Abstract
Several herpesviruses have acquired the gene for the CD200 membrane protein from their hosts and can downregulate myeloid activity through interaction of this viral CD200 orthologue with the host receptor for CD200, namely CD200R, which can give inhibitory signals. This receptor is a ‘paired receptor’, meaning proteins related to the inhibitory CD200R are present but differ in that they can give activating signals and also give a negligible interaction with CD200. We showed that the viral orthologues e127 from rat cytomegalovirus and K14 from human herpesvirus 8 do not bind the activating CD200R-like proteins from their respective species, although they do bind the inhibitory receptors. It is thought that the activating receptors have evolved in response to pathogens targeting the inhibitory receptor. In this case, the CD200 orthologue is not trapped by the activating receptor but has maintained the specificity of the host from which it was acquired, suggesting that the activating members of the CD200R family have evolved to protect against a different pathogen.
Introduction
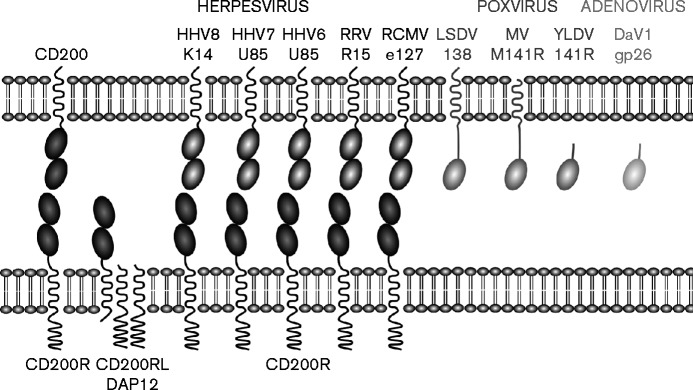
CD200 is a broadly distributed membrane protein that interacts with an inhibitory receptor, termed CD200R, that is restricted to leukocytes with particularly high levels of expression on basophils and macrophages (Barclay et al., 2002; Shiratori et al., 2005). Many viruses have acquired CD200 genes from their hosts and it has been shown that several herpesvirus CD200 orthologues interact with CD200R, as summarized in Fig. 1. Both CD200 and CD200R contain two Ig-like domains in their extracellular regions and they interact through their N-terminal domains (Hatherley et al., 2013). The viral orthologues of CD200 also have two extracellular domains, with the exception of the adenovirus and poxvirus orthologues (Fig. 1). In two cases, the K14 protein from human herpesvirus 8 (HHV8) (Foster-Cuevas et al., 2004) and the e127 protein from rat cytomegalovirus (RCMV) (Foster-Cuevas et al., 2011), the affinities of their interactions with CD200R have been determined and shown to be indistinguishable from the host interactions. The sequences of K14 and e127 have 41 and 87 % amino acid sequence identity with their host CD200, respectively (in the putative ligand binding domain), but the structure of the CD200/CD200R complex indicates that residues that are likely to be involved in binding CD200R are conserved (Hatherley et al., 2013). The CD200 orthologues from HHV6, HHV7 and HHV8 bind CD200R expressed on basophils (Shiratori et al., 2005). The engagement of the inhibitory receptor by the viral orthologue gives downregulation of the immune system as shown for HHV8 (Foster-Cuevas et al., 2004; Shiratori et al., 2005) and for the rhesus rhadinovirus (Estep et al., 2014). Expression of both host and HHV8 CD200 on antigen-presenting cells inhibited T-cell activation (Misstear et al., 2012).
Fig. 1.
Cartoon illustrating the organization of domains in CD200 and its viral orthologues. Ig-like domains are indicated by ovals. The herpesvirus orthologues have a similar topology to the mammalian CD200, whereas the poxviruses show less similarity. The herpesvirus CD200 orthologues shown are from human herpesvirus 8 (HHV8; NCBI Protein accession no. AAK53415), HHV7 (AAC54746), HHV6 (CAA58334), rhesus monkey rhadenovirus (RRV; AAF60072) and rat cytomegalovirus (RCMV; AAO45420). The poxvirus CD200 orthologues shown are from lumpy skin disease virus (LSDV; NP_150572), myxoma virus (MV; AAF15029) and yaba-like disease virus (YLDV; NP_073526). The duck adenovirus A CD200-like protein (DaV1gp26, NP_044723) is also shown.
