Abstract
The biosynthesis of structural and signaling molecules depends on intracellular concentrations of essential amino acids, which are maintained by a specific system of plasma membrane transporters. We identify a unique population of nutrient amino acid transporters (NATs) within the sodium-neurotransmitter symporter family and have characterized a member of the NAT subfamily from the larval midgut of the Yellow Fever vector mosquito, Aedes aegypti (aeAAT1, AAR08269), which primarily supplies phenylalanine, an essential substrate for the synthesis of neuronal and cuticular catecholamines. Further analysis suggests that NATs constitute a comprehensive transport metabolon for the epithelial uptake and redistribution of essential amino acids including precursors of several neurotransmitters. In contrast to the highly conserved subfamily of orthologous neurotransmitter transporters, lineage-specific, paralogous NATs undergo rapid gene multiplication/substitution that enables a high degree of evolutionary plasticity of nutrient amino acid uptake mechanisms and facilitates environmental and nutrient adaptations of organisms. These findings provide a unique model for understanding the molecular mechanisms, physiology, and evolution of amino acid and neurotransmitter transport systems and imply that monoamine and GABA transporters evolved by selection and conservation of earlier neuronal NATs.
Subsets of essential amino acids that are accumulated by the uptake of digested nutrients rather than by de novo synthesis are similar in all metazoans. For example, mammals and mosquito larvae cannot tolerate dietary deprivation of any one of the basic (arginine, lysine, and histidine), neutral (leucine, methionine, isoleucine, threonine, and valine), or aromatic (phenylalanine and tryptophan) L amino acids (1). Aromatic amino acids are exceptionally critical, being essential precursors for the synthesis of neurotransmitters and the polymerization of ectodermic components. Surprisingly, molecular mechanisms of accumulation and redistribution of essential amino acids are poorly understood in mammals and virtually unknown in other organisms, including insects. Metazoan Na+-dependent and -independent carrier systems transporting amino acids comprise B0,+, LAT1, LAT2, and TAT secondary transporters and ATP-binding cassette (ABC) pumps. Mammalian Na+-driven B0,+ amino acid systems deriving from initially characterized SLC6A14 members in Homo sapiens may contribute to primary transapical uptake of phenylalanine (2). Nevertheless, these transporters belong to a glycine–proline phylogenetic cluster within the sodium-neurotransmitter symporter family (SNF) and have a relatively low selectivity for phenylalanine and several other essential amino acids: IEC50 = 6 μM > L12 > M14 >F17 > other amino acids (3). The situation is similar for the recently cloned B0AT1, an amino acid transporter from mouse: QKm=522 μM > F589 > L630 (4). Obvious substrate competition for a substrate-docking site would render this carrier phenotype insufficient for the comprehensive accumulation of essential amino acids, especially from the short digestive tract, e.g., in mosquito larvae. Na+-independent LAT1 and LAT2 systems are represented by SLC7A5 and SLC7A8 members, respectively, in mammals (5). These transporters constitute functional heterodimers with an ancillary subunit (SLC3A2) (6) and act as an obligatory 1:1 amino acid exchange mechanism, which would be wasteful for nutrient accumulation. Indeed, SLC7A8 is a basolateral complement of unidentified apical transporters that together form a putative transepithelial pathway (7), and SLC7A5 is a plasma membrane transporter in actively metabolizing, nonepithelial cells (8). The Na+-independent TAT system of insects corresponds to SLC16A10, a unique member of the monocarboxylate transporter family in mammals (9). It constitutes a basolateral carrier that mediates downhill diffusion of aromatic amino acids. However, because of its narrow substrate specificity and diffusion mechanism, it is unable to satisfy the requirement for aromatic and other essential amino acids. ABC pumps are not suited for this high-throughput uptake because they are too slow. The uptake of peptides by proton motive force-driven transporters (SLC15), followed by hydrolysis to amino acids within the cells, comprises another possibility that may contribute to high-throughput and low-specificity uptake of protein digestion products, but may only partially contribute to the essential amino acid uptake and redistribution (10). This brief summary reveals a surprising knowledge gap in molecular mechanisms of essential amino acid accumulation in mammals, especially regarding their primary uptake across apical membranes of nutrient epithelia. It contrasts sharply with the extensive knowledge of Na+-dependent essential amino acid transport systems from brush border studies.
Molecular identities of essential amino acid transporters in other metazoans are virtually unknown (11). Two unusual transporters, which belong to the SNF but break electrochemical rules of this family, were cloned from the caterpillar of Manduca sexta. First, KAAT1 can use a caterpillar midgut-specific K+ gradient instead of the widespread Na+ gradient (12). Second, CAATCH1 (13) can switch between transporter and cation channel modes, depending on available substrates (14). Both of these transporters can be classified as nutrient amino acid transporters based on the prevalence of essential amino acids in their substrate spectra and their apical expression in midgut epithelia, the site of primary amino acid absorption in insect larvae.
