Abstract
As a member of a highly conserved family of NAD+-dependent histone deacetylases, Sirt6 is a key regulator of mammalian genome stability, metabolism, and life span. Previous studies indicated that Sirt6 is hardwired to remove histone acetylation at H3K9 and H3K56. However, how Sirt6 recognizes its nucleosome substrates has been elusive due to the difficulty of accessing homogenous acetyl-nucleosomes and the low activity of Sirt6 toward peptide substrates. Based on the fact that Sirt6 has an enhanced activity to remove long chain fatty acylation from lysine, we developed an approach to recombinantly synthesize histone H3 with a fatty acylated lysine, Nε-(7-octenoyl)-lysine (OcK), installed at a number of lysine sites and used these acyl-H3 proteins to assemble acyl-nucleosomes as active Sirt6 substrates. A chemical biology approach that visualizes OcK in nucleosomes and therefore allows directly sensitizes Sirt6 activities on its acyl-nucleosome substrates was also formulated. By combining these two approaches, we showed that Sirt6 actively removes acylation from H3K9, H3K18, and H3K27, has relatively low activities toward H3K4 and K3K23, but sluggishly removes acylation at H3K14, H3K36, H3K56, and H3K79. Overexpressing Sirt6 in 293T cells led to downregulated acetylation at H3K18 and K3K27, confirmed these two novel Sirt6-targeted nucleosome lysine sites in cells. Given that downregulation of H3K18 acetylation is correlated with poor prognosis of several cancer types and H3K27 acetylation antagonizes repressive gene regulation by di- and trimethylation at H3K27, our current study implies that Sirt6 may serve as a target for cancer intervention and regulatory pathway investigation in cells.
Introduction
As a founding member of NAD+-dependent histone deacetylases or sirtuins, Saccharomyces cerevisiae Sir2 removes lysine acetylation from histones H3 and H4 and subsequently impacts genome stability, gene silencing, and yeast lifespan.1, 2 There are seven Sir2 homologs in mammalian cells, Sirt1-7.3 Among mammalian sirtuins, Sirt1, Sirt6, and Sirt7 are primarily localized in the nucleus.4 Their functions have long been considered to remove lysine acetylation from chromatin. However, unlike Sirt1 that displays high deacetylation activities toward acetyl-peptide substrates, both Sirt6 and Sirt7 only sluggishly remove acetylation from acetyl-peptide substrates.5, 6 A recent discovery that Sirt6 has enhanced activities toward fatty acyl-peptide substrates and directly regulates lysine myristoylation of tumor necrosis factor alpha (TNF-α) has led to speculation that Sirt6 serves primarily to remove lysine fatty acylation instead of conventional lysine acetylation.7, 8 However, both knock-down/out and overexpressing Sirt6 in cells have shown clear acetylation pattern changes at H3K9 and H3K56, indicating deacetylation activities of Sirt6 at these two sites.9-11 The discrepancy between weak in vitro activities of Sirt6 on acetyl-peptide substrates and robust deacetylation of H3K9 and H3K56 by Sirt6 in cells may be due to additional cellular factors that contribute to Sirt6 activation but were missing in the in vitro analysis. Indeed, a recent report by Cohen et al. showed higher deacetylation activities of Sirt6 on nucleosomes assembled from bulk chicken histones than bulk chicken histones themselves, implying that the nucleosome structure may contribute to Sirt6 activity.12 However, the molecular details of this activation are complicated by the high heterogeneity of the bulk chicken histones used in this study, which contained multiple histone isoforms and many modification types and their combinations. To understand how Sirt6 recognizes its acetyl-lysine substrates on the nucleosome level and how precisely the nucleosome structure activates Sirt6 at molecular details, homogenous acetyl-nucleosome substrates are required. However, this has been hampered due to the formidable difficulty of their synthesis using traditional methods.13, 14 Despite the remaining doubts of Sirt6 biochemistry, mounting evidence has proven the critical functions of Sirt6 in metabolism, inflammation, and genome stability.15 Transgenic mice overexpressing Sirt6 have low LDL cholesterol and triglyceride levels, reduced body fat, improved glucose tolerance, and a long lifespan. However, Sirt6-deficient mice show a markedly degenerative phenotype and have a short lifespan, hypoglycemia, and defects in DNA repair.16 Given its role in regulating glycolysis, loss of Sirt6 leads to tumor formation independent of oncogene activation.16 This tumor-suppressing role is also supported by the fact that Sirt6 is downregulated in several human cancers including pancreatic and colorectal cancers.17, 18 However, other studies also suggest oncogenic effects of Sirt6. Expression of Sirt6 has been associated with poor prognosis and chemosensitivity in patients with non-small cell lung, breast, prostate, and skin cancers.19-22 Sirt6 also plays a role in inflammatory pathways, exerting anti-inflammatory effects at the transcriptional level and in bone metabolism, culminating in low-turnover osteopenia in Sirt6-deficient mice.23
It is obvious that Sirt6 plays a number of roles in cells, in strong contrast to the very few deacetylation targets that have been identified so far. Recently we demonstrated that both Sirt1 and Sirt2 do not have sequence preferentiality when they target nucleosomal lysines for their deacetylation.24 Due to its high structural similarity with Sirt1 and Sirt2, Sirt6's selective targeting of certain histone sites is of a surprise. There are many lysines in the four histones whose sequence contexts resemble H3K9 and H3K56. Potentially those are Sirt6-targeted deacetylation sites but they have not yet tested. As for H3K56, it is located at the nucleosome core region. Unlike H3K9, which is at the H3 N-terminal tail and flexible when binding Sirt6, the surrounding residues of H3K56 have rigid structures. Sirt6 recognition of H3K56 must be dramatically different from that of H3K9. This aspect needs to be addressed using nucleosome substrates that have acetylation site-specifically installed at H3K9 and H3K56, respectively. In the current study, we showed that Sirt6-targeted nucleosomal deacylation sites can be swiftly screened and studied using nucleosome substrates site-specifically modified with a fatty acyl-lysine that can be covalently and fluorescently labeled by a dye.