CD200R is a ‘paired receptor’, meaning that as well as the inhibitory receptor there is also one or more activating receptors that have similar extracellular regions. However, instead of motifs that lead to inhibitory signals, these activating receptors have very short cytoplasmic regions but they are associated with adapters such as DAP12 that can recruit kinases and lead to activation (Akkaya et al., 2013; Hatherley et al., 2013; Wright et al., 2003). These activating receptors are termed CD200Rlike (CD200RL) to distinguish them from the inhibitory receptors. The CD200RL proteins are highly diverse with only one member in humans, one in the one rat genome available but between one and at least four in mice according to the strain (Akkaya & Barclay, 2010; Wright et al., 2003). The affinity of the interaction of CD200 with CD200R in several species is typical for many cell surface protein interactions, with a KD of around 1 μM (van der Merwe & Barclay, 1994). No binding of CD200 was observed to the mouse CD200RLa or CD200RLb using cell-binding assays (Wright et al., 2003). There were contradictory data suggesting that CD200 did bind to these receptors in the mouse (Gorczynski et al., 2004), but further studies showed there is minimal binding of the CD200 ( < 2 % of that to CD200R) to the activating forms CD200RLa, CD200RLb, CD200RLc and CD200RLe (Hatherley et al., 2005) (an alternative nomenclature is also used, CD200R4, CD200R3, CD200R2 and CD200R5, respectively) (Gorczynski et al., 2004; Hatherley et al., 2005, 2013; Voehringer et al., 2004). Others also failed to show binding of CD200 to CD200RL (Kojima et al., 2007). However, definitive data were found when the structures of CD200R in complex with CD200 and CD200RLa alone were elucidated (Hatherley et al., 2013). This showed that the molecular basis of the failure of CD200 to bind to CD200RLa was due to changes in the structure of the contact site. Three residues were identified in CD200RLa, which, when mutated to those found in CD200R, restored binding to CD200. Thus, CD200 does not bind activating receptors to any significant extent.
There are limited data on the distribution of the activating receptors, but in general they are expressed on myeloid cells but to a lesser degree than the inhibitory receptor. CD200RLa was observed on NK cells, monocytes and a subset of NKT cells (Wright et al., 2003), while mCD200RLb is expressed on mast cells and basophils (Kojima et al., 2007). Expression of human CD200RLa protein has not been detected (Wright et al., 2003).
As the CD200 orthologues from herpesviruses are known to bind the inhibitory CD200R, a key question is whether they also bind the activating members, which might eliminate the downregulatory effects on myeloid cells of the orthologues. We found no detectable binding of two viral CD200 orthologues, the K14 protein from HHV8 and the e127 protein from RCMV, with the activating CD200R-like proteins. We discuss this with regard to mechanisms of viral evasion and evolution of paired receptors.
Results
RCMV CD200 orthologue (e127) does not bind activating rat CD200RLa
In order to test e127 binding to rat receptors, a CD200RL gene was identified in the rat genome. Examination of the rat genome sequence indicated that there is one likely CD200RL gene and recently, an entry in NCBI based on this and EST data has been published (NCBI Protein accession no. XP_006221193). Analysis of this sequence showed that the extracellular region showed the highest amino acid sequence similarity to rat CD200R (89 %) and around 70 % to the mouse CD200R family (CD200R, 70 %; CD200RLa, 68 %; CD200RLb, 47 %; CD200RLc, 75 %, and CD200RLe, 69 %). Thus, the single activating gene identified in rats is no closer to any particular activating gene in the mouse and is termed rCD200RLa. This is consistent with the activating genes evolving rapidly from the inhibitory gene (Wright et al., 2003). Thus, the ability of RCMV e127 protein to bind activating receptors was tested against the single rat activating rCD200RLa and also for cross-reaction on the mouse activating CD200RLs.
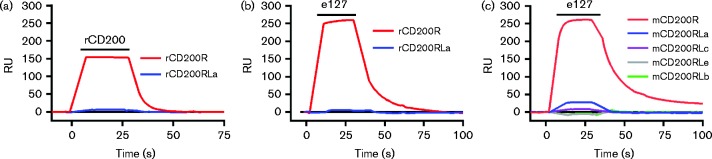
The viral orthologues were tested for direct binding to the receptors using surface plasmon resonance and recombinant proteins corresponding to the extracellular domains expressed by transient expression. The receptors were expressed as chimaeric proteins with domains 3+4 of rat CD4, which is an effective and widely used antigenic tag (Brown et al., 1998; Hatherley et al., 2005). This enabled the proteins to be immobilized directly onto the BIAcore chip to which CD4 mAb had been immobilized, from the spent tissue culture supernatant, minimizing the chances of anomalous binding due to denatured proteins arising during purification. Rat CD200 and the RCMV e127 clearly bound to the inhibitory rCD200R, but there was no binding to the activating rCD200RLa (Fig. 2a, b).