Here, we report the comprehensive analysis of a unique amino acid transporter (aeAAT1) from Aedes aegypti larvae. Like KAAT1 and CAATCH1, aeAAT1 belongs to the SNF but is unable to transport neurotransmitters. In contrast, it preferably mediates the uptake and redistribution of essential phenylalanine and, to a lesser extent, other essential amino acids. Our characterization of this transport phenotype and subsequent comparative phylogenetic analysis of aeAAT1 homologs in a comprehensive framework of SNF members from selected prokaryotic and metazoan genomes led to the identification of a unique population of lineage-specific nutrient amino acid transporters (NATs) that are distinct from orthologous neurotransmitter transporters (NTTs).
Materials and Methods
Bioinformatics. Transcript sequences were assembled by using seqman ii (DNASTAR, Madison, WI). Protein topology and hydropathy were predicted by using topped ii (15) through a remote server in the Pasteur Institute (http://bioweb.pasteur.fr). Phosphorylation sites were detected by using peptools 2.0 software (BioTools, Edmonton, Alberta, Canada). Homologous protein sequences, derived as best reciprocal matches to aeAAT1 and other SNF members, were acquired by using NCBI and EMBL databases and blast tools. Protein alignments were completed by using clustalx (16) and fine-tuned by using differential setting of alignment parameters for conserved and low scoring regions. Quartet-based maximum-likelihood (17) and neighbor-joining (18) methods were used for phylogenetic tree reconstruction.
Molecular Cloning. Degenerate primers (13) were used to screen a high-performance cDNA collection (19) from posterior midgut of A. aegypti larvae. Touchdown hot-start PCR (20) was performed by using TaqDNA Polymerase in 1.5 mM MgCl2 solution (Roche Molecular Biochemicals). The PCR product was cloned into a TOPO vector (Invitrogen) and sequenced. Exact primers, based on the partial sequences from PMG cDNA, were used in combination with cDNA-flanked TRsa and LU4 adapters for 3′/5′ RACE. ORF-flanked primers with nonredundant BamHI and NotI restriction sites (sense, cgGGATCCcgATGCCCGAGATAG CAACAATATCGTA; antisense, atagtttaGCGGCCGCattcttatATACTCACGTCAAAGC CTGGTACTAT; underlined are restriction sites with digestion enhancer in lowercase.) were used to amplify the coding region of aeAAT1 from PMG cDNA. BamHI/NotI-digested PCR product was separated by 1% agarose gel electrophoresis, purified with an Extraction kit (Qiagen), and ligated into a pXOOM-expression vector (21).
Heterologous Expression. cRNA was obtained by in vitro transcription of PmeI-linearized pXOOM-aeAAT1 with mMessage mMachine, a high-yield, capped RNA transcription kit (Ambion). The integrity and amount of the RNA was confirmed by agarose gel electrophoresis. Surgically isolated and collagenase-treated stage V-VI Xenopus laevis oocytes (Nasco, Fort Atkinson, WI) were injected with ≈40 ng of aeAAT1 cRNA and incubated for 3–7 days at 17°C in a sterile oocyte medium (98.0 mM NaCl/2.0 mM KCl/1.0 mM MgCl/1.8 mM CaCl/2.5 mM Na pyruvate/100 units/ml penicillin/100 μg/ml streptomycin/5% horse serum/10 mM Hepes, pH 7.4).
Electrophysiological Analysis. Oocytes were analyzed in a 50-μl permanent-flow perfusion chamber connected through two 3M KCl-1% agar-Ag/AgCl referencing electrodes to a GeneClamp 500 two-electrode voltage clamp amplifier via a VG-2A-x100 current monitor head stage (Axon Instruments). Microelectrodes (0.5–1 MΩ) were prepared with a Sutter-2000 puller from a 1.2-mm borosilicate glass capillary and filled with 1 M KCl/1 M K2SO4 electrolyte. Two acquisition devices, MP100 (World Precision Instruments) and DigiData1200 (Axon Instruments), were synchronized for simultaneous recording of slow pharmacological responses and fast IV tests (1 Hz and 2 kHz low-pass filter cutoff, respectively). pClamp-8 (Axon Instruments) and sigmaplot 8 software (SPSS, Chicago) were used for data analysis. Ion substitution solutions are given in Table 1.
Table 1. Composition of stock solutions for ion substitution assay in mM.
| Solution | 98Na | 100K | 98Na-Cl | 98K-CL | ChCl |
|---|---|---|---|---|---|
| NaCl | 98 | 0 | 0 | 0 | 1 |
| KCl | 2 | 100 | 0 | 0 | 1 |
| Na Gluconate | 0 | 0 | 98 | 2 | 0 |
| K Gluconate | 0 | 0 | 2 | 98 | 0 |
| Choline Cl | 0 | 0 | 0 | 98 | |
| MgSO4 7H2O | 0 | 0 | 0.5 | 0.5 | 0 |
| MgCl2 6H2O | 0.5 | 0.5 | 0 | 0 | 0.5 |
| CaCl2 2H2O | 0.5 | 0.5 | 0 | 0 | 0.5 |
| Ca Gluconate | 0 | 0 | 0.5 | 0.5 | 0 |
| Hepes (pH 7.2) | 10 | 10 | 10 | 10 | 10 |
Bold face indicates primary salt.