Results
A chemical biology approach to screen Sirt6-targeted nucleosomal deacylation sites
To search histone H3 lysines that can be targeted by Sirt6 for deacetylation, we initiated our study using recombinant mono-acetylated H3 (acetyl-H3) proteins as potential Sirt6 substrates. The synthesis of acetyl-H3 proteins was achieved using an evolved Methanosarcina mazei pyrrolysyl-tRNA synthetase (AcKRS) that selectively charges its cognate tRNAPyl with Nε-acetyl-lysine (AcK or Kac) for the delivery of AcK at an amber mutation site in histone H3 in Escherichia coli.25, 26 Using this approach, nine acetyl-H3 proteins with AcK installed at K4, K9, K14, K18, K23, K27, K36, K56, and K79 (H3K4-79ac) were synthesized and purified to homogeneity (Supplementary Figure 1). For a comparative study, activities of three sirtuin enzymes, Sirt1, Sirt2, and Sirt6 on these acetyl-H3 substrates were analyzed. The acetyl-H3 proteins (6.4 μM) were solubilized in buffer containing 0.5 M arginine and incubated with a sirtuin enzyme (0.4 μM) for 12 h followed by Western blot analysis of relative acetylation level changes (Supplementary Figure 2). We noticed strong Sirt1- and Sirt2-catalyzed deacetylation for most acetyl-H3 proteins. In striking contrast, there was no significant Sirt6-catalyzed deacetylation for the acetyl-H3 proteins. Weak deacetylation activity of Sirt6 toward peptide substrates has been well documented. Here we show that using acetyl-H3 proteins as substrates doesn't significantly improve Sirt6's deacetylation activity. Since not every specific acetylation has a commercially available antibody for its detection, the pan-Kac antibody from PTM Biolabs was used in our study. We noticed that this antibody has varied recognition abilities toward different acetylation sites and it recognizes acetylation at H3K36 weakly. This feature has been commonly observed for a pan antibody.
Inspired by the observation made by Lin, Denu, and their coworkers that Sirt6 has enhanced deacylation activities toward peptide substrates containing a long chain fatty acyl-lysine and the observation by Cohen et al. that the nucleosome structure may activate Sirt6,7, 8, 12 we sought to synthesize acyl-H3 proteins with a long chain fatty acyl-lysine and use them to assemble corresponding acyl-nucleosomes as active substrates for screening Sirt6-targeted nucleosomal deacylation sites, though long chain fatty acylation on histones is artificial. To analyze Sirt6-catalyzed deacylation on these acyl-nucleosome substrates, they could be directly transferred to membranes for Western blot analysis with corresponding fatty acyl-lysine detecting antibodies. It is critical for us to directly analyze deacylation levels on acyl-nucleosome substrates instead of on acyl-H3 proteins because the dissolution of acyl-nucleosomes during assays to acyl-H3 proteins or their tetramers with H4 will jeopardize our analysis and provide misleading results about what sites are targeted by Sirt6 at the nucleosome level. Unfortunately, no commercial antibodies for lysine fatty acylation detection are available. An alternative analytical approach is to use mass spectrometry (MS). However, doing MS analysis on intact nucleosomes is very challenging. To overcome this analytical obstacle, we designed a chemical biology approach as illustrated in Figure 1. Previously we have shown that a tetrazine undergoes inverse electron-demand Diels-Alder reaction readily with an aliphatic (non-strained) terminal olefin under physiological conditions and at ambient temperature. This reaction is bioorthogonal and has been applied to label proteins containing a terminal olefin in live cells.27 Given the high structural similarity between an aliphatic terminal olefin and an ethyl group, changing the ethyl group at the end of the fatty acyl chain of an acyl-lysine to an olefin is not expected to significantly alter Sirt6 deacylation activity toward this fatty acyl-lysine in the nucleosome context. However, the alkene handle will allow for direct labeling of the corresponding acyl-nucleosome with a fluorogenic tetrazine dye, followed by native PAGE analysis. The acyl-nucleosome in a native PAGE gel can also be stained with ethidium bromide (EtBr) to confirm its nucleosomal state. If Sirt6 is active toward this acyl-nucleosome for its deacylation, the resulting non-modified nucleosome cannot be labeled with a tetrazine dye but is still visible after staining with EtBr. Therefore by comparing tetrazine-based fluorescent intensities of the original acyl-nucleosome and its Sirt6-treated counterpart, Sirt6 deacylation activity toward this acyl-nucleosome can be obtained. In the following section, we will describe how we applied this approach to fish out Sirt6-targeted H3 lysine sites at the nucleosome level.
Figure 1. Probing Sirt6-targeted lysine deacylation sites in nucleosome using a chemical antibody approach.
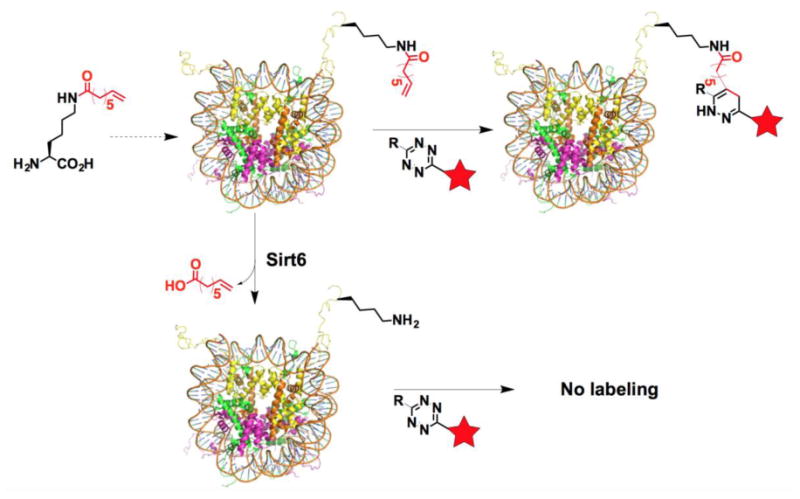
A terminal olefin-containing fatty acyl-lysine is site-specifically installed in a histone that is assembled into a nucleosome as an active Sirt6 substrate. A tetrazine probe can selectively react with the olefin in the installed fatty acyl-lysine to label the nucleosome. If Sirt6 can targets this particular fatty acylation site in nucleosome for its removal, incubating the assembled nucleosome with Sirt6 will remove the fatty acylation and therefore affords an unmodified nucleosome that can not be labeled with the tetrazine probe.
