS10 binding with LPIN1 overlaps with the U1 snRNA binding sites partly at the 5′ splice site.10 Accordingly, we can speculate that the modulation of SFRS10 to exon 6 of LPIN1 above mentioned is linked closely to U1 snRNA or hnRNPs.
Conclusion
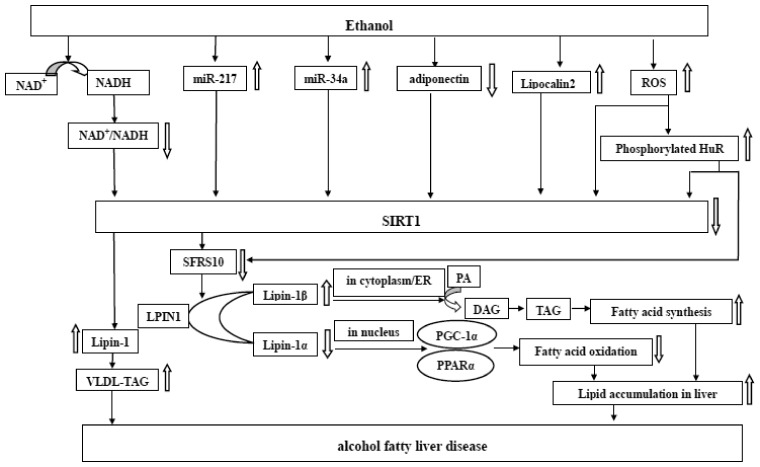
Ethanol dramatically induced the elevation of hepatic Lpin1β/α ratio by SFRS10 inhibition in cultured hepatocytes and in mice. Congruously, hepatic SIRT1 impairment remarkably increased LPIN1β/α via SFRS10 mediated by ethanol, suggesting that ethanol exposure may suppress SIRT1–SFRS10 in order in mice.6 The role of SIRT1–SFRS10–LPIN1 signaling axis plays in the pathogenesis of AFLD is still demanded to ascertain (Figure 3). Further study aimed at SIRT1–SFRS10–LPIN1 signaling system will possibly offer a more effective therapeutic target for AFLD.
Figure 3.
Roles of SIRT1–SFRS10–LPIN1 axis in the development of AFLD. Ethanol exposure impairs SIRT1 via several mechanisms, such as the increase of NADH, miR-217, miR-34a, Lipocalin2, ROS as well as the decrease of adiponectin and NAD+, proceeding to suppress the expression of SFRS10. The low level of SFRS10 and SIRT1 fosters the expression of total lipin-1 and lipin-1β in parallel with the moderate deregulation of lipin-1α. As a result, the enhanced fatty acid synthesis and the suppressed fatty acid oxidation urge lipid accumulation in liver, and ultimately progress to alcohol fatty liver disease
Acknowledgments
We thank the four foundations enclosed in “FUNDING” for supporting this project. This work was supported by HeBei Province Natural Science Funds (H2013206322), China Foundation for Hepatitis Prevention and Control TianQing Liver Disease Funds (TQGB20140244), China Foundation for Hepatitis Prevention and Control WangBaoen Liver Fibrosis Study Funds (CFHPC20151027) and the Government-Funded Provincial Talents of Clinical Medicine Project in 2016.
Authors’ contributions
JZ participated in the design and review of the manuscript. YL wrote the manuscript.
Declaration of Conflict Of Interest
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.You M, Jogasuria A, Lee K, Wu J, Zhang Y, Lee YK, Taylor C, Sadana P. Signal transduction mechanisms of alcoholic fatty liver disease: emerging role of lipin-1. Curr Mol Pharmacol 2016;9:1–11. [DOI] [PMC free article] [PubMed]
- 2.Orman ES, Odena G, Bataller R. Alcoholic liver disease: pathogenesis, management, and novel targets for therapy. J Gastroenterol Hepatol 2013; 28: 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams JA, Manley S, Ding WX. New advances in molecular mechanisms and emerging therapeutic targets in alcoholic liver diseases. World J Gastroenterol 2014; 20: 12908–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livero FA, Martins GG, Queiroz Telles JE, Beltrame OC, Petris Biscaia SM, Cavicchiolo Franco CR, Oude Elferink RP, Acco A. Hydroethanolic extract of Baccharis trimera ameliorates alcoholic fatty liver disease in mice. Chem Biol Interact 2016; 260: 22–32. [DOI] [PubMed] [Google Scholar]
- 5.Magdaleno F, Blajszczak CC, Nieto N. Key events participating in the pathogenesis of alcoholic liver disease. Biomolecules 2017;7:9.
