Abstract
The ability of the Ras oncogene to transform normal cells has been well established. One downstream effector of Ras is the lipid hydrolyzing enzyme phospholipase D. Recent evidence has emerged indicating a role for phospholipase D in cell proliferation, membrane trafficking, and migration. To study the potential importance of phospholipase D in the oncogenic ability of Ras, we used Rat-2 fibroblasts with reduced phospholipase D1 activity (Rat-2V25). Here, we show that H-Ras transformation of Rat-2 fibroblasts requires normal phospholipase D1 activity. WT Rat-2 fibroblasts transfected with the H-RasV12 oncogene grew colonies in soft agar and tumors in nude mice. However, Rat-2V25 cells when transfected with the H-RasV12 oncogene did not form colonies in soft agar or produce tumors when xenografted onto nude mice. Interestingly, in the presence of phosphatidic acid, the product of phospholipase D, growth in soft agar and tumor formation was restored. We also observed a dramatic increase in the expression of phospholipase D1 in colorectal tumors when compared with adjacent normal mucosa. Our studies identify phospholipase D1 as a critical downstream mediator of H-Ras-induced tumor formation.
Keywords: colorectal, Ras, phosphatidic acid, xenograft
Phospholipase D (PLD) is a ubiquitous enzyme that catalyzes the hydrolysis of phosphatidylcholine (PC) to phosphatidic acid (PA) and choline (1, 2). Two mammalian isozymes (PLD1 and PLD2) have been identified, and they are differentially regulated. PLD1 is activated by protein kinase C and small G proteins of the Rho and ADP-ribosylation factor (Arf) families, whereas PLD2 is not directly activated by these regulators and has a different subcellular localization (1). The Src oncogene can also activate PLD (3). However, the mechanism is indirect, involving the activation of Ras and other small G proteins (Ral and Arf) (1, 4–6). Although a great deal is known about the regulation of PLD1, its exact cellular function remains unclear. Recent evidence has supported a role for PLD in cell proliferation, survival signaling, apoptosis, and tumor progression (7, 8).
Mutations in the Ras oncogene are found in a variety of human malignancies, including those of the lung, breast, and colon (9). These mutations result in the constitutive activation of Ras and lead to the deregulated activation of important classical downstream signaling pathways, including Raf/MEK/extracellular signal-regulated kinase (10, 11) and PI3-kinase/Akt (12, 13). As noted above, recent evidence indicates that Ras can also induce the activation of Ral, and this small G-protein can synergize with Arf to activate PLD (14).
Because mutations in Ras can lead to the activation of PLD and Ras mutations occur in many cancers, this study was designed to explore the role of PLD1 in Ras-induced cellular transformation and tumor growth. The approach used a modified form of PLD1 that is catalytically inactive and inhibits the activity of endogenous PLD (15). The deletion or addition of a single amino acid at the C terminus of PLD1 renders it catalytically inactive (16). We therefore generated a PLD1 construct that was made inactive by adding a V5 epitope tag added to the C terminus. We then selected Rat-2 fibroblasts, which constitutively express this construct (termed Rat-2V25 cells). In the presence of lysophosphatidic acid or phorbol ester, these cells show >50% reduction in the stimulation of PLD activity versus WT Rat-2 cells (15). We used these cells to further evaluate the role of PLD1 in the Ras-induced transformation of normal fibroblasts.
Materials and Methods
Materials. wt Rat-2 cells were obtained from the American Type Culture Collection, and Rat-2V25 cells were generated as described in ref. 15. FBS, DMEM (powder and liquid), Opti-MEM, penicillin, streptomycin, and glutamine were from GIBCO (Invitrogen). SeaPlaque agarose was from BioWhittaker. FuGENE 6 was from Roche Applied Science (Indianapolis). Raf-1/Ras binding domain of Raf agarose and hPLD2 antibodies were obtained from Upstate Biotechnology (Charlottesville, VA). 1,2-Dilauroyl-glycero-3-phosphate (12:0 PA), 1,2-dipalmitoylglycero-3-phosphate (16:0 PA), and 1,2-lauroyl-glycero-3-phosphocholine lipids were from Avanti Polar Lipids. Polyvinylidene dif luoride membranes were from Millipore. Horseradish peroxidase-conjugated secondary antibodies were from Vector Laboratories. The primary Pan-Ras antibody was from Oncogene Research Products. Hygromycin B was from Clontech. H-RasV12 and K-RasV12 were obtained from Guthrie cDNA Resource Center. Hemagglutinin (HA) primary antibody was from Santa Cruz Biotechnology. Antibodies to PLD1 were raised in rabbits as described in ref. 17.