Fig. 2.
The e127 orthologue from rat cytomegalovirus binds the inhibitory CD200R, but not activating forms, in rats and mice. (a) Surface plasmon resonance experiments showing specific binding response of rat CD200 to rat CD200R and CD200RLa. (b, c) Binding response of RCMV e127 to immobilized rat (b) and mouse (c) CD200R family members. The black bar denotes the injection of either CD200 or e127 (at 10 μM) over the immobilized CD200R family members. Representative results from two (a), four (b) and three (c) experiments are shown. RU, response units.
RCMV CD200 orthologue (e127) does not bind activating mouse CD200RLa, CD200RLb, CD200RLc or CD200RLe
The possibility that the e127 protein might cross-react on mouse CD200RL receptors was tested, as more activating receptors had been defined in various mouse strains. The e127 protein was passed over mCD200R, mCD200RLa, mCD200RLb, mCD200RLc and mCD200RLe that had been immobilized on a BIAcore chip using CD4 mAb. The e127 bound to mCD200R, showing a cross-species interaction, but there was no binding to the activating forms apart from trace amounts on CD200RLa and CD200RLc (Fig. 2c), as was also found for the mouse CD200 (Hatherley et al., 2005).
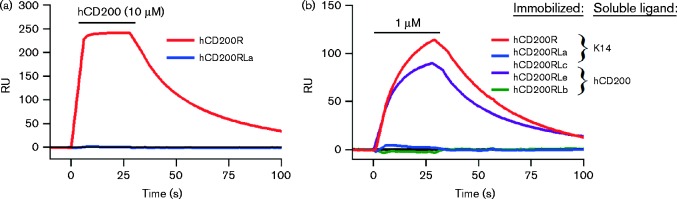
HHV8 CD200 orthologue does not bind the human CD200RLa activating receptor
Previous studies had established that the HHV8 K14 protein interacted with CD200R in humans (Foster-Cuevas et al., 2004) but it had not been tested for binding to the activating member of this paired receptor. There is only one putative activating receptor in humans, called hCD200RLa (Wright et al., 2003). First, we showed that human CD200 did not bind the activating human CD200RLa, in line with experiments on mouse proteins (Hatherley et al., 2005), even at a very high concentration (10 μM) (Fig. 3a). The HHV8 K14 protein was more difficult to express, but at around 1 μM there was no binding to CD200RLa and similar levels of binding to human CD200R (Fig. 3b), in accordance with previous results (Foster-Cuevas et al., 2004).
Fig. 3.
The herpesvirus HHV8 CD200 orthologue K14 binds inhibitory human CD200R but not the activating CD200RLa. (a) Specific binding response of soluble hCD200 (at 10 μM) to immobilized hCD200R and hCD200RLa. (b) Overlay of specific binding response of K14 and hCD200 (at 1 μM) over immobilized hCD200R and hCD200RLa. The black bar denotes the injection of either CD200 or K14 over the immobilized CD200R family members. Representative results from at least three experiments are shown. RU, response units.
Viral CD200 orthologues mimic host CD200
The viral orthologues tested showed very similar binding properties to those from the host, suggesting that they bind in the same manner. This was tested further by showing that the mAb which blocked the host interaction – OX108, OX131 and OX102 reacting against human, mouse and rat CD200R, respectively – also blocked the viral interactions (Akkaya et al., 2013; Foster-Cuevas et al., 2011). In addition, a mAb OX110 that recognizes mCD200R but does not block the interaction with host also did not block the viral interaction (Table 1).
Table 1. mAbs that block the host CD200/CD200R interaction also block the viral CD200/CD200R interaction.
