Abstract
Chronic obstructive pulmonary disease (COPD) is one of few chronic disorders with rising mortality and morbidity. It is a highly prevalent disorder, characterized by highly heterogeneous clinical symptoms, health status, and disease progression. COPD is also characterized by an inflammatory/immune response that persists despite smoking cessation and varies by the patient population, method of assessment, and timing of measurement. Bacterial colonization or infection is ubiquitous in patients with COPD and, until recently, has been predominantly assessed using culture-based methodologies. This colonization has been believed to be biologically relevant. It has been estimated that more than 70% of the bacterial species on body surfaces cannot be cultured by standard techniques. As such, advanced culture-independent techniques have been developed that target bacterial genes, such as the 16S ribosomal RNA gene, that function as molecular chronometers. Application of these techniques in patients with COPD has suggested microbial diversity that varies by age, disease severity, and medication use. All of these data provide unique and rapidly evolving insight into the potential role of the respiratory microbiome in disease genesis and expression.
Keywords: chronic obstructive pulmonary disease, microbiome, microbiota
Chronic obstructive pulmonary disease (COPD) is one of few chronic disorders in which mortality and morbidity are on the rise. It is highly prevalent and characterized by airflow obstruction that is not completely reversible (1). Therapeutic approaches have centered on bronchodilation and the use of relatively broad antiinflammatory therapy, including inhaled corticosteroids. Therapeutic approaches that modify disease course in COPD are few, with no pharmacotherapy conclusively altering disease progression. Acute exacerbations of COPD (AECOPD) account for many of the negative effects attributed to COPD (2).
COPD is a highly heterogeneous disorder with widely varying clinical symptoms, health status, and disease progression (3, 4). This heterogeneity impacts therapeutic approaches (4) and our understanding of disease pathogenesis (5). Distinct phenotypic groups include patients with predominance of cough and sputum production (6), those who experience recurrent AECOPD (7), and/or those with more rapid loss of lung function (8). Computed tomographic imaging has identified patients with worse emphysema, who experience increased dyspnea, decreased health status, and increased mortality (4). Airway structure is similarly quite heterogeneous, with increasing computed tomography abnormality associated with increased cough, breathlessness, worse health status, and more AECOPD (9). The biological nature underlying this heterogeneity remains unclear.
COPD Is an Inflammatory Disorder with both Local and Systemic Manifestations
The inflammatory response reflects a carefully orchestrated balance of innate and adaptive immune components (10). In the patient with COPD, the inflammatory/immune response persists despite smoking cessation and varies by the patient population, the method of assessment, and the timing of measurement (11). In addition, during AECOPD, inflammation increases (12). The genesis for the persistent inflammatory immune response in COPD remains controversial, with several theories advanced, including self-perpetuation of an immune response to autoantigens resulting from inflammatory and oxidative lung injury (5). Persistent or recurrent infection may play a particularly crucial role, as it may serve as a stimulator of specific immune responses or as a polyclonal activator (5, 13).
Multiple stimuli can increase airway and parenchymal inflammation (10), leading to increased bronchial tone, bronchial wall edema, and mucous hypersecretion (11). These concepts are best developed during the acutely increased inflammatory response characteristic of AECOPD. Numerous investigators have used various techniques to characterize the inflammatory response, including sputum analyses, bronchoalveolar lavage (BAL), bronchial biopsy, and blood markers. Several reports have confirmed an augmented inflammatory response in sputum, particularly during an AECOPD (14–19); neutrophilic and eosinophilic inflammation have been described. In addition, a multitude of inflammatory mediators have been implicated (14–17, 19, 20). Papi and colleagues examined sputum inflammatory markers and microbiology in 64 patients hospitalized during an AECOPD (21). The majority of infections were associated with sputum pathogens (bacterial in 29.7%, viral in 23.4%, and combined in 25%). Sputum neutrophils were noted in all exacerbations, whereas sputum eosinophils were increased in viral-associated exacerbations. Bathoorn and colleagues followed 114 patients with COPD, with 45 experiencing an AECOPD; sputum samples were obtained at the stable state and during the acute episode (19). Sputum total cells, neutrophils, eosinophils, and lymphocytes increased during the AECOPD compared with the stable state. The most recent data are those of Bafadhel and colleagues, who followed 145 patients with COPD for 1 year, examining sputum and blood samples in a stable state and during 182 exacerbations in 86 of the patients (22); 59% were associated with bacteria, 29% with virus, and 28% were associated with sputum eosinophilia. AECOPD associated with bacteria exhibited increased sputum and peripheral neutrophils, with sputum IL-1β being the most predictive biomarker.
Bronchoscopic data are limited, with one group confirming increased expression of regulated upon activation, normal T-cell expressed and secreted (RANTES) in both the surface epithelium and subepithelial lymphomononuclear cells during an AECOPD (23). Subsequently, increased numbers of neutrophils and expression of CXCL5, IL-8, and CXCR2 were seen with an AECOPD (24). Drost and colleagues confirmed elevation of bronchoalveolar lavage IL-8 levels from 14 subjects with COPD during an AECOPD (25). These data confirm an inflammatory (neutrophilic in most, eosinophilic in some) process during an AECOPD.