Cell Culture Conditions. Rat-2 cells were maintained in DMEM with 4 mM l-glutamine supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2. Rat-2V25 cells were maintained in DMEM with 4 mM l-glutamine supplemented with 10% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.5 mg/ml geneticin at 37°C in a humidified atmosphere of 5% CO2.
SDS/PAGE and Western Blotting. SDS gel electrophoresis was performed on 4–20% Tris/glycine-polyacrylamide gels at 100 V for 2.5 h, and the proteins were transferred onto polyvinylidene fluoride membranes by using a Novex wet transfer apparatus at 20 V for 2 h. The membranes were blocked overnight in 5% nonfat dry milk. The blots were then incubated for 1 h in the presence of the primary antibody, rinsed three times in Tris-buffered saline containing Tween 20, and incubated with the corresponding secondary antibody to horseradish peroxidase. Bands were visualized with the enhanced chemiluminescence kit from Amersham Pharmacia Biotech.
Ras Activation Assay. Cells were seeded to 70–80% confluency in six-well dishes and then transfected with H-RasV12 (Guthrie, Sayre, PA) by using FuGENE 6 according to the manufacturer's protocol. The transfections were allowed to proceed for 24 h. The cells were lysed in 0.6 ml of lysis buffer containing 25 mM Hepes (pH 7.5), 10 mM MgCl, 150 mM NaCl, 1 mM EDTA, 10 μg/ml leupeptin, 10 μg/ml antipain, 1 mM sodium orthovanadate, 25 mM β-glycerophosphate, 1% Nonidet P-40, and 10% glycerol. Cells were then diluted to 1 μg/μl with buffer. Then, 30 μl of Raf-1/Ras binding domain of Raf agarose beads were added to the lysate and allowed to rock at 4°C for 30 min. The beads were then washed three times with the lysis buffer and resuspended in 1× Laemmli sample buffer and boiled for 5 min. The relative amount of active Ras was determined by Western analysis.
Soft Agar Assay. Cells were seeded at 5 × 104 in 6-well dishes and transfected with H-RasV12 by using FuGENE 6 according to the manufacturer's protocol. The transfected cells were incubated for 48 h. Plates were then prepared by placing 1.5 ml of 2× DMEM 10% FBS and 1.6% sterile SeaPlaque (1:1) mixture into 35-mm sterile dishes. Agar was allowed to form for 1 h at room temperature. Transfected cells were then trypsinized and resuspended to a concentration of 5,000 cells per ml and mixed with 1.6% sterile Sea Plaque and 2× DMEM 10% FBS (2:1:1). One milliliter of the cell suspension was added to the agar plates, and then they were placed in a CO2 incubator for 24 h. Lipids were dried under nitrogen, resuspended in DMEM, and added dropwise to the plates at a final concentration of 200 μM.
Xenograph Studies. Stable WT Rat-2 and Rat-2V25 cells expressing H-RasV12 or vector were generated by treatment with 0.2 mg/ml hygromycin B and 0.05 mg/ml hygromycin B, respectively. A total of 5 × 106 cells were then sterilely injected intradermally into nude mice. Tumors were allowed to grow for 10 days and then measured by using the equation [V = (LW2) × 0.5], where V equals volume, L equals length, and W equals width.
Human Colorectal Tissue Analysis. Human colorectal carcinoma specimens were obtained from surgical resections with approval of the local Investigational Review Board. For each carcinoma sample, adjacent normal mucosa was collected for comparison. All samples were immediately frozen and stored in liquid nitrogen until use.
Northern Analysis. mRNAs were isolated and subjected to human PLD1 Northern analysis as described in refs. 18 and 19.