The genetic incorporation of Nε-(7-octenoyl)-lysine (OcK)
In order to genetically incorporate a terminal olefin-containing fatty acyl-lysine into H3, we exploited the amber suppression-based noncanonical amino acid mutagenesis approach similarly as in the synthesis of acetyl-H3 proteins. Denu et al. showed that Sirt6 has an enhanced deacylation activity toward lysine with a fatty acyl chain length of 8-14 carbons.7 Based on this observation, we synthesized two terminal olefin-containing fatty acyl-lysines, OcK (or Koc) (Figure 2A) and Nε-(9-decenoyl)-lysine (DeK). OcK can be dissolved in water up to 2 mM; however, DeK is barely soluble. We did not synthesize any acyl-lysine with a chain length long than 10 carbons given its insoluble tendency in water. Since we typically use 1 mM noncanonical amino acid for expressing noncanonical amino acid-containing proteins, OcK is a viable choice. For its incorporation, mutant M. mazei pyrrolysyl-tRNA synthetases that selectively charge tRNAPyl with OcK were first identified from a pyrrolysyl-tRNA synthetase gene library in which codons for four active site residues, Y306, L309, C348, and Y384, of pyrrolysyl-tRNA synthetase were randomized. A widely adopted and double-sieved selection protocol was applied for the selection.28, 29 The mutant, together with OcK and tRNAPyl, that displays the best amber suppression efficiency in E. coli has mutations as L309A/C348S and is coined as OcKRS. A plasmid pEVOL-OcKRS that contains genes encoding OcKRS and tRNAPyl was then constructed. Together with another plasmid pETDuet-UbK48Am that contains a gene encoding ubiquitin with an amber mutation at K48 and a C-terminal 6×His tag, was used to transform E. coli BL21(DE3) cells. Growing the transformed cells in LB supplemented with 1 mM OcK afforded the expression of full-length ubiquitin Ub-K48oc to a level of 15 mg L-1. In the absence of OcK, there was only a minimal amount of full-length ubiquitin expressed, confirming that OcKRS is specific for OcK (Figure 2C). The purified Ub-K48oc was analyzed by electrospray ionization-MS that gave a molecular weight of 9511.5 Da, agreeing well with the theoretical molecular weight at 9511.7 Da (Figure 3D, Supplementary Figure 4). To examine whether OcK in Ub-K48oc can be selectively labeled with a fluorogenic tetrazine, a previously synthesized tetrazine dye FITC-TZ (Figure 2B) was employed. After incubation with 100 μM FITC-TZ for 2 h, Ub-K48oc exhibited strong fluorescence in a SDS-PAGE gel, which was in striking contrast to the absence of fluorescence for wild type ubiquitin, confirming selective labeling of OcK in a protein by a tetrazine dye (Figure 2E).
Figure 2. The genetic incorporation of OcK.
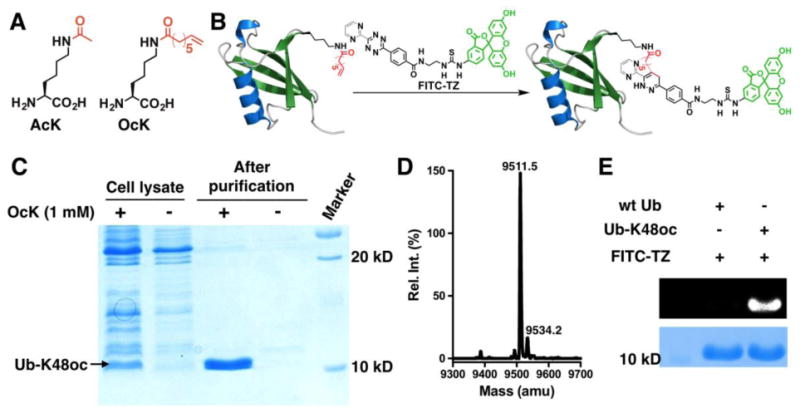
(A) Structures of AcK and OcK. (B) A diagram to illustrate fluorescent labeling of Ub-K48oc by a fluorogenic tetrazine dye, FITC-TZ. (C) The selective incorporation of OcK into ubiquitin at its K48 position. BL21(DE3) cells transformed with plasmid pEVOL-OcKRS coding OcKRS and tRNAPyl and plasmid pETDuet-UbK48Am coding ubiquitin with an amber mutation at K48 were grown in LB with or without 1 mM OcK. (D) The Deconvoluted ESI-MS spectrum of Ub-K48oc (calculated molecular weight: 9511.7 Da). (E) The selective labeling of Ub-K48oc by FITC-TZ. Proteins were incubated with 100 μM FITC-TZ for 2 h before they were analyzed by SDS-PAGE. The top panel shows the FITC-based fluorescent image of the gel and the bottom panel shows the same gel after it was stained with Coomassie Blue.
Figure 3. Sirt6 activities on oc-H3-H4 tetramers.
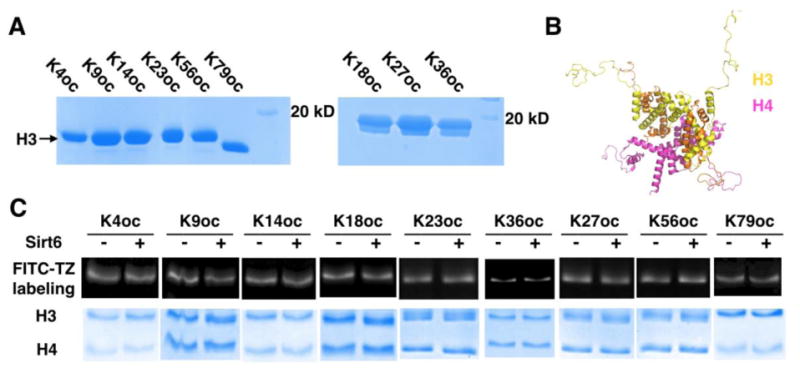
(A) SDS-PAGE analysis of 9 purified oc- H3 proteins. (B) The H3-H4 tetramer structure. (C) Sirt6 activities on 9 oc-H3-H4 tetramers. A tetramer (0.8 μM) was incubated with or without 0.4 μM Sirt6 for 3 h before it was labeled with 200 μM FITC-TZ for 3 h and then analyzed by SDS-PAGE.