The systemic inflammatory nature of COPD has become better defined. The ECLIPSE investigators have defined significant longitudinal variability in a series of blood-based biomarkers in patients with well-defined COPD; this may reflect biological variability or assay characteristics (26). A separate analysis of ECLIPSE examined a panel of six peripheral blood inflammatory markers; at baseline, 30% of subjects with COPD did not exhibit evidence of systemic inflammation, whereas persistent inflammation was noted in 16% (27). Importantly, those subjects with persistent inflammation had higher all-cause mortality and exacerbation frequency per year. The addition of inflammatory markers to clinical features improved mortality prediction in this cohort (28). The clinical implication of this systemic inflammatory response remains unclear (29).
Traditional Methods to Establish a Causative Role for Infection in COPD
Various infectious and noninfectious stimuli can provoke the inflammatory response seen in stable and exacerbated COPD. The methods used to establish the presence of colonizing bacteria or an etiologic role for bacteria have been best established for AECOPD and have included sputum culture, bronchoscopic sampling, molecular epidemiologic studies of bacterial pathogens, identifying an immune response to potential pathogens, and recording a response to antimicrobial therapy.
Sputum cultures have been the classic methodological approach to identifying potentially pathogenic organisms (PPM). Potentially pathogenic bacteria have been identified in 25 to 50% of patients with stable COPD (30). Bafadhel and colleagues (22) noted that colonization (Haemophilus influenzae, Moraxella catarrhalis, Streptococcus pneumoniae, Staphylococcus aureus, or Pseudomonas aeruginosa) was identified using standard culture techniques in 28% of patients with COPD at baseline; this number increased to 86% using quantitative polymerase chain reaction (qPCR). Although such data have been used to support the concept of colonization, this has been challenged, as the presence of bacteria has been associated with negative effects suggesting a host response to these organisms. The presence of bacterial pathogens in the sputum during the stable phase (“bacterial colonization”) has been associated with a greater AECOPD frequency (31), poorer health status (32), and a greater decline in lung function (33). Sputum (32, 34) and systemic (35) inflammatory markers are elevated in patients with COPD whose sputum is colonized with bacterial pathogens. This has been reported to occur even in subjects with milder airflow obstruction (36). Some investigators have correlated sputum bacterial load with an enhanced inflammatory response (31, 37). The limitations of sputum culture include methodological issues regarding sputum processing, culture techniques, contamination of the upper airway, and colonization.
Bronchoscopic studies have confirmed that PPM are identified in many patients with stable COPD and during AECOPDs (38–41). Two bronchoscopic studies have related the identification of PPM in bronchoalveolar lavage with increased markers of predominantly neutrophilic inflammation (36, 42). One group reviewed pooled data from six published studies that used a standardized protected brush bronchoscopic technique (43). Using a bacterial load threshold greater than 102 cfu, 29% of stable patients and 54% of patients with COPD experiencing an exacerbation exhibited bacterial colonization. A separate group identified intracellular H. influenzae in 87% of bronchial biopsy samples from acutely ill patients with chronic bronchitis compared with 33% of stable patients and 0% of healthy control subjects (44). The totality of these data support the role of bacteria in stable COPD and during an AECOPD with a vicious circle of innate lung disease failure, low-grade infection/colonization, and recurrent acute infection driving ongoing inflammation (30).
Studies examining genetic diversity among individual strains associated with AECOPD have provided additional insight. Sethi and colleagues demonstrated that acquisition of a bacterial strain with which the patient had not been previously infected was associated with a greater than twofold increase in the risk of an exacerbation (45). Importantly, in the majority of cases, new nontypeable H. influenzae strains were not associated with a symptomatic AECOPD. Subsequent work demonstrated inherent differences in the ability to provoke neutrophilic recruitment for new H. influenzae strains associated with symptomatic exacerbations compared with those not associated with such a clinical response (46). Similar work established the role of antigenic drift among M. catarrhalis isolates (47–51). The totality of these data provides compelling support for bacterial infection in a significant proportion of AECOPDs and strongly suggests a role for chronic bacterial colonization/low-grade infection in the genesis of COPD.
Novel Methods to Assess Lung Microbiology
Although it has been a long-held belief that the normal human lung is sterile, increasing evidence suggests that there is a distinct lung microbiome (the entire community of microorganisms that inhabit the lower respiratory tract) (52). This may be of particular relevance in obstructive lung diseases (53). It has been estimated that more than 70% of the bacterial species inhabiting bodily surfaces cannot be cultured by currently available techniques (54, 55), and that many of the remaining 20 to 30% of species are very difficult to culture (56). The ability to identify microorganisms in a complex community based on their unique genetic signatures has revolutionized our understanding of the indigenous microbiota that inhabits virtually every bodily organ (57). In particular, techniques that identify bacteria on the basis of ribosomal RNA (rRNA) sequences have provided insight into the depth and breadth of microbiota present in the normal and diseased lung.