Measurement of PLD Activity. PLD activity was measured as described in ref. 15. Briefly, cells were seeded to 50% confluency in 6-well dishes and then transfected with rat brain PLD1 (15) or vector control by using FuGENE 6 according to the manufacturer's protocol. The transfections were allowed to proceed for 24 h after serum starvation in serum-free DMEM for 18 to 24 h before the start of the assay. The cells were labeled with 1 μCi/ml (1 Ci = 37 GBq) of [9,10-3H]myristic acid for the final 16 h of serum starvation. The cells were washed three times with PBS and preincubated at 37°C in serum-free DMEM for 1 h. For the final 10 min of preincubation, 0.3% butan-1-ol was included. After treatment with 10 μg/ml lysophosphatidic acid for 5 min or 100 nM phorbol 12-myristate 13-acetate for 15 min, the cells were washed once with ice-cold PBS, and then ice-cold methanol was added. Cells were scraped off the plates, and the lipids were extracted and separated with methanol/chloroform/0.1 M HCl (1:1:1). The lower phase was dried under N2, resuspended in chloroform/methanol (2:1), and spotted on TLC plates of silica gel 60A. The plates were developed in the upper phase of the solvent system of ethyl acetate/isooctane/H2O/acetic acid (55:25:50:10), and the radioactivity of the bands corresponding to phosphatidylbutanol were measured.
Results and Discussion
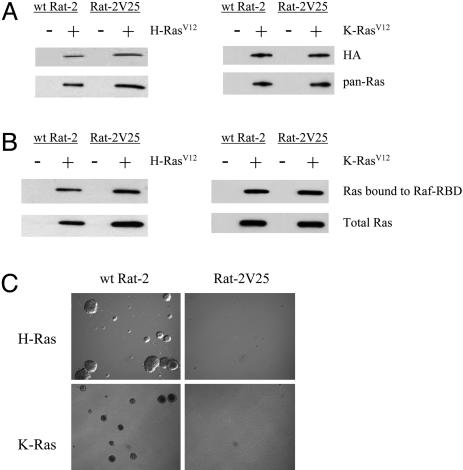
Many immediate downstream effectors of Ras have been identified, including Raf, Ral-guanine nucleotide dissociation stimulator, PI-3 kinase, and p120 GTPase-activating protein (20). Recent evidence indicates that the Ras-induced activation of Ral through the Ral guanine nucleotide dissociation stimulator converges with Arf in the synergistic activation of PLD (14). We therefore sought to determine what role PLD may play in the ability of Ras to transform cells. We transfected the constitutively active forms of H-Ras (H-RasV12) and K-Ras (K-RasV12) tagged with HA into WT Rat-2 and Rat-2V25 (expressing catalytically inactive V5-tagged PLD1) fibroblasts. Western analysis confirmed the expression of both HA-tagged H-RasV12 and K-RasV12 and showed that the level of endogenous Ras was extremely low (Fig. 1A). Only upon long exposure could WT Ras be detected (data not shown). To determine whether the expressed RasV12 constructs were indeed functional, we evaluated their activity in WT Rat-2 and Rat-2V25 fibroblasts. We incubated cell lysates in the presence of the Ras binding domain of Raf linked to agarose beads because only the active GTP bound form of Ras will associate with the Ras binding domain of Raf (21). These results indicate that H-Ras and K-Ras are equally active in both WT Rat-2 cells and in Rat-2V25 cells deficient in PLD1 activity (Fig. 1B).
Fig. 1.
Evaluation of H-RasV12 and K-RasV12 transformation in WT Rat-2 and Rat-2V25 fibroblasts. (A) Protein expression levels of HA-tagged H-RasV12, HA-tagged K-RasV12, and total Ras in WT Rat-2 and Rat-2V25 cells. (B) Ras activation assay in WT Rat-2 and Rat-2V25 cells expressing H-RasV12 or K-RasV12(C) Growth of H-RasV12- or K-RasV12-transformed cells in soft agar. These results are representative of three independent experiments.
The ability of an oncogene to induce the growth of colonies in soft agar is a classical in vitro experiment to determine its transforming potential. We observed, as expected, that the expression of H-RasV12 or K-RasV12 in WT Rat-2 fibroblasts induced the growth of colonies in soft agar (Fig. 1C). However, neither the expression of H-RasV12 nor K-RasV12 in Rat-2V25 fibroblasts produced colonies in soft agar. This observation is of interest because the exact role of PLD downstream of K-Ras activation is not clearly understood. Xu et al. (14) have shown that PLD activity is elevated in H-Ras transformed fibroblasts but not in K-Ras transformed cells. However, other studies have demonstrated that K-Ras-transformed cells induce PLD activation (22, 23). Our current data indicate that PLD1 activity is a required and necessary step in both the H-Ras- and K-Ras-induced transformation of normal fibroblasts.