Sirt6 activities on OcK-containing H3-H4 (oc-H3-H4) tetramers
We next proceeded to synthesize mono-acylated H3 (oc-H3) proteins with OcK incorporated at K4, K9, K14, K18, K23, K27, K36, K56, and K79, respectively. E. coli BL21(DE3) cells transformed with pEVOL-OcKRS and a pETDuet-H3 vector coding H3 with an amber mutation at a designated site were grown in 2YT supplemented with 1 mM OcK to produce an Oc-H3 protein. For OcK incorporation at K4-56 of H3 (H3K4-56oc), an N-terminal 6×His tag followed by a TEV protease digestion site was introduced for affinity purification with Ni-NTA resins and subsequent digestion by TEV protease to afford intact oc-H3 proteins. For OcK incorporation at K79 of H3 (H3K79oc), a truncation product with translation terminated at K79 predominated and therefore a C-terminal 6×His tag was introduced for easy separation of the full-length H3K79oc from its truncation product. Although we cannot remove the C-terminal 6×His tag from H3K79oc without leaving a scar, this C-terminal 6×His tag is not expected to affect assembling H3K79oc into either a tetramer with H4 or a nucleosome given that the H3 C-terminus is not directly involved in interactions with DNA and other histones. All nine oc-H3 proteins were expressed and purified to homogeneity (Figure 3A). They were then assembled together with H4 to form oc-H3-H4 tetramers (Figure 3B and Supplementary Figure 5). Activities of Sirt6 on these tetramers were then analyzed. We tested Sirt6 activities on oc-H3-H4 tetramers instead of oc-H3 proteins themselves due to a concern that the 0.5 M arginine that is required to solubilize H3 may affect Sirt6 function. After incubating 0.4 μM Sirt6 with an oc-H3-H4 tetramer (0.8 μM) for 3 h, the treated tetramer was labeled with FITC-TZ, analyzed by SDS-PAGE, and then fluorescently imaged. The original tetramer without Sirt6 treatment was labeled and analyzed by SDS-PAGE in parallel as a control. As shown in Figure 3C, there is no significant fluorescent intensity difference on H3 between Sirt6-treated and untreated samples for all tetramers except the tetramer assembled from H3K9oc. H3K9oc does show weaker fluorescence than its untreated counterpart, indicating that Sirt6 targets this site for deacylation although the activity is still weak.
Sirt6 activities on OcK-containing nucleosome (oc-nucleosome) substrates
We next moved on to assemble oc-nucleosomes from oc-H3 proteins and used them as substrates for Sirt6 in order to test our hypothesis that the nucleosome structure will activate Sirt6, revealing a clearer picture of Sirt6-targeted H3 sites. Following a standardized protocol developed by Luger et al.,30 we integrated all 9 oc-H3 proteins into their corresponding oc-nucleosomes (Supplementary Figure 6). A wild type nucleosome assembled from an unmodified H3 protein was also made as a control. Deacylation activities of Sirt6 on these oc-nucleosome substrates were then analyzed by incubating an oc-nucleosome (0.66 μM) with 0.33 μM Sirt6 for 3 h, followed by labeling with FITC-TZ and native PAGE analysis. FITC-TZ-labeled original oc-nucleosomes were analyzed in parallel (Figure 4). FITC fluorescence was imaged, the gels were stained with EtBr, and then EtBr fluorescence was imaged. As expected, for all 9 assembled oc-nucleosomes, incubation with Sirt6 did not cause their dissemblance. All oc-nucleosomes with and without treatment with Sirt6 and the control nucleosome showed strong EtBr-stained DNA bands around 500 bp, agreeing well with what the literature has reported about the migration of a mononucleosome in a native PAGE gel.30 All nucleosomes have two EtBr-stained bands, indicating one major and one minor nucleosome conformer, as previously observed.30 However, after incubation with Sirt6, H3K4oc-, H3K9oc-, H3K18oc-, H3K23oc-, and H3K27oc-nucleosomes displayed much lower FITC-labeling fluorescence than their original counterparts. H3K9oc- and H3K18oc-nucleosomes showed almost no FITC labeling after their reactions with Sirt6, indicating close to total removal of the fatty acyl group. However, H3K36oc-, H3K56oc-, and H3K79oc-nucleosomes that were exposed to Sirt6 showed no obvious FITC labeling difference from the original untreated nucleosomes. Thus, it is apparent that Sirt6 can target H3K4, H3K9, H3K18, H3K23, and H3K27 for their deacylation and that the nucleosome core structure significantly improves Sirt6's catalytic activities at these sites. In our in vitro setup, H3K56 is obviously not a preferential deacylation site for Sirt6.
Figure 4. Sirt6 activities on oc-nucleosomes.