The approaches used to study the microbial world originated more than 30 years ago with the work of Sogin and colleagues (58) and Carl Woese and colleagues (59, 60), who used rRNA sequence comparisons to reconstruct the evolutionary history of microbes. Subsequently, Pace and colleagues built molecular tools for assessing microorganism ecology (61, 62) that have been used to identify more than 100 novel bacterial phyla, most of which include currently unculturable organisms (53, 63). The majority of culture-independent techniques rely on PCR amplification of the 16S rRNA gene, a highly conserved locus of the bacterial genome (64, 65). The 16S rRNA gene possesses nine hypervariable regions that facilitate genus and species identification (66); amplification of conserved sequence stretches within the 16S rRNA gene, flanking these hypervariable regions, allows for the design of broad bacterial kingdom-specific primers to amplify bacterial sequences for subsequent analyses (53). Current molecular techniques can define entire microbial community fingerprints, locate and quantify bacteria geographically within the mucosa, quantify and estimate relative bacterial abundance, and identify the members of the community (53). In addition, by applying high-throughput shotgun sequencing it is possible to consider the microbial population as a whole “metagenome.” This approach, metagenomics, can provide additional insight by identifying the community members present and their genetically encoded functional capacity.
Up until very recently, culture-independent techniques either suffered from being low-throughput or relatively nonspecific. The most commonly used molecular technique to measure bacterial abundance is real-time qPCR of the 16S rRNA gene, which can provide kingdom-, genus-, and even species-level specificity (53). Data can be expressed and normalized as total bacterial gene copy number per unit sample or per single copy host gene, as a relative proxy of bacterial “abundance.” However, to identify the bacterial members within a mixed bacterial community, this method is very costly and low-throughput. Fluorescent in situ hybridization uses fluorescent oligonucleotide probes that bind to specific target sequences within the bacterial cell, allowing an assessment of the spatial distribution and number of organisms using a fluorescent microscope. As such, fluorescent in situ hybridization can visualize and quantify the abundance of bacteria in situ in their particular microenvironments. These techniques have been adapted for flow cytometry, permitting identification of individual bacteria (67). Molecular fingerprinting methods, such as denaturing- and temperature-gradient gel electrophoresis and terminal restriction fragment length polymorphisms, have for years been viewed as quick and simple techniques for determining shifts in community structure and composition (67); however, these methods are severely limited when the intention is to identify individual organisms. The construction of libraries of 16S rRNA gene amplicons from mixed bacterial populations is an excellent tool to identify community membership, because one amplifies the entire 16S rRNA gene for sequencing (using traditional Sanger sequencing) (53). Sequences of the clones can be compared using either algorithms that are classifier-based (giving the sequence a traditional taxonomic identity) or operational taxonomic unit–based (where sequences are clustered according to relative similarity to other sequences). The construction and analysis of these “clone libraries” to study a bacterial community sample has for years been the “gold standard,” but it suffers from the limitations of being costly and low-throughput—which often raised questions of sampling bias and inadequate sampling depth (53).
High-throughput sequencing provided a way to get beyond the limitations of previous techniques. Now a library of 16S rRNA gene amplicons could be sequenced at more than 10,000 times the depth of a traditional clone library, thus providing robust microbial community sampling (68–73). The addition of DNA “barcodes” during library construction means that many samples can be run simultaneously, thus reducing the cost of sequencing per sample (74). The 454-pyrosequencing platform has been the workhorse of most studies to date because it offers read lengths of more than 400 bp and output of greater than 106 high-quality sequence reads (74–76). As sequencing technology advances, other high-throughput sequencing platforms, such as the Illumina (Illumina Corporation, San Diego, CA) and PacBio RS (Pacific Biosciences, Menlo Park, CA) systems, are being adopted for microbial ecology studies (53), with the potential for even greater read depth (77) or discriminatory capacity. An alternate approach, which is particularly useful for identification of less abundant bacterial community members, is the 16S rRNA PhyloChip (designed at Lawrence Berkeley National Laboratory, manufactured by Affymetrix Corporation, Santa Clara, CA) (78, 79).
There are both strengths and limitations to the available techniques. Those based on amplified 16S rRNA genes detect only bacteria, whereas shotgun sequencing approaches can detect bacteria, fungi, and viruses. Some of the approaches are more useful for quantification; however, the frequent presence of multiple copies of 16S rRNA genes in different bacterial species and the potential of DNA-based methodologies to detect nonviable organisms make absolute quantification difficult. Methods involving the analysis of bacterial mRNA expression can circumvent the issue of viability, because this can only originate from viable organisms, but these methods are more costly, can be more technically difficult, and the advantage is not clear, because DNA from dead bacteria can still have a profound effect on immune responses. A final consideration of all these methods is the requirement for extensive computational analysis and method-specific analysis training.