In each experiment CD4 chimaeras of human, mouse or rat CD200R were immobilized on an OX68-coated chip, then, sequentially, host CD200, viral e127 and K14 proteins, saturating levels of OX110 (mouse non-blocking), OX131 (mouse blocking), OX108 (human blocking) and OX102 (rat blocking) were passed over the relevant flow cells. Then, the host CD200, viral e127 and K14 proteins were re-passed. The levels of ligand bound after antibody treatment are given as either ‘+’ (>70 % of pre-binding levels) or ‘ − ’ ( < 10 % pre-binding levels). Those mAbs that block the CD200/CD200R interaction also block the viral interaction (OX131, OX102 and OX108), while the non-blocking mouse mAb (OX110) does not block. The results are from two experiments.
| hCD200R | mCD200R | rCD200R | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| hCD200 | mCD200 | K14 | e127 | hCD200 | mCD200 | e127 | hCD200 | mCD200 | e127 | |
| OX108 | − | − | − | − | + | + | + | + | + | + |
| OX110 | + | + | + | + | + | + | + | + | + | + |
| OX131 | + | + | + | + | − | − | − | + | + | + |
| OX102 | + | + | + | + | + | + | + | − | − | − |
Discussion
The analysis showed no evidence of binding of viral CD200 orthologues to the CD200R like activating receptors. As there are no known ligands for these receptors, we carried out quantitative analysis using a variety of recombinant proteins, including activating receptors from three species and two viral orthologues. There are good data to suggest the CD200RL activating receptors are properly folded. For example, we have previously shown that the mouse recombinant CD200RL bind several of the CD200R antibodies, and when expressed at the cell surface can give activating signals (Akkaya et al., 2013); the constructs are similar to those used to determine the crystal structures of mouse CD200RLa, the closely related CD200R (known to be active as it binds CD200) and the CD200/CD200R complex (Hatherley et al., 2013). The results are all consistent with data showing that the viral orthologues are in fact very good mimics of the native CD200. Thus, it is unlikely that the failure of CD200 to bind the activating receptors is an artefact. The findings can be considered in relation to other paired receptor families, of which about 10 have been described on myeloid cells (Akkaya & Barclay, 2013). These are characterized by containing similar extracellular regions but different cytoplasmic regions giving different signalling mechanisms, suggesting two divergent functional outcomes. These paired receptors either bind the same ligand equally and there is a role in homeostasis or they bind different ligands and are utilized in different circumstances. It is probable that both mechanisms exist in different receptor families, for example, contrasting the CD200R here with Ly-49 on NK cells (see Akkaya & Barclay, 2013). Two other key features are that the ligands of the inhibitory receptors are often widely distributed proteins and they are involved in homeostasis, and the activating receptors are more diverse in number and sequence. The activating Ly49W and inhibitory Ly49G members of the Ly-49 NK cell receptor family have similar affinity for their ligands (MHC Antigens) (Ma et al., 2014), while other NK activating receptors have weaker binding for their MHC ligands such as NKG2D (Valés-Gomez et al., 1998; Winter et al., 1998). In the CD200/CD200R system discussed here and another macrophage receptor family, the SIRPs, there is virtually no binding of the ligand for the inhibitory receptor (CD200 and CD47, respectively) to the activating receptor, where subtle changes near the binding site have led to loss of activity (Hatherley et al., 2005, 2008, 2013).
If the evolution of the activating receptors is being driven by pathogen pressure, what are the interactions? The clearest evidence of a role for the activating receptors in pathogen defence is the Ly-49 system, where the m157 MHC like protein from mouse CMV binds some alleles of both activating and inhibitory receptors, with corresponding effects on survival and lethal infection, respectively. This led to the idea that activating receptors have evolved to protect against pathogens (Arase et al., 2002). There are some examples of pathogens binding both activating and inhibitory members of paired receptors, such as bacteria binding the macrophage PIR receptors (Akkaya & Barclay, 2013; Nakayama et al., 2007). As pathogens are able to evolve faster than the host, it is difficult to see how they retain binding to activating receptors when there is not an obvious advantage. An explanation may be that the pathogens target the inhibitory receptor and are willing to take some losses by being caught by the activating receptor – this ‘counterbalance theory’ is described in detail by Barclay & Hatherley (2008). For those activating receptors that do not bind the ligand of the inhibitory receptor, a viral homologue that exactly mimics the ligand will avoid the activating receptor and this is apparently the case for the viral orthologues of CD200 tested here. This is supported by structural studies that show that residues in the binding site of CD200 for CD200R are conserved in the herpesvirus orthologues (Hatherley et al., 2013). Thus the results are compatible with these orthologues being close mimics of the host and likewise distinguish the activating and inhibitory receptors. There are no other known ligands for CD200R, but it is possible that pathogens have interacted with both activating and inhibitory receptors during evolution and they have yet to be identified. Given the number and rapid evolution of these activating receptors in mice (Akkaya & Barclay, 2010), it is likely that such pathogens are still present. Further evidence that the CD200R pathway is important for herpesviruses is indicated by studies showing knocking out CD200R gave heightened responses despite the MCMV lacking a CD200 orthologue (Stack et al., 2015).