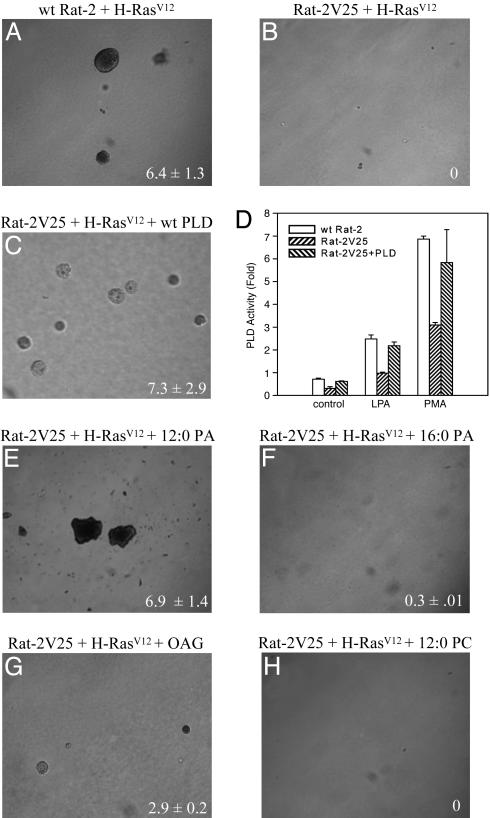
To determine whether the inhibition of transformation in Rat-2V25 cells is due to the reduced activity of PLD1 and not to impaired Ras signaling, we examined the effect of reintroduction of PLD1 on the ability of H-RasV12 to induce colony formation in soft agar. We transiently transfected Rat-2V25 cells with either PLD1 or vector control before growth in soft agar. The expression of PLD1 restored the ability of H-RasV12 to induce colony formation (Fig. 2 A–C). It also restored the level of PLD activity to nearly that of WT Rat-2 cells (Fig. 2D). We also examined what effect the product of PLD, PA, would have on the ability of H-RasV12 to induce transformation. We cultured Rat-2V25 cells in the presence of a cell permeable form of PA (12:0 PA, 200 μM), which induced the production of colonies of a similar rate as WT Rat-2 fibroblasts (Fig. 2E). Furthermore, the addition of 12:0 PA did not increase the rate or size of colony formation in H-RasV12 transformed WT Rat-2 fibroblasts (data not shown). The 16-carbon form of PA (16:0 PA, 200 μM), which is not readily taken up by cells, did not induce the growth of Rat-2V25 cells in soft agar (Fig. 2F). The formation of PA itself has been shown to induce cell proliferation and migration; however, PA can be metabolized into diacylglycerol (DAG). DAG is an activator of the conventional PKC isoforms that have been shown to play a role in Ras transformation (24). In the presence of 1-oleoyl-2-acetyl-glycerol (200 μM), a cell permeable analog of DAG, the H-RasV12-induced transformation of Rat-2V25 cells was restored, although to a lesser extent than with the addition of PA (Fig. 2G). The addition of PC (12:0 PC, 200 μM), the substrate of PLD, did not restore the ability of H-RasV12 to induce colony formation (Fig. 2H). Together these data indicate that the inability of H-RasV12 to transform Rat-2V25 fibroblasts is not due to aberrant Ras signaling, but is due to the specific loss of PLD1 activity.
Fig. 2.
The reintroduction of PLD1 or its lipid product (PA) can restore H-RasV12-induced transformation of Rat-2V25 fibroblasts in soft agar. (A) Growth of WT Rat-2 H-RasV12-transformed cells in soft agar. Growth of Rat-2V25 H-RasV12-transformed cells (B) transfected with PLD1 (C), in the presence of 200 μM 12:0 PA (E), 200 μM 16:0 PA (F), 200 μM 1-oleoyl-2-acetyl-glycerol (G), or 200 μMPC(H). (D) PLD activation on WT Rat-2, Rat-2V25, and Rat-2V25 cells transfected with PLD1. The results are representative of three independent experiments performed in triplicate. Numbers at lower right are the number of colonies per field ± SEM.