The Lung Microbiome in Health, Smoke Exposure, and COPD
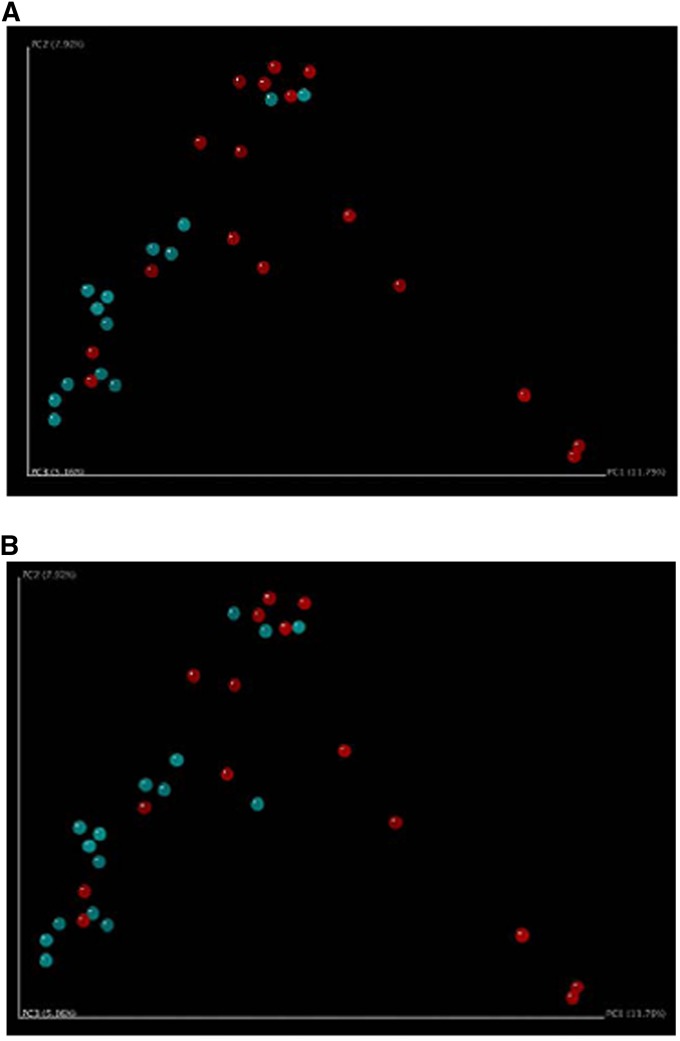
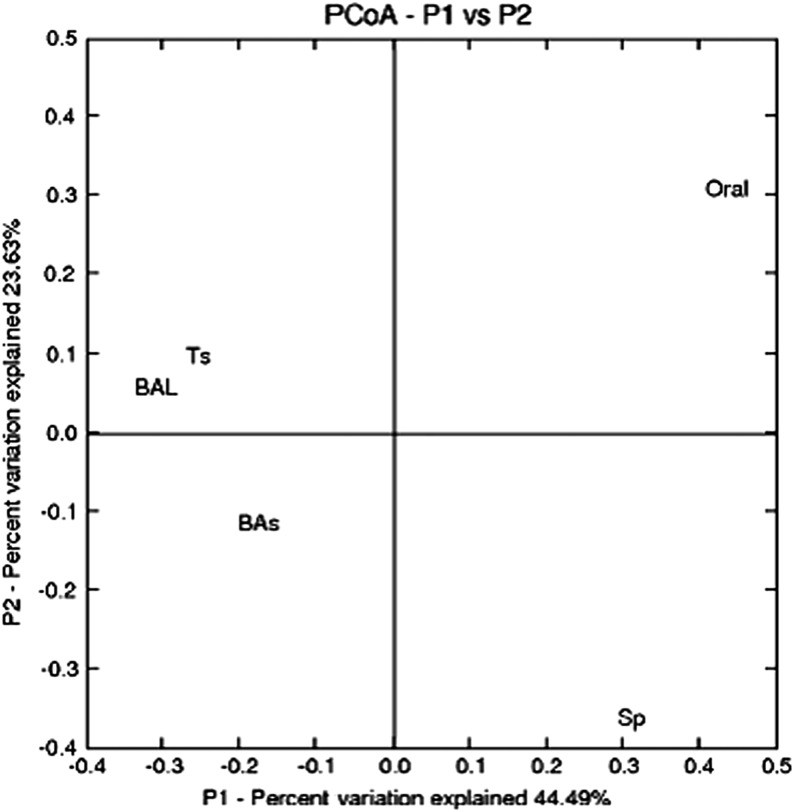
Until recently, modern molecular techniques had been infrequently applied to characterize the oral and/or lung microbiota in healthy nonsmokers, smokers without COPD, or patients with COPD. Some investigative groups have suggested that smoking alters the composition of the oral microbial flora (52, 80), whereas others have found less impressive differences (81). The effect of smoking status on the lung microbial species has been less consistent. Bronchoscopic studies have documented intriguing findings from the lungs of patients with COPD, although the studies have been limited by small numbers of subjects and lack of longitudinal sampling (Table 2). In early work, Hilty and colleagues collected nasal, oral, and bronchoscopic brushings from the left upper lobe in 10 patients with asthma, 8 healthy control subjects, and 5 patients with COPD (82). In the five subjects with COPD and eight control subjects, airway microbiota was similar (82). Members of the phylum Proteobacteria were more likely to be seen, and Bacteroidetes members were less likely to be seen in the patients with COPD compared with control subjects. In eight patients with COPD undergoing mechanical ventilation during an AECOPD, a diverse microbial community was identified, despite recent or ongoing broad-spectrum antibiotic therapy (83). More than 1,200 bacterial taxa were noted, including bacterial families typical of AECOPD (H. influenzae, P. aeruginosa, S. pneumoniae, M. catarrhalis) and a multitude of other families with potentially pathogenic members. A core microbiota consisting of 75 taxa present in all patients was noted. Erb-Downward and colleagues collected BAL from seven smokers with normal lung function, four with COPD, and three healthy nonsmokers (73). Significant levels of bacterial 16S gene signal were detected in all subjects, with no significant differences between the three study groups in the number of bacterial copies. The microbiome of the patients with moderate COPD was notable for decreased bacterial community diversity, although there was extensive overlap in membership between the bacterial communities of the groups. These investigators also suggested that there may be a “core” pulmonary microbiome. Candidate genera that were found in greater than 75% of the subjects included Pseudomonas, Streptococcus, Prevotella, and Fusobacteria. Haemophilus, Veillonella, and Porphyromonas were also identified in more than half of the samples. Pragman and colleagues examined banked, frozen BAL from 22 participants in the NHLBI–sponsored Feasibility of Retionids for the Treatment of Emphysema (FORTE) study and obtained BAL from 10 healthy control subjects (6 nonsmokers) (84). The larger sample size and heterogeneity of the cohort allowed additional inferences to be made. Overall, the investigators believed that microaspiration of the oral flora was the source of the lung microbiome. Interestingly, COPD was associated with increasing microbial diversity, although the major contributor was the increased age of the subjects. No major difference was noted between moderate and severe COPD, although principal coordinate analyses demonstrated clustering by the use of inhaled corticosteroids (Figure 1A) and bronchodilators (Figure 1B). These data suggest the importance of accounting for subject age and concomitant medication use in the interpretation of future COPD studies. A more recent elegant investigation in healthy individuals with variable smoking history suggested that microaspiration of supraglottic organisms was the source of lower airway microbial flora; a subgroup of individuals exhibited a relative abundance of Veillonella and