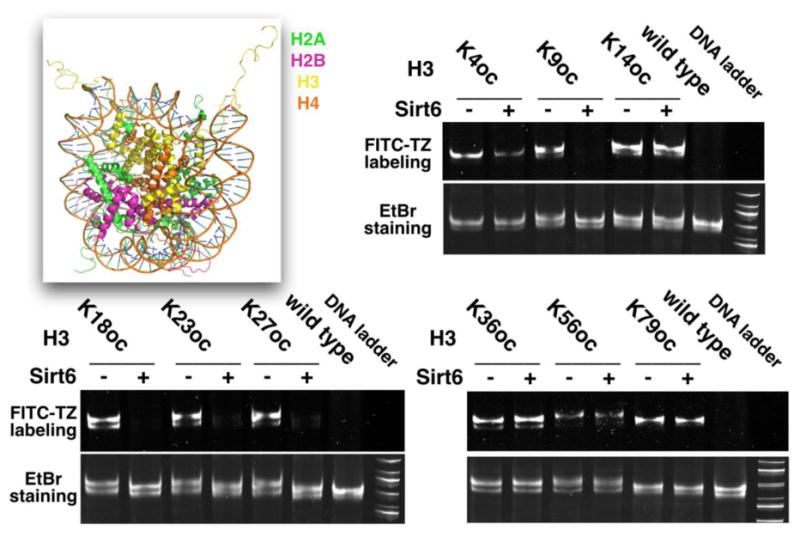
The image in the top left corner shows the structure of a nucleosome. For all gel images, the top panel shows the FITC-based fluorescent imaging and the bottom panel shows the EtBr-stained DNA from the same gel.
Sirt6-targeted lysine sites
To compare catalytic deacylation activities of Sirt6 on the five detected H3 sites, we also carried out a time-based analysis. H3K4oc-, H3K9oc-, H3K18oc-, H3K23oc-, and H3K27oc-nucleosomes were incubated with Sirt6. Sirt6-catalyzed deacylation was stopped through addition of nicotinamide (10 mM) at different times and oc-nucleosome substrates were labeled with FITC-TZ, followed by native PAGE analysis. Their FITC-labeling intensities were then integrated and plotted for comparison. As shown in Figure 5A, Sirt6 has similar catalytic efficiencies in removing fatty acylation from H3K9 and H3K18. After H3K9oc- and H3K18oc-nucleosomes (0.66 μM) reacted with 0.33 μM Sirt6 for 2 h, the fatty acyl group was quantitatively removed from the two nucleosomes. The deacylation activity of Sirt6 on H3K27 is slightly lower, but still comparable to H3K9 and H3K18. Residual fatty acylation was observed on the H3K27oc-nucleosome (0.66 μM) after incubation with Sirt6 (0.33 μM) for 2 h. For the two sites at H3K4 and H3K23, Sirt6 displays similar deacylation activities although Sirt6 activities on these two sites are much weaker than on H3K9, H3K18, and H3K27. After H3K4oc- and H3K23oc-nucleosomes (0.66 μM) reacted with 0.33 μM Sirt6 for 2 h, a significant amount of these two nucleosomes were still fatty acylated. This time-based assay allows us to divide Sirt6-targeted H3 lysine sites into two groups with the first group including H3K9, H3K18, and H3K27 and the second group including H3K4 and H3K23. Sirt6 preferentially targets the first group for deacylation but is also catalytically active toward the second group.
Figure 5. A time-based Sirt6 activity analysis.
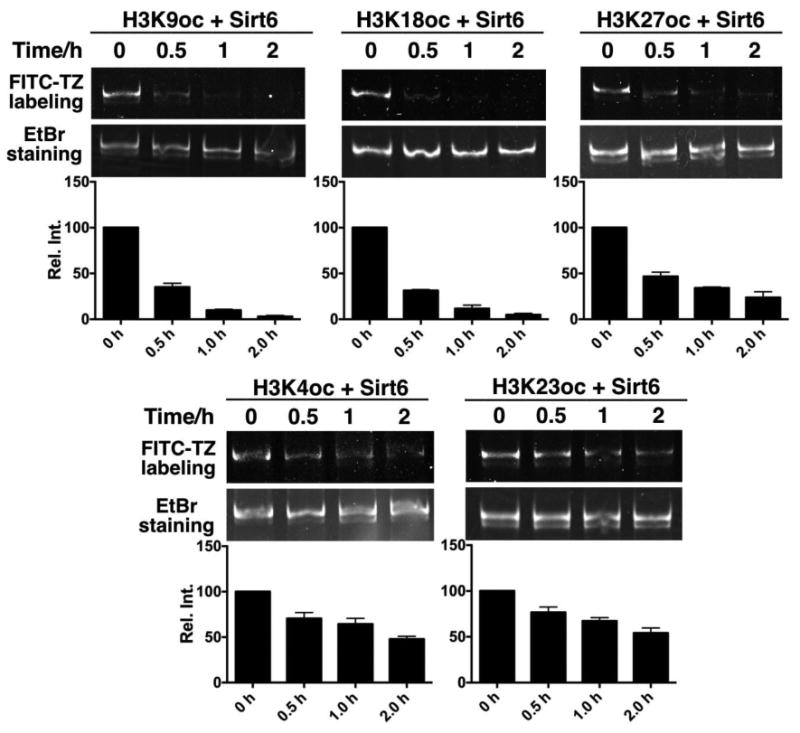
Relative catalytic activities of Sirt6 on five oc-nucleosome substrates. An oc-nucleosome substrate (0.66 μM) was incubated with 0.33 μM Sirt6. The reaction was stopped at 0.5, 1, and 2 h and oc-nucleosome samples at these different times were labeled with FITC-TZ and analyzed in a native PAGE gel together with the original oc- nucleosome (0 h). Top panels show FITC-based images, middle panels show same gels based on EtBr staining, and bottom panels show relative integrated FITC-based fluorescent intensities at different reaction times. All reactions were repeated three times.