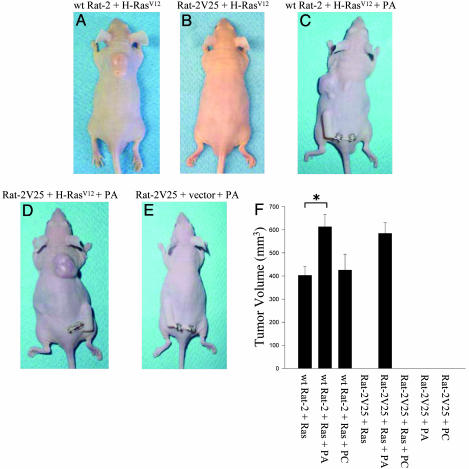
The introduction of the H-RasV12 oncogene cannot only stimulate growth in semisolid media but can also induce tumor formation in immunodeficient mice (25). Therefore, we determined what effect the loss of PLD1 function may have on the ability of H-Ras to induce tumor growth. We transfected Rat-2V25 and WT Rat-2 fibroblasts with the H-RasV12 oncogene and injected them into the dorsal flank of nude mice. Within 7–10 days large tumors were visible in mice injected with WT Rat-2 cells expressing H-RasV12 (Fig. 3A). Interestingly, Rat-2V25 cells (with reduced PLD1 activity) transfected with H-RasV12 did not produce any tumors (Fig. 3B). We maintained these mice beyond 45 days without any tumor development. Furthermore, we also observed what effect PA may have on the production of tumors in vivo. We continuously administered PA (12:0 PA) through the use of a microosmotic pump inserted s.c. at a dose of 5.25 nmol/h. The infusion of PA increased tumor growth when mice were injected with WT Rat-2 fibroblasts transfected with H-RasV12 (Fig. 3 C and F). Interestingly, PA-treated mice injected with Rat-2V25 cells transfected with H-RasV12 (Fig. 3 D and F) grew tumors at the same rate and size as control mice with WT Rat-2 cells (Fig. 3 C and F). Rat-2V25 cells transfected with empty vector did not form tumors in nude mice in the presence of PA, indicating that the growth of xenografts observed depends on H-RasV12 and the activation of PLD1 and is not induced by PA alone (Fig. 3E). When compared with mice not treated with PA, the infusion of PA induced an increase in tumor growth in mice injected with H-RasV12 transfected WT Rat-2 cells, whereas the infusion of PC had no effect on growth (Fig. 3F). Furthermore, in the presence of PA, mice injected with Rat-2V25 cells transformed with H-RasV12 showed a similar rate of tumor growth as mice injected with WT Rat-2 cells transformed with H-RasV12. Together, these data indicate a role for PLD1 in the transforming ability of H-Ras in vivo.
Fig. 3.
Xenograft growth of H-RasV12-transformed WT Rat-2 and Rat-2V25 fibroblasts. (A and B) Xenograft growth of WT Rat-2 (A) and Rat-2V25 (B) cells transformed with H-RasV12 on day 10. (C–E) Effect of PA on xenograft growth of H-RasV12-transformed WT Rat-2 cells (C), H-RasV12-transformed Rat-2V25 cells (D), and vector-transfected Rat-2V25 cells (E). (F) Summary graph of tumor size from H-RasV12-transformed cells (*, P < 0.05). Data represents the average ± SEM of three independent experiments.