Prevotella, which was associated with increased inflammatory markers (81). Cabrera-Rubio and colleagues sampled the respiratory system via multiple mechanisms (Table 1) (85). High diversity was noted, with Streptococcus, Prevotella, Moraxella, Haemophilus, Acinetobacter, Fusobacterium, and Neisseria being the most common genera identified. Sputum samples showed significantly lower diversity than the other sample types, whereas the lower bronchial samples (BAL and bronchial biopsies) showed similar bacterial composition (Figure 2). These data provide insight regarding the possible approaches to lung sampling and their limitations in future COPD studies.
Table 2.
Summary of available studies examining the microbial flora of patients with chronic obstructive pulmonary disease using molecular techniques
| Citation | Cohort Size | Age (yr) | FEV1 % Predicted | Lung Sampling Technique | Molecular Methods |
|---|---|---|---|---|---|
| Hilty et al (82) | Asthma (n =10) | 37.6 | 81.0 | Bronchoscopic brushes from LUL in all, RUL also in some | 454 |
| COPD (n = 5) | 57.0 | 51.2 | |||
| Healthy control subjects (n = 8) | 52.9 | 100.8 | |||
| Huang et al (83) | COPD (n = 8) | 74.1 | NA (all intubated for AECOPD) | Banked endotracheal aspirates | PhyloChip |
| Erb-Downward et al (73) | Healthy smokers (n = 7) | 50.7 | 103.1 | BAL | 454 |
| Healthy nonsmokers (n = 3) | 61.3 | 110.0 | BAL | ||
| COPD (n = 4) | 54.0 | 65.3 | BAL | ||
| COPD (n = 6) | 61.2 | 16.0 | Endobronchial tissue | ||
| Pragman et al (84) | Healthy control subjects (n = 10) | 39.5 | NA | BAL | 454 |
| Moderate COPD (n = 14) | 69.7 | Frozen BAL | |||
| Severe COPD (n = 8) | 62.6 | Frozen BAL | |||
| Cabrera-Rubio et al (85) | COPD (n = 6) | NA | 60.5 | Sputum | 454 |
| BAL | |||||
| Bronchial biopsy | |||||
| Bronchial aspirate | |||||
| Sze et al (86) | Healthy nonsmokers (n = 8) | 56.3 | 88.8 | Distal parenchymal tissue | 454 |
| Healthy smokers (n = 8) | 56.9 | 94.3 | |||
| COPD (n = 8) | 58.8 | 15.4 | |||
| Cystic fibrosis (n = 8) | 32.6 | 35.0 |
Definition of abbreviations: 454 = 454 pyrosequencing platform; AECOPD = acute exacerbation of COPD; BAL = bronchoalveolar lavage; COPD = chronic obstructive pulmonary disease; LUL = left upper lobe; NA = not available; RUL = right upper lobe.
Figure 1.
Principal coordinate analysis of subjects with chronic obstructive pulmonary disease clustered by (A) inhaled corticosteroid use (blue users, red nonusers), and (B) bronchodilator use (blue users, red nonusers). Reprinted by permission from Reference 84.
Table 1.
Comparison of molecular methods to assess the lung microbiota
| Method | Advantages | Disadvantages |
|---|---|---|
| DGGE or TGGE | Bands can be isolated for taxonomic identification | Technically difficult; results are variable; highly operator dependent |
| T-RFLP | High-throughput, inexpensive method for pattern identification | Taxonomy should not be inferred without extensive machine standardization |
| Clone libraries | Full-length 16S rRNA gene; most accurate taxonomy for 16S rRNA-based methods | Costly; labor intensive; very low throughput; highly operator dependent |
| 454-Pyrosequencing | Long reads span multiple variable regions; high-throughput; very good taxonomic classification; inexpensive with DNA barcoding | Expensive unless samples are barcoded; requires extensive post-run analysis |
| MiSeq | Long enough read length for good taxonomic classification; dual barcoding for extremely high throughput; very inexpensive per run cost; excellent sequencing depth | Sequencing of amplicons uses nonstandard protocol; extensive post-run analysis |
Definition of abbreviations: DGGE = denaturing-gradient gel electrophoresis; TGGE = temperature-gradient gel electrophoresis; T-RFLP = terminal restriction fragment length polymorphisms.