Uptake Assay. aeAAT1i-expressing oocytes were exposed to a 100 μM, 1/100 3H-labeled/unlabeled mixture of selected l-amino acids (Amersham Pharmacia) in 98Na medium at 17°C for specified time intervals. The uptake was terminated by three washes in 1 ml of N98. For each assay, four oocytes were placed in 3.5 ml of scintillation liquid (SintiVerse, Fisher) in a polypropylene vial and were analyzed with a liquid scintillation spectrometer/counter (1214 RackBeta instrument, Pharmacia LKB). Uptake values were measured as dpm and were normalized and converted to molar quantities by using the appropriate isotope-specific activity value for each 3H-labeled amino acid (Amersham Pharmacia).
In Situ Hybridization. A. aegypti larvae were raised as described (22) from eggs supplied by Malaria Research and Reference Reagent Center (MRA-112; American Type Culture Collection). Fourth-instar larvae were immobilized in ice-cold 0.1 M PBS, opened by a lateral incision, and fixed in 4% paraformaldehyde/PBS overnight. Preparations were dehydrated/rehydrated by passing through a PBS/methanol gradient set (100% PBS, 1:3/1:1/3:1, 100% methanol), 10 min for each mixture, pretreated with proteinase K/detergent solution (0.1% Tween in PBS supplemented with 10 μg/ml Proteinase K) for 10 min and denatured by incubation in the hybridization saline (50% formamide/5 mM EDTA in 1× SSC) at 70°C for 10 min. Digoxigenin-labeled SP6-antisense and T7-sense run-off transcripts were obtained from an NcoI- and NotI-linearized pGEMT-aeAAT1 clone by using a digoxigenin-UTP kit (Roche Diagnostics). A 1:10,000 probe in hybridization saline mix was denatured by incubation at 70°C for 10 min. Preparations were hybridized at 60°C overnight, washed in PBS, labeled with phosphatase-conjugated digoxigenin antibodies according to the manufacturer's protocol (Roche Diagnostics), and stained in nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate alkaline buffer solution (Boehringer Mannheim) overnight. Stained preparations were fixed in cold 1:1 methanol/4% paraformaldehyde/PBS mixture for 1 h and embedded in 1:1 PBS glycerol. Whole mount preparations were photographed by using an Olympus stereomicroscope and Pixera charge-coupled imaging device. This technique appears to be specific for both aeAAT1 and aeAAT1i transcripts.
Real-Time PCR. cDNA samples were produced from isolated, 70% methanol-fixed tissues and gut regions by using a Cells-to-cDNA kit and protocol (Ambion). A. aegypti 18S ribosomal RNA (GenBank accession ni. M95126) was selected as a template to optimize concentrations and to normalize data. Primers were designed by using primer express software (Applied Biosystems). Reactions were monitored by using ABI Prism 7000 Fluorescent Sequence Detection System and SYBR Green PCR Master mix (Applied Biosystems). Two-step PCR was used (50°C for 2 min, 95°C for 10 min and 40× (95°C for 15 s, 60°C for 1 min). Control runs included a subset of PCR components lacking the cDNA template. Relative expression levels were calculated as (threshold cycle number - intercept)/slope (primer efficiency, according to an Ambion protocol). The average log transcript quantities were normalized vs. an average value for 18S RNA. Data represent two averaged replicates of three independent experiments, each of which was carried out on a separate set of tissue samples. excel software (Microsoft) was used to analyze data and to generate representative graphs.
Results
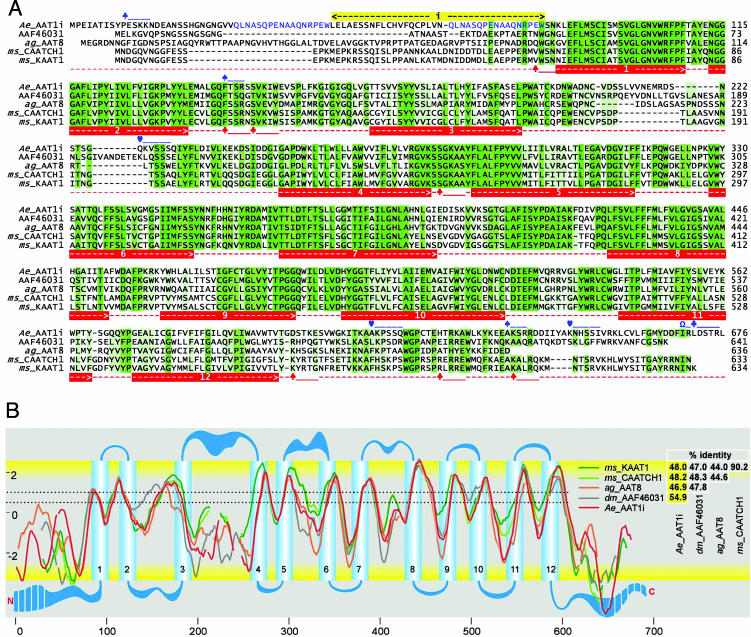
A long transcript, aeAAT1i (2,034 bp, GenBank accession no. AAR08269) and a short transcript, aeAAT1 (1923 bp, NCBI accession no. AAN40410) of the new transporter were cloned from midgut-specific cDNA of fourth-instar A. aegypti larvae. aeAAT1i encodes a 678-aa ORF with a SNF-specific PFAM domain (pfam00209; E = 3e-130 from the conserved domain database). In contrast to the short isoform, aeAAT1, long aeAAT1i has a 111-nt insert at the 5′ end (Fig. 1A). Each transcript encodes a protein with secondary structures similar to those of amino acid transporters KAAT1 and CAATCH1 from the caterpillar midgut (Fig. 1A). The secondary structure includes 12 predicted transmembrane domains (Fig. 1 A and B), 31 putative enzyme-specific phosphorylation sites, and six sites that could be phosphorylated by more than one kinase (partially present in Fig. 1A). Structural differences between insect homologs of aeAAT1 are most prominent in the cytosolic domains of the N and C terminals and the extracellular loops between the third/fourth and the fifth/sixth transmembrane domains (Fig. 1B).