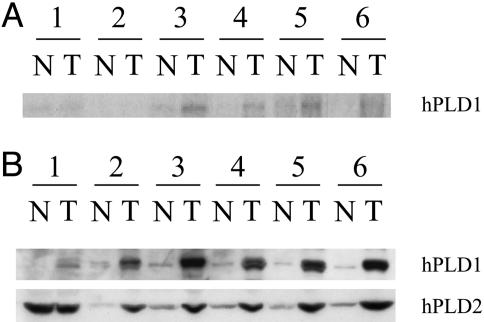
As mentioned earlier, mutations in the Ras oncogene occur in many cancers and have been shown to play a role in the initiation of tumor development. For example, the initiation of colon cancer can occur through a defined pathway. As colonic tissue progresses from hyperplasia through adenocarcinoma to carcinoma, consistent alterations of signaling pathways emerge. This alteration first involves deregulation of the APC/β-catenin pathway, followed by mutations in the Ras oncogene, eventually resulting in the loss of p53 function (26). Because the prevalence of Ras mutations in colorectal cancer is high (from 20% to 60%; refs. 27 and 28), we investigated the expression of PLD1 in colorectal carcinomas. Colorectal tumors paired with the corresponding adjacent normal mucosa were submitted to Northern and Western analysis. We observed an increase in PLD1 mRNA in the colorectal tumors versus the adjacent normal mucosa (Fig. 4A). Furthermore, we observed a dramatic coregulated increase of PLD1 and PLD2 protein expression in five of six colorectal tumor tissues when compared with normal adjacent mucosa (Fig. 4B). These data are consistent with recent findings that demonstrate that PLD activity is up-regulated in breast, renal, colorectal, and gastric cancers (29–32). Mutations in Ras signaling accompanied by an increase in PLD expression indicate that PLD may serve an important role in the Ras-induced colorectal carcinogenesis. Although we have shown the importance of PLD1 in Ras transformation, we cannot exclude any possible effects of PLD2 because both isoforms are overexpressed in colorectal cancer, and we could detect low levels of PLD2 in Rat-2 fibroblasts (data not shown).
Fig. 4.
Comparison of PLD1 expression between colorectal carcinomas and adjacent normal mucosa. (A) Northern analysis of hPLD1 mRNA. (B) Western analysis of hPLD1 and hPLD2 protein expression.
Our data indicate the requirement of PLD1 activity in the ability of H-Ras and K-Ras to induce transformation in vitro and in vivo. The activation of PLD1 by Ras has been established in many cell systems. Ras activates RalA through the direct activation of Ral-guanine nucleotide dissociation stimulator (33). The activation of RalA and Arf (Arf6) converge in the synergistic activation of PLD1 (14). The activation of PLD has also been shown to play an important role in Ras-mediated signaling through the generation of PA (6). Another important event in Ras-induced transformation is the activation of the Raf/MEK/mitogen-activated protein kinase (MAPK) pathway. This pathway is also critical to Ras signaling. A requirement of PA has been shown for the recruitment of Raf-kinase to the membrane where Ras-induced activation of Raf occurs (34). The endocytosis of signaling receptors resulting in signaling endosomes is also required for the activation of MAPK by MEK, both important downstream mediators of Ras (35). The formation of signaling endosomes has been reported to depend on the production of PA through the activation of PLD1 (36). Furthermore, the activation of Ras as well as the production of PA has been shown to activate the mammalian target of rapamycin (mTOR) (37–40). mTOR is an important kinase that can induce both an increase in cell growth and cell division (41), thus indicating an important role for PLD1 in Ras-induced transformation.
Interestingly, recent work has implicated PLD in other oncogene-induced transformation and growth. The over-expression of PLD2 with c-Src or PLD1 with the epidermal growth factor receptor transforms Rat 3Y1 fibroblasts (42), and the overexpression of PLD1 or PLD2 with c-Src enhances cell proliferation (43). PLD2 may also play a role in v-Src-mediated formation of cell protrusions that are involved in the invasive properties of v-Src-transformed cells (44). Here we report evidence that clearly demonstrates a direct requirement of PLD1 and its enzymatic product, PA, downstream of the Ras pathway leading to oncogenesis. Furthermore, the up-regulation of PLD1 in colorectal tumors coupled with a prevalence of Ras activation in colorectal carcinomas indicate PLD1 as an attractive antitumor target.
Acknowledgments
This work was supported in part by the Howard Hughes Medical Institute (J.H.E.) and U.S. Public Health Services Grants DK 47297, P30CA-68485, DK 62112, and PO1CA-77839 (to R.N.D.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ha, hemagglutinin; PA, phosphatidic acid; PC, phosphatidylcholine; PLD, phospholipase D; Arf, ADP-ribosylation factor.