- 6.You M, Jogasuria A, Taylor C, Wu J. Sirtuin 1 signaling and alcoholic fatty liver disease. Hepatobiliary Surg Nutr 2015; 4: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.European Association for the Study of L. EASL clinical practical guidelines: management of alcoholic liver disease. J Hepatol 2012; 57: 399–420. [DOI] [PubMed] [Google Scholar]
- 8.Bi L, Jiang Z, Zhou J. The role of lipin-1 in the pathogenesis of alcoholic fatty liver. Alcohol Alcohol 2015; 50: 146–51. [DOI] [PubMed] [Google Scholar]
- 9.Yin H, Hu M, Liang X, Ajmo JM, Li X, Bataller R, Odena G, Stevens SM, Jr, You M. Deletion of SIRT1 from hepatocytes in mice disrupts lipin-1 signaling and aggravates alcoholic fatty liver. Gastroenterology 2014; 146: 801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pihlajamaki J, Lerin C, Itkonen P, Boes T, Floss T, Schroeder J, Dearie F, Crunkhorn S, Burak F, Jimenez-Chillaron JC, Kuulasmaa T, Miettinen P, Park PJ, Nasser I, Zhao Z, Zhang Z, Xu Y, Wurst W, Ren H, Morris AJ, Stamm S, Goldfine AB, Laakso M, Patti ME. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab 2011; 14: 208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nogueiras R, Habegger KM, Chaudhary N, Finan B, Banks AS, Dietrich MO, Horvath TL, Sinclair DA, Pfluger PT, Tschop MH. Sirtuin 1 and sirtuin 3: physiological modulators of metabolism. Physiol Rev 2012; 92: 1479–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin H, Hu M, Zhang R, Shen Z, Flatow L, You M. MicroRNA-217 promotes ethanol-induced fat accumulation in hepatocytes by down-regulating SIRT1. J Biol Chem 2012; 287: 9817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu X, Jogasuria A, Wang J, Kim C, Han Y, Shen H, Wu J, You M. MitoNEET deficiency alleviates experimental alcoholic steatohepatitis in mice by stimulating endocrine adiponectin-Fgf15 axis. J Biol Chem 2016; 291: 22482–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Y, Jogasuria A, Yin H, Xu MJ, Hu X, Wang J, Kim C, Wu J, Lee K, Gao B, You M. The detrimental role played by lipocalin-2 in alcoholic fatty liver in mice. Am J Pathol 2016; 186: 2417–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louvet A, Mathurin P. Alcoholic liver disease: mechanisms of injury and targeted treatment. Nat Rev Gastroenterol Hepatol 2015; 12: 231–42. [DOI] [PubMed] [Google Scholar]
- 16.Akaike Y, Masuda K, Kuwano Y, Nishida K, Kajita K, Kurokawa K, Satake Y, Shoda K, Imoto I, Rokutan K. HuR regulates alternative splicing of the TRA2beta gene in human colon cancer cells under oxidative stress. Mol Cell Biol 2014; 34: 2857–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ceolotto G, De Kreutzenberg SV, Cattelan A, Fabricio AS, Squarcina E, Gion M, Semplicini A, Fadini GP, Avogaro A. Sirtuin 1 stabilization by HuR represses TNF-alpha- and glucose-induced E-selectin release and endothelial cell adhesiveness in vitro: relevance to human metabolic syndrome. Clin Sci 2014; 127: 449–61. [DOI] [PubMed] [Google Scholar]
- 18.Yoon JH, Abdelmohsen K, Srikantan S, Guo R, Yang X, Martindale JL, Gorospe M. Tyrosine phosphorylation of HuR by JAK3 triggers dissociation and degradation of HuR target mRNAs. Nucleic Acids Res 2014; 42: 1196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell 2007; 25: 543–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin H, Liang X, Jogasuria A, Davidson NO, You M. miR-217 regulates ethanol-induced hepatic inflammation by disrupting sirtuin 1-lipin-1 signaling. Am J Pathol 2015; 185: 1286–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng F, Glaser SS, Francis H, Yang F, Han Y, Stokes A, Staloch D, McCarra J, Liu J, Venter J, Zhao H, Liu X, Francis T, Swendsen S, Liu CG, Tsukamoto H, Alpini G. Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am J Pathol 2012; 181: 804–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab 2009; 9: 327–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamros MA, Aubol BE, Keshwani MM, Zhang Z, Stamm S, Adams JA. Intra-domain cross-talk regulates serine-arginine protein kinase 1-dependent phosphorylation and splicing function of transformer 2beta1. J Biol Chem 2015; 290: 17269–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradley T, Cook ME, Blanchette M. SR proteins control a complex network of RNA-processing events. RNA 2015; 21: 75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeo K, Kawai T, Nishida K, Masuda K, Teshima-Kondo S, Tanahashi T, Rokutan K. Oxidative stress-induced alternative splicing of transformer 2beta (SFRS10) and CD44 pre-mRNAs in gastric epithelial cells. Am J Physiol Cell Physiol 2009; 297: C330–8. [DOI] [PubMed] [Google Scholar]