Fig. 1.
Comparison of insect NAT structures. (A) A protein sequence alignment of selected insect NATs that includes conceptual translations of A. aegypti aeAAT1i (AAR08269), a putative homologous protein from the Drosophila sequencing project (AAF46031 or NP_572219) cloned in our laboratory, Anopheles gambiae ag_AAT8 (AAN40409, prediction XP_309840), and M. sexta KAAT1 and CAATCH1 (AAC24190 and AAF18560 respectively). Sequence similarities are shown by the intensities of the green background. aeAAT1i-specific (blue) and NAT universal (red) features are shown as follows: numbered red bars, transmembrane domains; blue bars, two internal repeats, yellow bar, insert that is absent in short isoform (NCBI no. AAN40410); ♣, protein kinase C phosphorylation sites; ♠, cGMP-dependent protein kinase phosphorylation sites; ♡, myosin 1 heavy chain kinase phosphorylation sites; Ω, protein kinase C phosphorylation sites of all types. (B) An alignment of hydropathy profiles and predicted transmembrane topologies (y and x axes are hydropathy and sequence positions, respectively). Gaps and transmembrane domains plot were generated electronically by using protein sequence alignment and TOPPED2 coordinates of transmembrane domains. Dotted lines correspond to lower (0.5) and upper (1) cutoff values. Reciprocal identity of amino acid sequences is shown on the right.
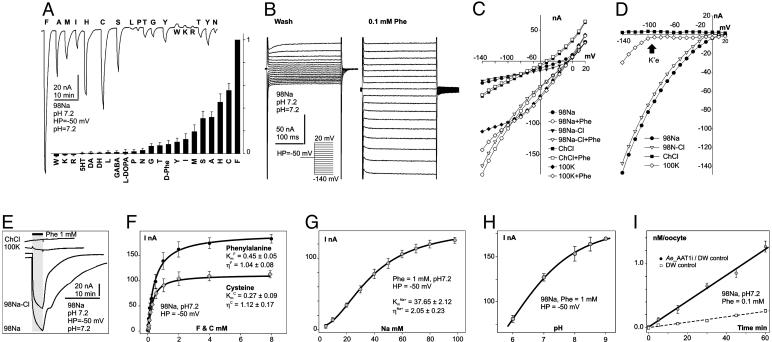
Application of amino acids did not induce electrical responses either from short isoform-injected Xenopus oocytes or from deionized water-injected controls. In contrast, application of several amino acids to long isoform-expressing oocytes induced large inward currents that resulted from asymmetric charge transfer during substrate/carrier ion translocations (Fig. 2A Upper and B). The amplitude of the substrate-induced current decreased according to the following amino acid order: F >> C > H > A ≈ S > M > I > Y > T ≈ G > N ≈ P > l-DOPA > GABA > L >> DA ≈ Octopamine ≈ 5HT (Fig. 2A Lower) and depended on transmembrane voltage (Fig. 2B). Ae_ATT1i appears to be isomer-specific, expressing weak responses to d-enantiomers of phenylalanine (Fig. 2A Lower), histidine, and alanine. aeAAT1i is a Na+-driven and Cl- modulated transporter, because replacement of Na+ by K+ or choline+ eliminated ligand-induced inward currents, and replacement of Cl- by gluconate- modified response kinetics and weakly reduced amplitudes of substrate-induced currents (Fig. 2 C–E). In addition, aeAAT1i enables substrate-induced K+ currents to flow at transmembrane voltages that exceed the K+ reversal potential for Xenopus oocytes (arrow at Fig. 2D) suggesting that, under certain electrochemical conditions, aeAAT1i may use K+ gradients as well as Na+ gradients. aeAAT1i exhibits saturable kinetics with apparent dissociation constants 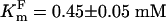 ,
, 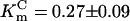 , and
, and 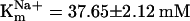 , and Hill coefficients (ηF) = 1.04 ± 0.08, ηC = 1.12 ± 0.17, and ηNa+ = 2.05 ± 0.23 for Phe, Cys, and Na+, respectively (Fig. 2 F and G). The amplitude of the ligand induced currents depends on pH and increases in alkaline media (Fig. 2H). Together with the voltage dependency of responses, these data suggest that aeAAT1i mediates an electrophoretic symport of amino acids and Na+ with apparent stoichiometry 1(aa):2(Na+). In contrast, basic amino acids and tryptophan induced small Na+-dependent outward currents (Fig. 2A). Such currents might result from a ligand-induced block of a Na+ leak current or an increase of a K+ leak. Millimolar concentrations of the acidic amino acids, glutamic and aspartic acid, did not produce notable responses at the midgut-specific alkalinity (pH 7–9).