Figure 2.
Principal component analysis (PCoA) of four respiratory tract sample types (n = 6 for each sample type) and samples from the oral cavity of healthy individuals (n = 16) according to the microbial composition, as inferred by pyrosequencing of the 16S rRNA gene. BAL = bronchoalveolar lavage; Bas = bronchial aspirate; Oral = supragingival dental plaques; Sp = sputum; Ts = tissue. Reprinted by permission from Reference 85.
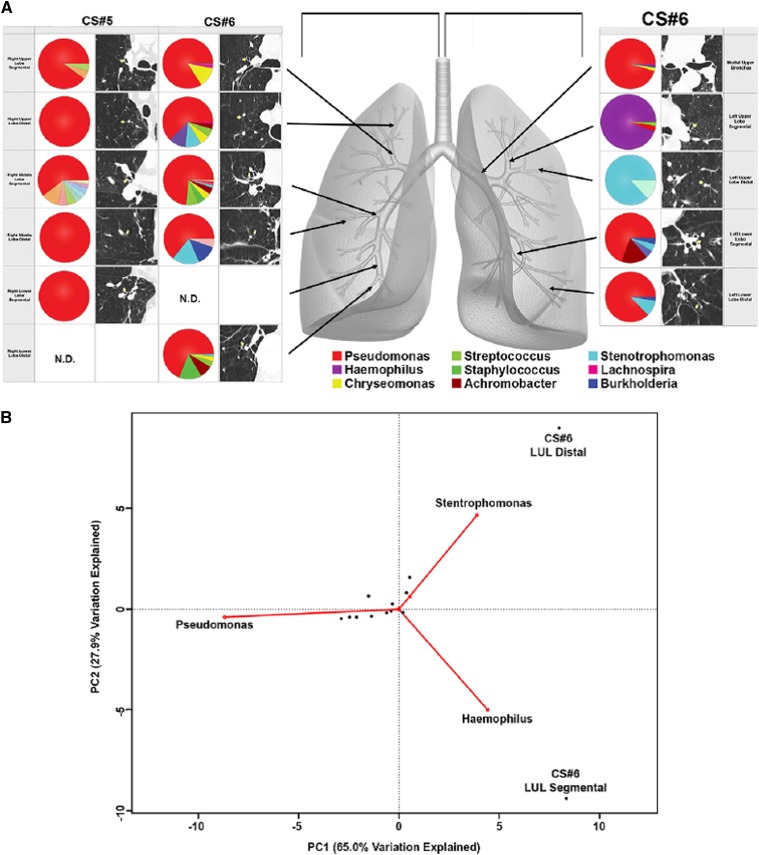
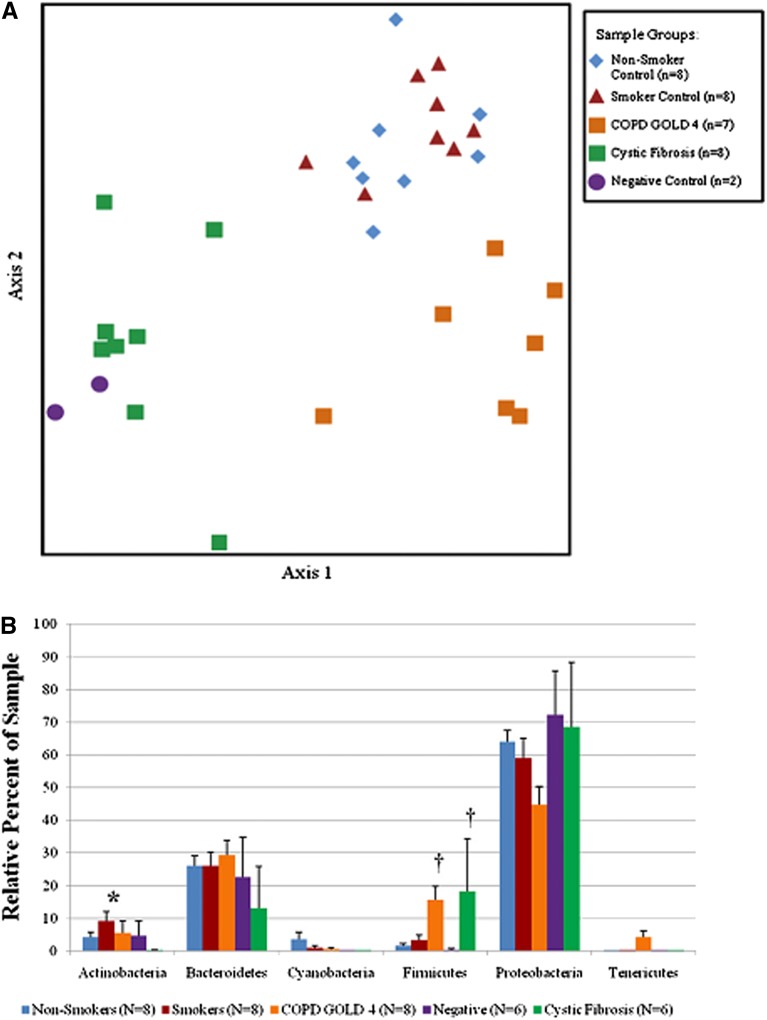
Some studies have provided additional data using tissue sampling. Cabrera-Rubio and colleagues noted similar bacterial composition between endobronchial biopsies and BAL (Figure 2) (85). Erb-Downward and colleagues sampled explants removed at lung transplantation in six patients with severe COPD (Table 1); significant differences in bacterial community composition within the same lung were seen (Figure 3). Ordination by principal component analysis demonstrated that the microanatomic variation in the samples was driven by either the dominance of Pseudomonas, Haemophilus, or Stenotrophomonas at the sampled site. Significant numeric differences in the number of operational taxonomic units were noted in tissue sites within the same lung and even within the same lobe. As such, marked microanatomic differences can exist in bacterial communities even within the same lung of subjects with advanced COPD. These findings may have important biological impact regarding disease pathogenesis. Sze and colleagues examined distal lung parenchymal tissue from a group of nonsmokers, smokers without COPD, patients with severe COPD, and patients with cystic fibrosis (Table 1) (86). Total bacterial populations were small, but greater in diseased subjects than in control subjects. Three distinct bacterial communities were identified, including one common to the patients with severe COPD (Figure 4A); a significantly greater abundance of the phylum Firmicutes was noted and mainly driven by the Lactobacillus genus (Figure 4B).