Sirt6 catalyzes deacetylation at H3K9, H3K18 and H3K27 in cells
With knowing that H3K9, H3K18, and H3K27 are preferential Sirt6-targeted sites for deacylation of fatty acids, we next move on to demonstrate that Sirt6 also catalyzes deacetylation at these three sites in an in vitro biochemistry setup and in cells. Three corresponding acetyl-nucleosomes were assembled from H3K9ac, H3K18ac, and H3K27ac, respectively, together with H2A, H2B, H4, and 601 DNA (Figure 6A and Supplementary Figure 6). These acetyl-nucleosomes (0.66 μM) were then reacted with 0.33 μM Sirt6 for 3 h, followed by native PAGE analysis, EtBr staining, and the transfer to a membrane for probing with a pan anti-Kac antibody (Figure 6B). In the EtBr stained gel, parallel samples that were incubated with and without Sirt6 showed similar intensities, indicating that Sirt6 did not dissemble any acetyl-nucleosome substrate. In the Western blot analysis, the pan anti-Kac antibody showed very different detection abilities toward three acetylation sites. Although all three original acetyl-nucleosomes were detected by the pan anti-Kac antibody, signals for H3K9ac- and H3K27ac-nucleosomes were much stronger than that for H3K18ac-nucleosome. This observation supports the use of our chemical biology approach that results in almost equal detection levels for nucleosomes incorporated with OcK. Notwithstanding different detection abilities of the pan anti-Kac antibody toward three acetylation sites, all three acetyl-nucleosome samples that reacted with Sirt6 displayed much decreased acetylation levels in comparison to their original acetylated counterparts. For H3K9ac- and H3K18ac-nucleosomes, no signal was detected, indicating total removal of acetylation; For H3K27ac-nucleosome, there was residual acetylation after its reaction with Sirt6 but much weaker than acetylation of the original nucleosome. For comparison, acetyl-nucleosomes were also assembled from H3K14ac, H3K23ac, and H3K79ac and used as substrates to react with Sirt6 in a same setup. As expected, there were no significant acetylation changes for all three acetyl-nucleosomes after their reactions with Sirt6 (Figure 6B). To demonstrate that Sirt6 targets H3K9, H3K18 and H3K27 for deacetylation in cells, a Sirt6-coding pEGFP vector was constructed and transiently transfected into 293T cells for overexpressing Sirt6. Cell lysates from Sirt6-transfected and control cells were then separated and probed by specific antibodies raised for H3K9ac, H3K18ac, H3K27ac, and H3K23ac. As shown in Figure 6C, Sirt6-transfected cells have much weaker acetylation at H3K9, H3K18, and H3K27 than control cells. This result confirms that all three sites are indeed targeted by Sirt6 for deacetylation in cells. However, the acetylation level change for H3K23 in Sirt6-transfected cells was minimal.
Figure 6. Sirt6 catalyzes deacetylation at H3K9, H3K18, and H3K27.
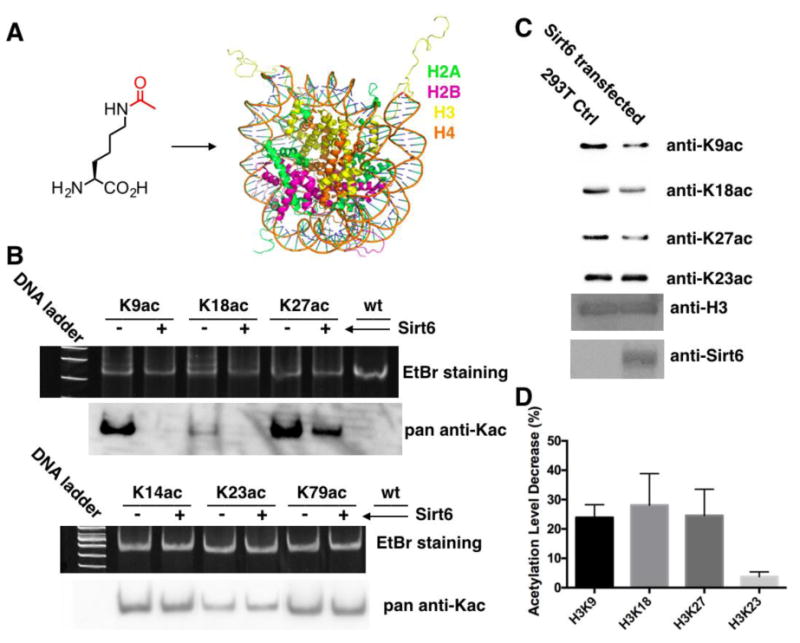
(A) The incorporation of AcK into a nucleosome. (B) Sirt6 activities on H3K9ac-, H3K18ac-, H3K27ac, H3K14ac, H3K23ac, and H3K79ac-nucleosomes. Acetyl-nucleosomes (0.66 μM) were incubated with or without 0.33 μM Sirt6 for 3 h before they was analyzed by a native PAGE gel. The top panel shows the EtBr stained native PAGE gel and the bottom panel shows the pan anti-Kac antibody detected acetylation levels. (C) Acetylation levels at H3K9, H3K18, H3K27, and H3K23 in Sirt6-transfected and control cells. (D) Quantified acetylation level decrease at H3K9, H3K18, H3K27, and H23 in Sirt6-transfected cells with respect to control cells.
Discussion
As a chromatin-associated protein and a NAD+-dependent sirtuin enzyme, Sirt6 has long been recognized as a histone deacetylase but with puzzlingly low activities toward peptide substrates. We show in this report that Sirt6 has close to undetectable deacetylation activities toward all 9 tested monomeric acetyl-H3 substrates. Although it was recently discovered that Sirt6 has enhanced activity toward a peptide lysine with long chain fatty acylation, our efforts to make oc-H3 proteins and fold them to corresponding oc-H3-H4 tetramers did not yield active substrates for Sirt6 either. However, deacylation activities of Sirt6 toward several H3 lysines were immensely improved when corresponding acetyl- or oc-H3 proteins were assembled into their nucleosomal forms. Although the focus of our current study is to identify Sirt6-targeted H3 lysines for deacetylation, our results raise an interesting question as to what are the real substrates of Sirt6 in cells? Are they free histones or their nucleosomal forms? In fact, in active transcribed gene regions where Sirt6 is primarily found, chromatin is in a dynamic process of assembly and disassembly.31, 32 Sirt6 has the chance to interact with both free histones and nucleosomes. Our results imply that nucleosomes are the real substrates of Sirt6 but do not rule out that Sirt6, as part of a large protein complex, catalyzes efficient deacetylation of free histones. As a matter of fact, a lot of histone epigenetic modifiers are found in large multi-subunit complexes that contain various histone or DNA recognition domains, though nothing has been reported in the context of Sirt6.33, 34 Another interesting question raised by the current study is how the nucleosome structure activates Sirt6. Sirt6 has a highly positively charged C-terminus. Chua et al. showed that the cleavage of this C-terminus from Sirt6 does not deactivate the enzyme but abolishes its binding to chromatin.35 One explanation of this observation is that the C-terminus is involved in nonspecific electronic interactions with negatively charged DNA for recruiting Sirt6 to chromatin. However, DNA may also neutralize the positively charged C-terminus and therefore allows Sirt6 to interact with positively charged histone tails for deacetylation. This aspect needs to be explored further. Another important feature of Sirt6, revealed in this study, is the preferential sequence contexts around its targeted lysines. Unlike Sirt1, Sirt2, and Sirt3 that are more or less universal deacetylases,5, 36 Sirt6 prefers to deacetylate certain lysine sites. This selective targeting feature of Sirt6 needs to be further explored.