, and Hill coefficients (ηF) = 1.04 ± 0.08, ηC = 1.12 ± 0.17, and ηNa+ = 2.05 ± 0.23 for Phe, Cys, and Na+, respectively (Fig. 2 F and G). The amplitude of the ligand induced currents depends on pH and increases in alkaline media (Fig. 2H). Together with the voltage dependency of responses, these data suggest that aeAAT1i mediates an electrophoretic symport of amino acids and Na+ with apparent stoichiometry 1(aa):2(Na+). In contrast, basic amino acids and tryptophan induced small Na+-dependent outward currents (Fig. 2A). Such currents might result from a ligand-induced block of a Na+ leak current or an increase of a K+ leak. Millimolar concentrations of the acidic amino acids, glutamic and aspartic acid, did not produce notable responses at the midgut-specific alkalinity (pH 7–9).
Fig. 2.
Representative electrochemical properties of aeAAT1i expressed in Xenopus oocytes. (A Upper) Amino acid-induced currents (all amino acids were presented at 1 mM final concentration for 30 sec). (A Lower) Normalized amino acid-induced responses (bars are means of normalized values ± SE; number of oocytes tested n ≥ 3 for each presented value). (B) Voltage clamp currents after application of 0.1 mM phenylalanine. (C) Representative collection of I/V plots from ion substitution experiments. Symbols with similar shapes represent pairs of a “control” (no phenyl alanine) vs. 0.1 mM phenylalanine added. (D) “Control” subtracted I/V plots. Arrow indicates a K+ equilibrium potential for Xenopus oocytes under physiological conditions. (E) Superimposed set of phenylalanine-induced currents upon ion substitutions. (F–H) Phenylalanine, cysteine, Na+, and pH dependency of substrate induced currents respectively (means of current ± SE; n ≥ 3 for each point). Data were fitted by nonlinear regression with three parameter sigmoidal function 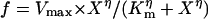 , where Km is the apparent Michaelis–Menten constant and η is the Hill coefficient; values are shown on the graphs. (I) Phenylalanine uptake by aeAAT1i-expressing oocytes vs. water-injected “controls” (values are means ± SE, n = 3).
, where Km is the apparent Michaelis–Menten constant and η is the Hill coefficient; values are shown on the graphs. (I) Phenylalanine uptake by aeAAT1i-expressing oocytes vs. water-injected “controls” (values are means ± SE, n = 3).
aeAAT1i expression significantly increased isotope-labeled phenylalanine uptake in Xenopus oocytes (4.35 ± 0.15 times; n ≥ 3; Fig. 2I, pared t test, P > 0.05). No significant increase in the uptake of the isotope-labeled acidic amino acids glutamate and aspartate was found (data not shown). The neurotransmitters 5-HT, dopamine, octopamine, and GABA induced no responses in aeAAT1i-expressing oocytes.
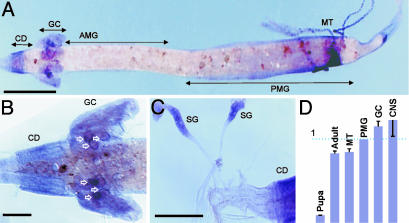
No signals were detected with control sense probes (data not shown). Antisense aeAATi mRNA probes produced specific hybridization signals in the specific parts of alimentary canal of A. aegypti larvae including cardia, posterior midgut, salivary glands, specific cells in the proximal part of the Malpighian tubules, and a group of large epithelial cells with specific locations in the proximal part of gastric caeca (Fig. 3). No antisense hybridization was detected in the anterior midgut and the middle region between the anterior and posterior midgut. Spatial hybridization patterns were similar in earlier and late fourth-instar larvae. Developmental changes of aeAAT1 expression include an intensity increase and dispersion of labeling from the perinuclear region of individual epithelial cells following transition from the earlier to late instars. Spatial and developmental patterns from whole mount in situ hybridization were confirmed by quantitative real-time PCR analysis (Fig. 3D). In addition, a high level of aeAAT1 transcript was found in the isolated CNS tissue, which included isolated brain and visceral ganglia.
Fig. 3.