Figure 3.
(A) Bacterial communities present in individual lung airways. Multiple samples were taken from lung explants (right lung, subject CS#5; both lungs, subject CS#6) at the time of elective transplantation. Samples were harvested from the regions of lung indicated by the arrows on the gray lung schematic. Pie diagrams depict the genus level classification of 16S sequences, and the computed tomography images demonstrate the absence of bronchiectasis in the airways adjacent to where samples were obtained. The key for the nine most abundant organisms is provided below the lung schematic. (B) Cluster analysis of the bacterial communities sampled from sites in the left upper lobe (LUL). Biplot of the principle components analysis of the normalized bacterial communities from multiple anatomic sites in the lung explants. Reprinted by permission from Reference 73.
Figure 4.
(A) Nonmetric multidimensional scaling analysis of the terminal restriction fragment length polymorphism profiles providing adequate number of amplicons. Three distinct groups can be identified, including one with the patients with severe chronic obstructive pulmonary disease (COPD). (B) Comparison among the nonsmoker control subjects, the smokers without COPD, the patients with severe COPD (Global Initiative for Chronic Obstructive Lung Disease [GOLD] 4), the patients with cystic fibrosis, and the negative control subjects. Histograms reflect the average + SEM. *P < 0.007 versus all other groups; †P < 0.003 versus nonsmoking, smoking, and negative control groups. Reprinted by permission from Reference 86.
Future Directions
A recent report from a National Institutes of Health workshop provides insight and an investigative direction regarding the application of novel genomic techniques to define and characterize the lung microbiome in smokers with and without COPD (87). It is apparent that despite the relatively young nature of this field, there is great potential for these approaches to provide crucial insight into disease pathogenesis and disease heterogeneity and yield novel therapeutic avenues. Further investigative considerations include:
-
1.Methodological
-
A.Work needs to be done to identify the contribution of nonbacterial microbes (including viruses and fungi) in patients with COPD.
-
B.Work is required to refine the complex statistical approaches required to analyze the large amount of data generated from high-throughput sequencing methods.
-
C.Development of biomarkers, including in vivo imaging, that address lung microbial burden and community structure will be required to obviate difficulties with sampling and to increase clinical relevance.
-
A.
-
2.Clinical
-
A.Detailed studies characterizing the relationship between the microbiome of the upper and lower airways in patients with highly characterized COPD, smokers without COPD, and healthy subjects are required.
-
B.An appreciation of the relationship between the altered microbiome of the COPD lung and the heterogeneous clinical manifestations and ultrastructural abnormalities seen in patients with COPD should provide crucial biological insight.
-
C.Longitudinal assessment of the respiratory microbiome will provide insight regarding the heterogenous clinical course typical of the patient with COPD.
-
D.As COPD is characterized by episodic exacerbations, research is required to examine how bacterial communities change around the time of an AECOPD and whether changes in the bacterial community structure might predict exacerbation, possibly allowing early or preemptive intervention.
-
E.Will manipulation of the lung microbiome allow improvement in disease activity and/or alter disease progression?
-
A.
-
3.Translational
-
A.A better understanding of the relationship between enhanced culture-dependent and molecular microbiological techniques is required. This approach has provided intriguing data in the cystic fibrosis community, where expanded use of culture-enriched molecular profiling has identified the majority of bacteria identified via culture-independent methods (56).
-
B.The use of metagenomics should provide insight into gene function; metatranscriptomics provides collective gene expression profiles, including those of the microbes and the host; and metabolomics provides additional functional information (87).
-
C.Defining the host response to alterations in the respiratory microbiome should provide insight regarding the local and systemic inflammatory responses seen in the patient with COPD. Such an approach has provided intriguing insight in the gastrointestinal tract (53) and has already proven fruitful in identifying divergent proinflammatory responses of respiratory dendritic cells to airway microbes (88).