Besides revealing important biochemical features of Sirt6, the current study also has strong biological implications. Reports have indicated that Sirt6 is frequently recruited to relevant gene promoters and represses gene transcription via removing acetylation at H3K9 and H3K56.37-40 Our results show that H3K18 and H3K27 are targeted by Sirt6 as well for deacetylation. H3K18ac and H3K27ac are associated with active gene expression.41 Targeting these two sites for deacetylation is expected to reinforce the transcriptional repression role of Sirt6. Functional redundancy of histone acetylation on chromatin regulation has been observed. For example, a single mutation of an N-terminal H3 lysine to a glycine has little repression effect on gene expression in yeast. However, simultaneous mutations of all N-terminal lysines to glycine significantly reduces yeast gene expression.42 Thus, it is reasonable to assume that Sirt6 removes acetylation from multiple lysines to fulfill a strong gene transcription suppression role. By removing acetylation from H3K9, H3K18, and H3K27, Sirt6 may also regulate functions of proteins that recognize acetylation at these three lysines. One bromodomain YEATS recognizes acetylation at H3K9, H3K18, and H3K27 with similar binding affinities.43 It is highly possible that Sirt6 regulates functions of YEATS-containing proteins such as AF9 and ENL by modulating acetylation at H3K9, H3K18, and H3K27. Di- and trimethylation at H3K27 (H3K27me2/3) has long been known for shutting down transcription.44 H3K27me3 is an important mark for inactivated X chromosome. H3K27ac is associated with active transcription and is apparently antagonistic to the repression of gene transcription by H3K27me2/3.45 Sirt6 may serve as an important mediator to switch H3K27 between these two antagonistic states.
In cancer biology, H3K18ac is an important prognostic mark. Downregulation of H3K18ac has been found correlated with poor prognosis of pancreatic adenocarcinoma and low grade prostate cancer.21, 46 Patients with high H3K18ac show low resurgence of prostate cancer after treatment.47 Incidentally overexpression of Sirt6 is observed in prostate cancer cell line PC-3 and inhibition of Sirt6 leads to apoptosis of PC-3.21 Our study provides the missing link between downregulation of H3K18ac and high expression of Sirt6 in PC-3. It is highly possible that Sirt6 changes prostate cancer development through its deacetylation of H3K18 and Sirt6 itself can be a potential anti-cancer drug target.
Previous studies showed that Sirt6 catalyzes deacetylation at H3K56 on purified H3 and Sirt6 knockout mouse tissue had enhanced H3K56ac. Our results detected sluggish removal of long chain fatty acylation from H3K56 by Sirt6. According to current nucleosomal structures, H3K56 directly interacts with the wrapping DNA. In this nucleosomal format we have also adopted in our study, H3K56 is blocked from access by Sirt6. However, in cells Sirt6 may involve additional cellular factors for direct deacetylation of H3K56 on free H3 or positioning of DNA wrapping could be different and expose H3K56 for deacetylation.48 It is also possible that H3K56-targeting histone acetyltransferases/deacetylases are regulated by Sirt6 in cells.
In summary, we have developed a chemical biology approach and used it to successfully reveal several Sirt6-targeted deacetylation sites in histone H3. Our study indicates that Sirt6 preferentially targets acetyl-nucleosomes for deacetylation and is selective for particular nucleosomal lysine sites. Two novel Sirt6-targeted deacetylation sites H3K18 and H3K27 were confirmed in cells. Our study offers critical information for revaluating Sirt6's functional importance and rebuilding Sirt6's connection with other epigenetic players.
Methods
The synthesis of OcK, selection of OcKRS, expression of acetyl- and Oc-H3 proteins, assembly of acetyl- and oc-nucleosomes, and Sirt6-catalyzed deacylation on H3 proteins and nucleosomes are provided in the supporting information.
Supplementary Material
Acknowledgments
Support of this work was provided from National Institute of Health (grant CA161158), National Science Foundation (grant CHE-1148684), and Welch Foundation (grant A-1715).
Footnotes
Author Contributions: W. R. Liu conceived the design of the study; W. W. Wang, Y. Zeng, and Bo Wu carried out the experiments; W. R. Liu and W. W. Wang prepared the manuscript.
Associated content: The supporting information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.xxxxxxx.