Spatial/temporal profiles of aeAAT1 expression. (A) In situ hybridization of whole-mount alimentary canal from fourth instar A. aegypti larvae with Ae_ATT1 probes. (B and C) Magnified images of gastric caeca and salivary gland, respectively. CD, cardia (esophageal invaginations which are associated with synthesis of peritrophic membranes); GC, gastric caeca; AMG, anterior midgut; PMG, posterior midgut; MT, Malpighian tubules; SG, salivary glands. White outline arrows indicate strong hybridization signal in large epithelial cells of posterior caeca which are associated with synthesis of caecal membranes. (Scale bars, 1 mm in A and 200 μmin B and C.) (D) Relative expression of Ae_AAT transcripts in different tissues and developmental stages by quantitative real-time PCR analysis (n ≥ 3; bars are relative expression ± SE).
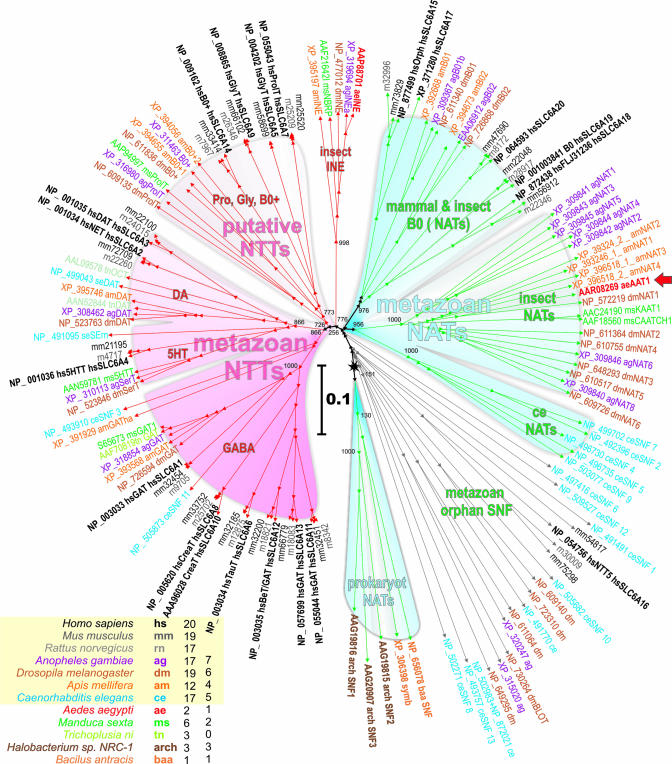
Screening for aeAAT1 homologs in selected eukaryotic and prokaryotic genomic databases and among cloned insect transporters (Fig. 4 Inset) retrieved 136 established and predicted gene products which belong to the SNF [also known as the neurotransmitter sodium symporter family 2.22 (23) and as SCL6 in mammals (2)]. A neighbor-joining tree of the selected transporters reveals two major clusters that represent specific physiological functions by consensus of transport phenotypes. They are labeled in the dendrogram as NTTs and NATs (Fig. 4). In addition several members of bacteria and archaean SNF form a cluster which is labeled as a “prokaryote NATs” because these transporters supply essential carbon in several prokaryotes by Na+-driven accumulation of specific amino acids (24). Several metazoan SNF members differ by out-branching points between prokaryote and metazoan NATs, and participating lineages are labeled as a “metazoan orphan SNF” because the transport phenotype or other functions of these transporters remain unidentified, except for “orphan BLOT” transporters, whose essential role in cell differentiation and epithelial morphogenesis has been inferred from morphological mapping and knockout analysis (25).
Fig. 4.
Neighbor-joining dendrogram of 136 selected SNF members (bootstrap value was 1,000; low values are shown at nodes). Unified accession numbers, species abbreviations, and specific names based on defined or tentative functions are shown. Truncated GenBank accession numbers for mm and rn lineages can be recovered by replacing the species abbreviations with ENSRNOP000000 string. 5HT, serotonin; 5HTT and SerT, high-affinity serotonin transporters; DA, dopamine; DAT, dopamine transporter; OAT, octopamine transporter; GlyT, glycine transporter; GAT, GABA transporter; INE, inebriated protein; simb, symbiotic contamination in the ag genome. (Inset) Include color code, species abbreviations, numbers of identified SNF and NATs members in each species. The red arrow shows a characterized aeAAT1 transporter. The scale bar shows evolutionary distance in numbers of substitutions per site.
The “metazoa NTTs” cluster includes well characterized neurotransmitter transporters from all selected lineages except for prokaryotes. Its members form clearly orthologous segments which reflect a substrate specificity for specific neurotransmitters including GABA, monoamines serotonin (5HT) and dopamine (DA), and putative neurotransmitter Gly/Pro/B0+ for glycine, proline, and some neutral/cationic amino acids, respectively (Fig. 4). The “Gly/Pro/B0+” cluster comprises both mammalian and insect lineages; however, there are no insect transporters proximal to the mammalian SLC6A14 B0+ system transporter that participates in intestinal uptake of neutral and cationic amino acids (3).
The “insect INE” cluster represents a transition between metazoan NTTs and NATs and includes relatively conserved insect-specific proteins (a single protein per lineage). The transport phenotype of “insect INE” proteins remains unknown; however, their role in brain functions has been recognized.