In poxviruses, the CD200 orthologues are more divergent in sequence and domain organization in that they contain only one domain (Fig. 1). There is evidence from the myxoma virus that they inhibit pro-inflammatory cytokine responses, but binding to the CD200R (mouse) was not convincing (Cameron et al., 2005; Zhang et al., 2009). The recent structure of the CD200/CD200R complex (Hatherley et al., 2013) reveals that key residues in the herpesvirus and host CD200 for ligand binding are not present in the poxvirus CD200, and it seems likely that the poxvirus CD200-related proteins function in a different manner.
Thus, it is clear that viral CD200 orthologues, at least in some herpesviruses, are highly effective at mimicking the host protein and hence have a role in downregulating immune activation of the innate system, where the CD200R is mainly expressed. The failure of these proteins to bind the activating members means that there is no limitation of the effect by these receptors.
Methods
Expression of recombinant CD200R soluble proteins
The extracellular domains of all the CD200Rs of the human, rat and mouse, including the activating forms, were expressed with rat CD4 domains 3 and 4 (CD4 d3+4) and a biotinylation (bio) site at the C terminus, using the pEFBOS vector as described previously (Hatherley et al., 2005; Mizushima & Nagata, 1990). Recombinant rat CD200RLa (NCBI Protein accession no. XP_006221193; residues 1–235) and human CD200RLa (using the rCD4 leader, NCBI Protein accession no. P05540, residues 1–27, followed by SerThr link to human CD200RLa, NCBI Protein accession no. NP_001008784, residues 1–212) were expressed in the same way with a CD4d3+4 bio tag. Soluble recombinant proteins were produced by transfecting 293T cells using polyethylenimine (PEI), and supernatants were harvested and quantified by ELISA using the OX68 CD4 mAb as previously described (Hatherley et al., 2005; Mizushima & Nagata, 1990). The supernatants were concentrated using Amicon Ultra 15 columns (10K MWCO) following the manufacturer's instructions, to a final volume of 500 μl.
Expression of recombinant host and viral CD200 soluble proteins
The extracellular domains of human CD200 (NCBI Protein accession no. EAW79672, residues 1–232) and rat CD200 (NCBI Protein accession no. P04218; residues 1–232), followed by a C-terminal hexahistidine tag, were stably expressed by CHO.K1 and CHO Lec3.2.8.1 cells, respectively, using the pEE14 expression vector (Davis et al., 1990; Hatherley & Barclay, 2004; Wright et al., 2000). The extracellular region of mouse CD200 (NCBI Protein accession no. NP_034948; residues 1–232) was expressed with a C-terminal hexahistidine tag by CHO Lec3.2.8.1 (Hatherley et al., 2013). The extracellular domains of the viral orthologues K14 (NCBI Protein accession no. P0C788; residues 1–225) and e127 (NCBI Protein accession no. AFX83435; residues 1–238) were cloned into the pHL-Avitag3 vector with the amino acid linker of AlaSerArg for K14 and SerArg for e127, preceding the biotin ligase recognition site, to allow biotinylation followed by a LysHis6 tag (Aricescu et al., 2006). The viral orthologue recombinant proteins were produced by transfection of 293T cells using either PEI or Fugene HD. His-tagged proteins were purified by nickel affinity chromatography and fractionated by gel filtration in 10 mM HEPES, pH 7.5, 150 mM NaCl.
Surface plasmon resonance
Surface plasmon resonance experiments were performed using a BIAcore 3000 at 25 °C. A rat CD4 mAb (OX68) was amine coupled to a CM5 chip allowing immobilization of each recombinant CD200R protein containing the CD4 d3+4 tag. CD4 d3+4 was coupled as a control protein. Freshly prepared protein was used to minimize non-specific binding due to denatured protein. In all experiments, around 500 response units of recombinant protein were immobilized. Rat CD200 and the RCMV CD200 orthologue e127 were passed over the immobilized proteins at 10 μM, at a flow rate of 10 μl min− 1. Human CD200 and HHV8 K14 were passed over immobilized CD200R proteins at 1 μM. Specific binding responses were obtained by subtracting the response units obtained for the immobilized control protein (CD4 d3+4).