References
- 1.Exton, J. H. (1999) Biochim. Biophys. Acta 1439, 121–133. [DOI] [PubMed] [Google Scholar]
- 2.McDermott, M., Wakelam, M. J. & Morris, A. J. (2004) Biochem. Cell Biol. 82, 225–253. [DOI] [PubMed] [Google Scholar]
- 3.Song, J. G., Pfeffer, L. M. & Foster, D. A. (1991) Mol. Cell. Biol. 11, 4903–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang, H., Luo, J. Q., Urano, T., Frankel, P., Lu, Z., Foster, D. A. & Feig, L. A. (1995) Nature 378, 409–412. [DOI] [PubMed] [Google Scholar]
- 5.Jiang, H., Alexandropoulos, K., Song, J. & Foster, D. A. (1994) Mol. Cell. Biol. 14, 3676–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang, H., Lu, Z., Luo, J. Q., Wolfman, A. & Foster, D. A. (1995) J. Biol. Chem. 270, 6006–6009. [DOI] [PubMed] [Google Scholar]
- 7.Foster, D. A. & Xu, L. (2003) Mol. Cancer Res. 1, 789–800. [PubMed] [Google Scholar]
- 8.Nozawa, Y. (2002) Biochim. Biophys. Acta 1585, 77–86. [DOI] [PubMed] [Google Scholar]
- 9.Bos, J. L. (1989) Cancer Res. 49, 4682–4689. [PubMed] [Google Scholar]
- 10.McCormick, F. (1994) Curr. Opin. Genet. Dev. 4, 71–76. [DOI] [PubMed] [Google Scholar]
- 11.Robinson, M. J. & Cobb, M. H. (1997) Curr. Opin. Cell Biol. 9, 180–186. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez-Viciana, P., Warne, P. H., Khwaja, A., Marte, B. M., Pappin, D., Das, P., Waterfield, M. D., Ridley, A. & Downward, J. (1997) Cell 89, 457–467. [DOI] [PubMed] [Google Scholar]
- 13.Osada, M., Tolkacheva, T., Li, W., Chan, T. O., Tsichlis, P. N., Saez, R., Kimmelman, A. C. & Chan, A. M. (1999) Mol. Cell. Biol. 19, 6333–6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu, L., Frankel, P., Jackson, D., Rotunda, T., Boshans, R. L., D'Souza-Schorey, C. & Foster, D. A. (2003) Mol. Cell. Biol. 23, 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kam, Y. & Exton, J. H. (2001) Mol. Cell. Biol. 21, 4055–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie, Z., Ho, W. T. & Exton, J. H. (2000) Eur. J. Biochem. 267, 7138–7146. [DOI] [PubMed] [Google Scholar]
- 17.Min, D. S., Kim, E. G. & Exton, J. H. (1998) J. Biol. Chem. 273, 29986–29994. [DOI] [PubMed] [Google Scholar]
- 18.Hammond, S. M., Jenco, J. M., Nakashima, S., Cadwallader, K., Gu, Q., Cook, S., Nozawa, Y., Prestwich, G. D., Frohman, M. A. & Morris, A. J. (1997) J. Biol. Chem. 272, 3860–3868. [DOI] [PubMed] [Google Scholar]
- 19.Gibbs, T. C. & Meier, K. E. (2000) J. Cell. Physiol. 182, 77–87. [DOI] [PubMed] [Google Scholar]
- 20.Campbell, S. L., Khosravi-Far, R., Rossman, K. L., Clark, G. J. & Der, C. J. (1998) Oncogene 17, 1395–1413. [DOI] [PubMed] [Google Scholar]
- 21.de Rooij, J. & Bos, J. L. (1997) Oncogene 14, 623–625. [DOI] [PubMed] [Google Scholar]
- 22.Ramirez de Molina, A., Rodriguez-Gonzalez, A., Penalva, V., Lucas, L. & Lacal, J. C. (2001) Biochem. Biophys. Res. Commun. 285, 873–879. [DOI] [PubMed] [Google Scholar]
- 23.Oliva, J. L., Zarich, N., Martinez, N., Jorge, R., Castrillo, A., Azanedo, M., Garcia-Vargas, S., Gutierrez-Eisman, S., Juarranz, A., Bosca, L., et al. (2004) J. Biol. Chem. 279, 33480–33491. [DOI] [PubMed] [Google Scholar]