The “metazoan NATs” cluster is characterized by a common out branching point and obvious lineage segregation except for the “mammalian and insect B0 (NATs)” cluster. The role of these transporters in the nutrient amino acid uptake is supported by characterization of transporters from M. sexta (12, 13), mammals (4, 26), and aeAAT1 from mosquito larvae (presented here).
Discussion
Our cloning and characterization of a NAT phenotype and comparative phylogenetic analysis revealed two fundamentally different clusters in the SNF (“metazoan NTTs” and “metazoan NATs”) and led to the identification of an insect-specific cluster of nutrient amino acid transporters, “insect NATs” (Fig. 4). At present, the “insect NATs” cluster comprises 20 transporters from four insect lineages including five cloned and 16 predicted gene products. aeAAT1 is clearly a NAT on the basis of its substrate uptake spectrum and its expression pattern. It mediates both primary apical uptake in nutrient epithelia and the redistribution of a specific subset of nutrient amino acids through both basal and plasma membrane uptake mechanisms with phenylalanine being a preferred substrate. The role of insect NATs in the uptake and redistribution of essential amino acids is strongly supported by the functional consensus of the five cloned and characterized transporters (Fig. 4, underlined members of insect NATs), namely A. aegypti aeAAT1, M. sexta KAAT1 and CAATCH1, and Anopheles gambiae proteins agNAT8 and agNAT6, which were characterized in our laboratory as tyrosine-phenylalanine and tryptophan transporters, respectively (GenBank accession nos. AAN40409 and AY536865). From the observed phylogenetic pattern, we may predict a “nematode NATs” cluster that fits between “prokaryote NATs” and “insect NATs” but currently includes only uncharacterized nematode gene products (Fig. 4). Similarly, mammal and insect NATs, which in general comprise mammalian transporters but also include two orthologous group of insect transporters, can be tentatively designated to a NAT phenotype. Recently characterized Hartnup disorder-associated B0 amino acid transporters from mouse SLC6A18 (4) and human homolog SLC6A19 (26) (Fig. 4) provide additional collateral support to our hypothesis.
NATs encode a cation-driven, high-throughput uptake mechanism similar to the B0 system from brush border studies, but with varied affinities and selectivity for particular groups of nutrient amino acids that are encoded by specific genes in the NAT group. With minor reservations regarding alternative transport systems (11), we may postulate that NAT genes constitute a previously illusive essential amino acid uptake metabolon, at least in insects and most likely in other species. In addition to its well understood apical expression in absorptive sites of the insect alimentary canal, aeAAT1 is expressed in secretory regions, e.g., anterior midgut, gastric caeca, and salivary glands, the functions of which depend on basal absorption of amino acids from the haemolymph and are consistent with a basal location of the coupled transporter. Potentially, a basal vs. apical anchoring motif may be expressed by alternative splicing as in neurotransmitter transporters; however, the exact anchoring mechanism for NATs remains to be analyzed. aeAAT1 is also expressed in CNS, where it supplies primary substrates and appear to be a rate limiting component in catecholamine synthesis pathways (23). Therefore, we may expect that individual NAT genes encode several isoforms that produce morphologically distinct phenotypes for integration into apical or basal membranes of polarized epithelial cells or plasma membrane of neuronal cells.
The homology of bacterial NATs with SNF members and evolutionary path observed from the SNF tree reveals that NAT populations existed some time before the origin of metazoans and neuronal functions. This fact, together with established phylogenetic, structural, and physiological correlations among SNF members, suggests strongly that NTTs evolved from specific NATs that supplied essential amino acids in primordial neuronal cells. In contrast to highly conserved and clearly orthologous NTTs, NATs changed rapidly during evolution. NAT plasticity and NTT conservation are essential in evolution and imply different selection mechanisms (27). Indeed, the NTT group is stabilized by adaptation to particular substrates (neurotransmitters) and electrochemical homeostasis of the neuronal environ. In contrast, NAT populations undergo extensive substitution as a result of multiplication and selections for members that can perform better upon transition to new ecological niches. Plasticity of NAT phenotypes may include tuning of electrochemical properties, substrate affinity, and selectivity of NATs in different species. It also implies that archaic NATs will rapidly become extinct and in parallel with gene duplication will lead to the formation of linage-specific paralogous clusters. Among factors that can impact NAT populations are sophistication of the absorptive epithelia and alimentary canal, which increase the efficiency of protein digestion and amino acid absorption, and the evolution of sensory and cognitive functions that are beneficial for the acquisition of desired food sources. Multiple NATs with specific substrate preferences appear to be obligatory to avoid reciprocal substrate inhibition of transapical carriers in the short and rapidly absorbing insect gut.
In summary, functionally explicit NATs and NTTs provide a unique model for the study of the evolution and structural adaptation of membrane transport proteins. Essential and lineage-specific NATs appear to be better suited than the highly conserved NTTs as targets for selective and environmentally safe management of disease vector mosquitoes and agriculture pest insects.
Acknowledgments
We thank Dr. T. Jespersen (University of Copenhagen, Copenhagen) for expression plasmids; Drs. M. H. Saier, M. J. Greenberg, B. C. Donly, M. V. Matz, D. A. Price, and Y. V. Bobkov for helpful discussions; and Ms. E. V. Bobkova for preparing Xenopus oocytes. This work was supported in part by National Institutes of Health Grants AI030464 and AI405022 and the Whitney Laboratory (P. A. V. Anderson, director).