- 26.Grellscheid S, Dalgliesh C, Storbeck M, Best A, Liu Y, Jakubik M, Mende Y, Ehrmann I, Curk T, Rossbach K, Bourgeois CF, Stevenin J, Grellscheid D, Jackson MS, Wirth B, Elliott DJ. Identification of evolutionarily conserved exons as regulated targets for the splicing activator tra2beta in development. PLoS Genet 2011; 7: e1002390–e1002390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grellscheid SN, Dalgliesh C, Rozanska A, Grellscheid D, Bourgeois CF, Stevenin J, Elliott DJ. Molecular design of a splicing switch responsive to the RNA binding protein Tra2beta. Nucleic Acids Res 2011; 39: 8092–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karunakaran DK, Banday AR, Wu Q, Kanadia R. Expression analysis of an evolutionarily conserved alternative splicing factor, Sfrs10, in age-related macular degeneration. PloS One 2013; 8: e75964–e75964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li SJ, Qi Y, Zhao JJ, Li Y, Liu XY, Chen XH, Xu P. Characterization of nuclear localization signals (NLSs) and function of NLSs and phosphorylation of serine residues in subcellular and subnuclear localization of transformer-2beta (Tra2beta). J Biol Chem 2013; 288: 8898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoilov P, Daoud R, Nayler O, Stamm S. Human tra2-beta1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum Mol Genet 2004; 13: 509–24. [DOI] [PubMed] [Google Scholar]
- 31.Matsuo N, Ogawa S, Imai Y, Takagi T, Tohyama M, Stern D, Wanaka A. Cloning of a novel RNA binding polypeptide (RA301) induced by hypoxia/reoxygenation. J Biol Chem 1995; 270: 28216–22. [DOI] [PubMed] [Google Scholar]
- 32.Kiryu-Seo S, Matsuo N, Wanaka A, Ogawa S, Tohyama M, Kiyama H. A sequence-specific splicing activator, tra2beta, is up-regulated in response to nerve injury. Brain Res Mol Brain Res 1998; 62: 220–3. [DOI] [PubMed] [Google Scholar]
- 33.Tsukamoto Y, Matsuo N, Ozawa K, Hori O, Higashi T, Nishizaki J, Tohnai N, Nagata I, Kawano K, Yutani C, Hirota S, Kitamura Y, Stern DM, Ogawa S. Expression of a novel RNA-splicing factor, RA301/Tra2beta, in vascular lesions and its role in smooth muscle cell proliferation. Am J Pathol 2001; 158: 1685–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nayler O, Cap C, Stamm S. Human transformer-2-beta gene (SFRS10): complete nucleotide sequence, chromosomal localization, and generation of a tissue-specific isoform. Genomics 1998; 53: 191–202. [DOI] [PubMed] [Google Scholar]
- 35.Rong S, Cortes VA, Rashid S, Anderson NN, McDonald JG, Liang G, Moon YA, Hammer RE, Horton JD. Expression of SREBP-1c requires SREBP-2-mediated generation of a sterol ligand for LXR in livers of mice. eLife 2017;6:e25015. [DOI] [PMC free article] [PubMed]
- 36.Smulan LJ, Ding W, Freinkman E, Gujja S, Edwards YJ, Walker AK. Cholesterol-independent SREBP-1 maturation is linked to ARF1 inactivation. Cell Rep 2016; 16: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Zhang J, Qiu W, Han GS, Carman GM, Adeli K. Lipin-1gamma isoform is a novel lipid droplet-associated protein highly expressed in the brain. FEBS Lett 2011; 585: 1979–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pawlak M, Lefebvre P, Staels B. Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol 2015; 62: 720–33. [DOI] [PubMed] [Google Scholar]
- 39.Peng CH, Yang MY, Yang YS, Yu CC, Wang CJ. Antrodia cinnamomea prevents obesity, dyslipidemia, and the derived fatty liver via regulating AMPK and SREBP signaling. Am J Chin Med 2017; 45: 67–83. [DOI] [PubMed] [Google Scholar]
- 40.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011; 146: 408–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu M, Yin H, Mitra MS, Liang X, Ajmo JM, Nadra K, Chrast R, Finck BN, You M. Hepatic-specific lipin-1 deficiency exacerbates experimental alcohol-induced steatohepatitis in mice. Hepatology 2013; 58: 1953–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang P, Verity MA, Reue K. Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab 2014; 20: 267–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meana C, Pena L, Lorden G, Esquinas E, Guijas C, Valdearcos M, Balsinde J, Balboa MA. Lipin-1 integrates lipid synthesis with proinflammatory responses during TLR activation in macrophages. J Immunol 2014; 193: 4614–22. [DOI] [PubMed] [Google Scholar]