- 24.Kampfer, S., Windegger, M., Hochholdinger, F., Schwaiger, W., Pestell, R. G., Baier, G., Grunicke, H. H. & Uberall, F. (2001) J. Biol. Chem. 276, 42834–42842. [DOI] [PubMed] [Google Scholar]
- 25.Clark, G. J., Cox, A. D., Graham, S. M. & Der, C. J. (1995) Methods Enzymol. 255, 395–412, 395–412. [DOI] [PubMed] [Google Scholar]
- 26.Fearon, E. R. & Vogelstein, B. (1990) Cell 61, 759–767. [DOI] [PubMed] [Google Scholar]
- 27.Bos, J. L., Fearon, E. R., Hamilton, S. R., Verlaan-de Vries, M., van Boom, J. H., van der Eb, A. J. & Vogelstein, B. (1987) Nature 327, 293–297. [DOI] [PubMed] [Google Scholar]
- 28.Forrester, K., Almoguera, C., Han, K., Grizzle, W. E. & Perucho, M. (1987) Nature 327, 298–303. [DOI] [PubMed] [Google Scholar]
- 29.Noh, D. Y., Ahn, S. J., Lee, R. A., Park, I. A., Kim, J. H., Suh, P. G., Ryu, S. H., Lee, K. H. & Han, J. S. (2000) Cancer Lett. 161, 207–214. [DOI] [PubMed] [Google Scholar]
- 30.Zhao, Y., Ehara, H., Akao, Y., Shamoto, M., Nakagawa, Y., Banno, Y., Deguchi, T., Ohishi, N., Yagi, K. & Nozawa, Y. (2000) Biochem. Biophys. Res. Commun. 278, 140–143. [DOI] [PubMed] [Google Scholar]
- 31.Uchida, N., Okamura, S. & Kuwano, H. (1999) Anticancer Res. 19, 671–675. [PubMed] [Google Scholar]
- 32.Oshimoto, H., Okamura, S., Yoshida, M. & Mori, M. (2003) Oncol. Res. 14, 31–37. [DOI] [PubMed] [Google Scholar]
- 33.Urano, T., Emkey, R. & Feig, L. A. (1996) EMBO J. 15, 810–816. [PMC free article] [PubMed] [Google Scholar]
- 34.Rizzo, M. A., Shome, K., Vasudevan, C., Stolz, D. B., Sung, T. C., Frohman, M. A., Watkins, S. C. & Romero, G. (1999) J. Biol. Chem. 274, 1131–1139. [DOI] [PubMed] [Google Scholar]
- 35.Roy, S., Wyse, B. & Hancock, J. F. (2002) Mol. Cell. Biol. 22, 5128–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andresen, B. T., Rizzo, M. A., Shome, K. & Romero, G. (2002) FEBS Lett. 531, 65–68. [DOI] [PubMed] [Google Scholar]
- 37.Shao, J., Evers, B. M. & Sheng, H. (2004) Cancer Res. 64, 229–235. [DOI] [PubMed] [Google Scholar]
- 38.Chen, Y., Zheng, Y. & Foster, D. A. (2003) Oncogene 22, 3937–3942. [DOI] [PubMed] [Google Scholar]
- 39.Fang, Y., Vilella-Bach, M., Bachmann, R., Flanigan, A. & Chen, J. (2001) Science 294, 1942–1945. [DOI] [PubMed] [Google Scholar]
- 40.Kam, Y. & Exton, J. H. (2004) FASEB J. 18, 311–319. [DOI] [PubMed] [Google Scholar]
- 41.Schmelzle, T. & Hall, M. N. (2000) Cell 103, 253–262. [DOI] [PubMed] [Google Scholar]
- 42.Joseph, T., Wooden, R., Bryant, A., Zhong, M., Lu, Z. & Foster, D. A. (2001) Biochem. Biophys. Res. Commun. 289, 1019–1024. [DOI] [PubMed] [Google Scholar]
- 43.Ahn, B. H., Kim, S. Y., Kim, E. H., Choi, K. S., Kwon, T. K., Lee, Y. H., Chang, J. S., Kim, M. S., Jo, Y. H. & Min do, S. (2003) Mol. Cell. Biol. 23, 3103–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen, Y., Zheng, Y. & Foster, D. A. (2002) Biochem. Biophys. Res. Commun. 293, 201–206. [DOI] [PubMed] [Google Scholar]