Acknowledgements
This work was supported by the Medical Research Council, grant number G9826026.
References
- Akkaya M., Barclay A. N. (2010). Heterogeneity in the CD200R paired receptor family Immunogenetics 6215–22 10.1007/s00251-009-0415-6. [DOI] [PubMed] [Google Scholar]
- Akkaya M., Barclay A. N. (2013). How do pathogens drive the evolution of paired receptors? Eur J Immunol 43303–313 10.1002/eji.201242896. [DOI] [PubMed] [Google Scholar]
- Akkaya M., Aknin M. L., Akkaya B., Barclay A. N. (2013). Dissection of agonistic and blocking effects of CD200 receptor antibodies PLoS One 8e63325. 10.1371/journal.pone.0063325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arase H., Mocarski E. S., Campbell A. E., Hill A. B., Lanier L. L. (2002). Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors Science 2961323–1326 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- Aricescu A. R., Lu W., Jones E. Y. (2006). A time- and cost-efficient system for high-level protein production in mammalian cells Acta Crystallogr D Biol Crystallogr 621243–1250 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- Barclay A. N., Hatherley D. (2008). The counterbalance theory for evolution and function of paired receptors Immunity 29675–678 10.1016/j.immuni.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay A. N., Wright G. J., Brooke G., Brown M. H. (2002). CD200 and membrane protein interactions in the control of myeloid cells Trends Immunol 23285–290 10.1016/S1471-4906(02)02223-8. [DOI] [PubMed] [Google Scholar]
- Brown M. H., Boles K., van der Merwe P. A., Kumar V., Mathew P. A., Barclay A. N. (1998). 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48 J Exp Med 1882083–2090 10.1084/jem.188.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron C. M., Barrett J. W., Liu L., Lucas A. R., McFadden G. (2005). Myxoma virus M141R expresses a viral CD200 (vOX-2) that is responsible for down-regulation of macrophage and T-cell activation in vivo J Virol 796052–6067 10.1128/JVI.79.10.6052-6067.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S. J., Ward H. A., Puklavec M. J., Willis A. C., Williams A. F., Barclay A. N. (1990). High level expression in Chinese hamster ovary cells of soluble forms of CD4 T lymphocyte glycoprotein including glycosylation variants J Biol Chem 26510410–10418. [PubMed] [Google Scholar]
- Estep R. D., Rawlings S. D., Li H., Manoharan M., Blaine E. T., O'Connor M. A., Messaoudi I., Axthelm M. K., Wong S. W. (2014). The rhesus rhadinovirus CD200 homologue affects immune responses and viral loads during in vivo infection J Virol 8810635–10654 10.1128/JVI.01276-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster-Cuevas M., Wright G. J., Puklavec M. J., Brown M. H., Barclay A. N. (2004). Human herpesvirus 8 K14 protein mimics CD200 in down-regulating macrophage activation through CD200 receptor J Virol 787667–7676 10.1128/JVI.78.14.7667-7676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster-Cuevas M., Westerholt T., Ahmed M., Brown M. H., Barclay A. N., Voigt S. (2011). Cytomegalovirus e127 protein interacts with the inhibitory CD200 receptor J Virol 856055–6059 10.1128/JVI.00064-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczynski R., Chen Z., Kai Y., Lee L., Wong S., Marsden P. A. (2004). CD200 is a ligand for all members of the CD200R family of immunoregulatory molecules J Immunol 1727744–7749 10.4049/jimmunol.172.12.7744. [DOI] [PubMed] [Google Scholar]
- Hatherley D., Barclay A. N. (2004). The CD200 and CD200 receptor cell surface proteins interact through their N-terminal immunoglobulin-like domains Eur J Immunol 341688–1694 10.1002/eji.200425080. [DOI] [PubMed] [Google Scholar]
- Hatherley D., Cherwinski H. M., Moshref M., Barclay A. N. (2005). Recombinant CD200 protein does not bind activating proteins closely related to CD200 receptor J Immunol 1752469–2474 10.4049/jimmunol.175.4.2469. [DOI] [PubMed] [Google Scholar]