References
- 1.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 2.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 3.Herskovits AZ, Guarente L. Sirtuin deacetylases in neurodegenerative diseases of aging. Cell Res. 2013;23:746–758. doi: 10.1038/cr.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blander G, Olejnik J, Krzymanska-Olejnik E, McDonagh T, Haigis M, Yaffe MB, Guarente L. SIRT1 shows no substrate specificity in vitro. J Biol Chem. 2005;280:9780–9785. doi: 10.1074/jbc.M414080200. [DOI] [PubMed] [Google Scholar]
- 6.Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. J Biol Chem. 2011;286:14575–14587. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, Hang HC, Hao Q, Lin H. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TLA, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–U416. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwer B, Schumacher B, Lombard DB, Xiao C, Kurtev MV, Gao J, Schneider JI, Chai H, Bronson RT, Tsai LH, Deng CX, Alt FW. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc Natl Acad Sci U S A. 2010;107:21790–21794. doi: 10.1073/pnas.1016306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gil R, Barth S, Kanfi Y, Cohen HY. SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic Acids Res. 2013;41:8537–8545. doi: 10.1093/nar/gkt642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shogren-Knaak MA, Fry CJ, Peterson CL. A native peptide ligation strategy for deciphering nucleosomal histone modifications. J Biol Chem. 2003;278:15744–15748. doi: 10.1074/jbc.M301445200. [DOI] [PubMed] [Google Scholar]
- 14.Holt M, Muir T. Application of the protein semisynthesis strategy to the generation of modified chromatin. Annu Rev Biochem. 2015;84:265–290. doi: 10.1146/annurev-biochem-060614-034429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kugel S, Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem Sci. 2014;39:72–81. doi: 10.1016/j.tibs.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu PF, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 17.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined Gene Expression Analysis of Whole-Tissue and Microdissected Pancreatic Ductal Adenocarcinoma identifies Genes Specifically Overexpressed in Tumor Epithelia. Hepato-Gastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 18.Anders M, Fehlker M, Wang Q, Wissmann C, Pilarsky C, Kemmner W, Hocker M. Microarray meta-analysis defines global angiogenesis-related gene expression signatures in human carcinomas. Mol Carcinog. 2013;52:29–38. doi: 10.1002/mc.20874. [DOI] [PubMed] [Google Scholar]
- 19.Azuma Y, Yokobori T, Mogi A, Altan B, Yajima T, Kosaka T, Onozato R, Yamaki E, Asao T, Nishiyama M, Kuwano H. SIRT6 expression is associated with poor prognosis and chemosensitivity in patients with non-small cell lung cancer. J Surg Oncol. 2015;112:231–237. doi: 10.1002/jso.23975. [DOI] [PubMed] [Google Scholar]
- 20.Khongkow M, Olmos Y, Gong C, Gomes AR, Monteiro LJ, Yague E, Cavaco TB, Khongkow P, Man EP, Laohasinnarong S, Koo CY, Harada-Shoji N, Tsang JW, Coombes RC, Schwer B, Khoo US, Lam EW. SIRT6 modulates paclitaxel and epirubicin resistance and survival in breast cancer. Carcinogenesis. 2013;34:1476–1486. doi: 10.1093/carcin/bgt098. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Xie QR, Wang B, Shao J, Zhang T, Liu T, Huang G, Xia W. Inhibition of SIRT6 in prostate cancer reduces cell viability and increases sensitivity to chemotherapeutics. Protein Cell. 2013;4:702–710. doi: 10.1007/s13238-013-3054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ming M, Han W, Zhao B, Sundaresan NR, Deng CX, Gupta MP, He YY. SIRT6 promotes COX-2 expression and acts as an oncogene in skin cancer. Cancer Res. 2014;74:5925–5933. doi: 10.1158/0008-5472.CAN-14-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugatani T, Agapova O, Malluche HH, Hruska KA. SIRT6 deficiency culminates in low-turnover osteopenia. Bone. 2015;81:168–177. doi: 10.1016/j.bone.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu WW, Wu B, Liu WR. Sirtuins 1 and 2 Are Universal Histone Deacetylases. ACS Chem Biol. 2016;11:792–799. doi: 10.1021/acschembio.5b00886. [DOI] [PubMed] [Google Scholar]
- 25.Umehara T, Kim J, Lee S, Guo LT, Soll D, Park HS. N-acetyl lysyl-tRNA synthetases evolved by a CcdB-based selection possess N-acetyl lysine specificity in vitro and in vivo. FEBS Lett. 2012;586:729–733. doi: 10.1016/j.febslet.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 26.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 27.Lee YJ, Kurra Y, Yang Y, Torres-Kolbus J, Deiters A, Liu WR. Genetically encoded unstrained olefins for live cell labeling with tetrazine dyes. Chem Commun (Camb) 2014;50:13085–13088. doi: 10.1039/c4cc06435f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 29.Wang YS, Wu B, Wang Z, Huang Y, Wan W, Russell WK, Pai PJ, Moe YN, Russell DH, Liu WR. A genetically encoded photocaged Nepsilon-methyl-L-lysine. Mol Biosyst. 2010;6:1557–1560. doi: 10.1039/c002155e. [DOI] [PubMed] [Google Scholar]
- 30.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 31.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, Durham T, Zhang X, Donaghey J, Epstein CB, Regev A, Bernstein BE. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lalonde ME, Cheng X, Cote J. Histone target selection within chromatin: an exemplary case of teamwork. Genes Dev. 2014;28:1029–1041. doi: 10.1101/gad.236331.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tennen RI, Berber E, Chua KF. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech Ageing Dev. 2010;131:185–192. doi: 10.1016/j.mad.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith BC, Settles B, Hallows WC, Craven MW, Denu JM. SIRT3 substrate specificity determined by peptide arrays and machine learning. ACS Chem Biol. 2011;6:146–157. doi: 10.1021/cb100218d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Etchegaray JP, Chavez L, Huang Y, Ross KN, Choi J, Martinez-Pastor B, Walsh RM, Sommer CA, Lienhard M, Gladden A, Kugel S, Silberman DM, Ramaswamy S, Mostoslavsky G, Hochedlinger K, Goren A, Rao A, Mostoslavsky R. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol. 2015;17:545–557. doi: 10.1038/ncb3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1 alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, D'Urso A, Naar AM, Kingston R, Rippe K, Mostoslavsky R. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin AM, Pouchnik DJ, Walker JL, Wyrick JJ. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics. 2004;167:1123–1132. doi: 10.1534/genetics.104.026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li YY, Wen H, Xi YX, Tanaka K, Wang HB, Peng DN, Ren YF, Jin QH, Dent SYR, Li W, Li HT, Shi XB. AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation. Cell. 2014;159:558–571. doi: 10.1016/j.cell.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 45.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manuyakorn A, Paulus R, Farrell J, Dawson NA, Tze S, Cheung-Lau G, Hines OJ, Reber H, Seligson DB, Horvath S, Kurdistani SK, Guha C, Dawson DW. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J Clin Oncol. 2010;28:1358–1365. doi: 10.1200/JCO.2009.24.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 48.Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