Abbreviations: SNF, sodium-neurotransmitter symporter family; NAT, nutrient acid transporter; NTT, neurotransmitter transporter.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AAR08269 and AAN40410).
References
- 1.Clements, A. N. (1992) The Biology of Mosquitoes (Chapman & Hall, London).
- 2.Chen, N. H., Reith, M. E. & Quick, M. W. (2004) Pflugers Arch. 447, 519-531. [DOI] [PubMed] [Google Scholar]
- 3.Sloan, J. L. & Mager, S. (1999) J. Biol. Chem. 274, 23740-23745. [DOI] [PubMed] [Google Scholar]
- 4.Broer, A., Klingel, K., Kowalczuk, S., Rasko, J. E., Cavanaugh, J. & Broer, S. (2004) J. Biol. Chem. 279, 24467-24476. [DOI] [PubMed] [Google Scholar]
- 5.Verrey, F. (2003) Pflugers Arch. 445, 529-533. [DOI] [PubMed] [Google Scholar]
- 6.Palacin, M. & Kanai, Y. (2004) Pflugers Arch. 447, 490-494. [DOI] [PubMed] [Google Scholar]
- 7.Rossier, G., Meier, C., Bauch, C., Summa, V., Sordat, B., Verrey, F. & Kuhn, L. C. (1999) J. Biol. Chem. 274, 34948-34954. [DOI] [PubMed] [Google Scholar]
- 8.Meier, C., Ristic, Z., Klauser, S. & Verrey, F. (2002) EMBO J. 21, 580-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim, D. K., Kanai, Y., Chairoungdua, A., Matsuo, H., Cha, S. H. & Endou, H. (2001) J. Biol. Chem. 276, 17221-17228. [DOI] [PubMed] [Google Scholar]
- 10.Fei, Y. J., Kanai, Y., Nussberger, S., Ganapathy, V., Leibach, F. H., Romero, M. F., Singh, S. K., Boron, W. F. & Hediger, M. A. (1994) Nature 368, 563-566. [DOI] [PubMed] [Google Scholar]
- 11.Palacin, M., Estevez, R., Bertran, J. & Zorzano, A. (1998) Physiol. Rev. 78, 969-1054. [DOI] [PubMed] [Google Scholar]
- 12.Castagna, M., Shayakul, C., Trotti, D., Sacchi, V. F., Harvey, W. R. & Hediger, M. A. (1998) Proc. Natl. Acad. Sci. USA 95, 5395-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldman, D. H., Harvey, W. R. & Stevens, B. R. (2000) J. Biol. Chem. 275, 24518-24526. [DOI] [PubMed] [Google Scholar]
- 14.Quick, M. & Stevens, B. R. (2001) J. Biol. Chem. 276, 33413-33418. [DOI] [PubMed] [Google Scholar]
- 15.Claros, M. G. & von Heijne, G. (1994) Comput. Appl. Biosci. 10, 685-686. [DOI] [PubMed] [Google Scholar]
- 16.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt, H. A., Strimmer, K., Vingron, M. & von Haeseler, A. (2002) Bioinformatics 18, 502-504. [DOI] [PubMed] [Google Scholar]
- 18.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406-425. [DOI] [PubMed] [Google Scholar]
- 19.Matz, M. V. (2003) in Generation of cDNA Libraries: Methods and Protocols, ed. Ying, S.-Y. (Humana, Totowa, NJ), pp. 103-116.
- 20.Hecker, K. H. & Roux, K. H. (1996) BioTechniques 20, 478-485. [DOI] [PubMed] [Google Scholar]
- 21.Jespersen, T., Grunnet, M., Angelo, K., Klaerke, D. A. & Olesen, S. P. (2002) BioTechniques 32, 536-538, 540. [DOI] [PubMed] [Google Scholar]
- 22.Boudko, D. Y., Moroz, L. L., Linser, P. J., Trimarchi, J. R., Smith, P. J. S. & Harvey, W. R. (2001) J. Exp. Biol. 204, 691-699. [DOI] [PubMed] [Google Scholar]
- 23.Saier, M. H. (2000) J. Bacteriol. 182, 5029-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Androutsellis-Theotokis, A., Goldberg, N. R., Ueda, K., Beppu, T., Beckman, M. L., Das, S., Javitch, J. A. & Rudnick, G. (2003) J. Biol. Chem. 278, 12703-12709. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, K., Knust, E. & Skaer, H. (1999) Dev. Biol. 212, 440-454. [DOI] [PubMed] [Google Scholar]
- 26.Seow, H. F., Broer, S., Broer, A., Bailey, C. G., Potter, S. J., Cavanaugh, J. A. & Rasko, J. E. (2004) Nat. Genet. 36, 1003-1007. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu, T., Mitsuke, H., Noto, K. & Arai, M. (2004) J. Mol. Biol. 339, 1-15. [DOI] [PubMed] [Google Scholar]