- Hatherley D., Graham S. C., Turner J., Harlos K., Stuart D. I., Barclay A. N. (2008). Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47 Mol Cell 31266–277 10.1016/j.molcel.2008.05.026. [DOI] [PubMed] [Google Scholar]
- Hatherley D., Lea S. M., Johnson S., Barclay A. N. (2013). Structures of CD200/CD200 receptor family and implications for topology, regulation, and evolution Structure 21820–832 10.1016/j.str.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T., Obata K., Mukai K., Sato S., Takai T., Minegishi Y., Karasuyama H. (2007). Mast cells and basophils are selectively activated in vitro and in vivo through CD200R3 in an IgE-independent manner J Immunol 1797093–7100 10.4049/jimmunol.179.10.7093. [DOI] [PubMed] [Google Scholar]
- Ma B. J., Craveiro Salvado C. M., Kane K. P. (2014). The activating Ly49W and inhibitory Ly49G NK cell receptors display similar affinities for identical MHC class I ligands Immunogenetics 66467–477 10.1007/s00251-014-0777-2. [DOI] [PubMed] [Google Scholar]
- Misstear K., Chanas S. A., Rezaee S. A., Colman R., Quinn L. L., Long H. M., Goodyear O., Lord J. M., Hislop A. D., Blackbourn D. J. (2012). Suppression of antigen-specific T cell responses by the Kaposi's sarcoma-associated herpesvirus viral OX2 protein and its cellular orthologue, CD200 J Virol 866246–6257 10.1128/JVI.07168-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima S., Nagata S. (1990). pEF-BOS, a powerful mammalian expression vector Nucleic Acids Res 185322. 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M., Underhill D. M., Petersen T. W., Li B., Kitamura T., Takai T., Aderem A. (2007). Paired Ig-like receptors bind to bacteria and shape TLR-mediated cytokine production J Immunol 1784250–4259 10.4049/jimmunol.178.7.4250. [DOI] [PubMed] [Google Scholar]
- Shiratori I., Yamaguchi M., Suzukawa M., Yamamoto K., Lanier L. L., Saito T., Arase H. (2005). Down-regulation of basophil function by human CD200 and human herpesvirus-8 CD200 J Immunol 1754441–4449 10.4049/jimmunol.175.7.4441. [DOI] [PubMed] [Google Scholar]
- Stack G., Jones E., Marsden M., Stacey M. A., Snelgrove R. J., Lacaze P., Jacques L. C., Cuff S. M., Stanton R. J., other authors (2015). CD200 receptor restriction of myeloid cell responses antagonizes antiviral immunity and facilitates cytomegalovirus persistence within mucosal tissue PLoS Pathog 11e1004641. 10.1371/journal.ppat.1004641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valés-Gómez M., Reyburn H. T., Erskine R. A., Strominger J. (1998). Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors Proc Natl Acad Sci U S A 9514326–14331 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe P. A., Barclay A. N. (1994). Transient intercellular adhesion: the importance of weak protein-protein interactions Trends Biochem Sci 19354–358 10.1016/0968-0004(94)90109-0. [DOI] [PubMed] [Google Scholar]
- Voehringer D., Rosen D. B., Lanier L. L., Locksley R. M. (2004). CD200 receptor family members represent novel DAP12-associated activating receptors on basophils and mast cells J Biol Chem 27954117–54123 10.1074/jbc.M406997200. [DOI] [PubMed] [Google Scholar]
- Winter C. C., Gumperz J. E., Parham P., Long E. O., Wagtmann N. (1998). Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition J Immunol 161571–577. [PubMed] [Google Scholar]
- Wright G. J., Puklavec M. J., Willis A. C., Hoek R. M., Sedgwick J. D., Brown M. H., Barclay A. N. (2000). Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function Immunity 13233–242 10.1016/S1074-7613(00)00023-6. [DOI] [PubMed] [Google Scholar]
- Wright G. J., Cherwinski H., Foster-Cuevas M., Brooke G., Puklavec M. J., Bigler M., Song Y., Jenmalm M., Gorman D., other authors (2003). Characterization of the CD200 receptor family in mice and humans and their interactions with CD200 J Immunol 1713034–3046 10.4049/jimmunol.171.6.3034. [DOI] [PubMed] [Google Scholar]
- Zhang L., Stanford M., Liu J., Barrett C., Jiang L., Barclay A. N., McFadden G. (2009). Inhibition of macrophage activation by the myxoma virus M141 protein (vCD200) J Virol 839602–9607 10.1128/JVI.01078-09. [DOI] [PMC free article] [PubMed] [Google Scholar]