- 44.Palmisano BT, Le TD, Zhu L, Lee YK, Stafford JM. Cholesteryl ester transfer protein alters liver and plasma triglyceride metabolism through two liver networks in female mice. J Lipid Res 2016; 57: 1541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Z, Norris JY, Finck BN. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) stimulates VLDL assembly through activation of cell death-inducing DFFA-like effector B (CideB). J Biol Chem 2010; 285: 25996–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Z, Gropler MC, Norris J, Lawrence JC, Jr, Harris TE, Finck BN. Alterations in hepatic metabolism in fld mice reveal a role for lipin 1 in regulating VLDL-triacylglyceride secretion. Arterioscler Thromb Vasc Biol 2008; 28: 1738–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu M, Wang F, Li X, Rogers CQ, Liang X, Finck BN, Mitra MS, Zhang R, Mitchell DA, You M. Regulation of hepatic lipin-1 by ethanol: role of AMP-activated protein kinase/sterol regulatory element-binding protein 1 signaling in mice. Hepatology 2012; 55: 437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun X, Luo W, Tan X, Li Q, Zhao Y, Zhong W, Sun X, Brouwer C, Zhou Z. Increased plasma corticosterone contributes to the development of alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol 2013; 305: G849–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris TE, Finck BN. Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol Metab 2011; 22: 226–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu GH, Gerace L. Sumoylation regulates nuclear localization of Lipin-1a in neuronal cells. PLoS One 2009;4:e7031. [DOI] [PMC free article] [PubMed]
- 51.Steiner JL, Lang CH. Alcohol, adipose tissue and lipid dysregulation. Biomolecules 2017;7:16.
- 52.You M, Rogers CQ. Adiponectin: a key adipokine in alcoholic fatty liver. Exp Biol Med 2009; 234: 850–9. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Kim C, Jogasuria A, Han Y, Hu X, Wu J, Shen H, Chrast R, Finck BN, You M. Myeloid cell-specific Lipin-1 deficiency stimulates endocrine adiponectin-FGF15 axis and ameliorates ethanol-induced liver injury in mice. Sci Rep 2016; 6: 34117–34117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu J, Ren T, Zhou M, Cheng M. The combination of blueberry juice and probiotics reduces apoptosis of alcoholic fatty liver of mice by affecting SIRT1 pathway. Drug Des Dev Ther 2016; 10: 1649–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jang CH, Kim KM, Yang JH, Cho SS, Kim SJ, Shin SM, Cho IJ, Ki SH. The role of Lipin-1 in the regulation of fibrogenesis and TGF-beta signaling in hepatic stellate cells. Toxicol Sci 2016; 153: 28–38. [DOI] [PubMed] [Google Scholar]
- 56.Tang LY, Chen Y, Rui BB, Hu CM. Resveratrol ameliorates lipid accumulation in HepG2 cells, associated with down-regulation of lipin1 expression. Can J Physiol Pharmacol 2016; 94: 185–189. [DOI] [PubMed] [Google Scholar]
- 57.Bourgeois CF, Lejeune F, Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol 2004; 78: 37–88. [DOI] [PubMed] [Google Scholar]
- 58.Kaminska D, Pihlajamaki J. Regulation of alternative splicing in obesity and weight loss. Adipocyte 2013; 2: 143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010; 463: 457–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Medina MW, Krauss RM. Alternative splicing in the regulation of cholesterol homeostasis. Curr Opin Lipidol 2013; 24: 147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clery A, Jayne S, Benderska N, Dominguez C, Stamm S, Allain FH. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1. Nat Struc Mol Biol 2011; 18: 443–50. [DOI] [PubMed] [Google Scholar]
- 62.Elliott DJ, Best A, Dalgliesh C, Ehrmann I, Grellscheid S. How does Tra2beta protein regulate tissue-specific RNA splicing? Biochem Soc Trans 2012; 40: 784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Csaki LS, Reue K. Lipins: multifunctional lipid metabolism proteins. Annu Rev Nutr 2010; 30: 257–72. [DOI] [PMC free article] [PubMed] [Google Scholar]