-
D.Addressing the location of organisms within the respiratory tract and their relation to the normal and altered mucus should provide insight as to the local effects of altered microbial communities.
-
E.Research focusing on the gut–lung axis may provide unique information regarding the impact of gut microbial populations on the development and nature of respiratory disease.
-
F.Animal models should be used effectively to provide additional mechanistic understanding of respiratory microbiome and host response interactions.
-
A.
Footnotes
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Vestbo J, Hurd SS, Agusti AG, Jones PW, Vogelmeier C, Anzueto A, Barnes PJ, Fabbri LM, Martinez FJ, Nishimura M, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2013;187:347–365. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- 2.Anzueto A. Impact of exacerbations on COPD. Eur Respir Rev. 2010;19:113–118. doi: 10.1183/09059180.00002610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agusti A, Sobradillo P, Celli B. Addressing the complexity of chronic obstructive pulmonary disease: from phenotypes and biomarkers to scale-free networks, systems biology, and P4 medicine. Am J Respir Crit Care Med. 2011;183:1129–1137. doi: 10.1164/rccm.201009-1414PP. [DOI] [PubMed] [Google Scholar]
- 4.Han M, Agusti A, Calverley P, Celli B, Criner G, Curtis J, Fabbri L, Goldin J, Jones P, Macnee W, et al. Chronic obstructive pulmonary disease phenotypes: the future of COPD. Am J Respir Crit Care Med. 2010;182:598–604. doi: 10.1164/rccm.200912-1843CC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis J, Freeman C, Hogg J. The immunopathogenesis of chronic obstructive pulmonary disease. Insights from recent research. Proc Am Thorac Soc. 2007;4:512–521. doi: 10.1513/pats.200701-002FM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim V, Criner GJ. Chronic bronchitis and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;187:228–237. doi: 10.1164/rccm.201210-1843CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurst JR, Vestbo J, Anzueto A, Locantore N, Mullerova H, Tal-Singer R, Miller B, Lomas DA, Agusti A, Macnee W, et al. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med. 2010;363:1128–1138. doi: 10.1056/NEJMoa0909883. [DOI] [PubMed] [Google Scholar]
- 8.Vestbo J, Edwards LD, Scanlon PD, Yates JC, Agusti A, Bakke P, Calverley PM, Celli B, Coxson HO, Crim C, et al. Changes in forced expiratory volume in 1 second over time in COPD. N Engl J Med. 2011;365:1184–1192. doi: 10.1056/NEJMoa1105482. [DOI] [PubMed] [Google Scholar]
- 9.Han MK, Kazerooni EA, Lynch DA, Liu LX, Murray S, Curtis JL, Criner GJ, Kim V, Bowler RP, Hanania NA, et al. Chronic obstructive pulmonary disease exacerbations in the COPDGene study: associated radiologic phenotypes. Radiology. 2011;261:274–282. doi: 10.1148/radiol.11110173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brusselle GG, Joos GF, Bracke KR. New insights into the immunology of chronic obstructive pulmonary disease. Lancet. 2011;378:1015–1026. doi: 10.1016/S0140-6736(11)60988-4. [DOI] [PubMed] [Google Scholar]
- 11.Macario C, Tajes J, Lanus E. Epoc: Inflamacion bronquial y sistemica. Arch Bronconeumol. 2010;46:9–15. doi: 10.1016/S0300-2896(10)70027-2. [DOI] [PubMed] [Google Scholar]
- 12.Koutsokera A, Stolz D, Loukides S, Kostikas K. Systemic biomarkers in exacerbations of COPD: the evolving clinical challenge. Chest. 2012;141:396–405. doi: 10.1378/chest.11-0495. [DOI] [PubMed] [Google Scholar]
- 13.Martinez F, Han M, Flaherty K, Curtis J. Role of infection and antimicrobial therapy in acute exacerbations of chronic obstructive pulmonary disease. Expert Rev Anti Infect Ther. 2006;4:101–124. doi: 10.1586/14787210.4.1.101. [DOI] [PubMed] [Google Scholar]
- 14.Murphy T, Sethi S, Niederman M. The role of bacteria in exacerbations of COPD. A constructive view. Chest. 2000;118:204–209. doi: 10.1378/chest.118.1.204. [DOI] [PubMed] [Google Scholar]
- 15.Gompertz S, O’Brien C, Bayley D, Hill S, Stockley R. Changes in bronchial inflammation during acute exacerbations of chronic bronchitis. Eur Respir J. 2001;17:1112–1119. doi: 10.1183/09031936.01.99114901. [DOI] [PubMed] [Google Scholar]
- 16.White A, Gompertz S, Bayley D, Hill SL, O'Brien C, Unsal I, Stockley RA. Resolution of bronchial inflammation is related to bacterial eradication following treatment of exacerbations of chronic bronchitis. Thorax. 2003;58:680–685. doi: 10.1136/thorax.58.8.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill AT, Bayley D, Stockley RA. The interrelationship of sputum inflammatory markers in patients with chronic bronchitis. Am J Respir Crit Care Med. 1999;160:893–898. doi: 10.1164/ajrccm.160.3.9901091. [DOI] [PubMed] [Google Scholar]
- 18.Aaron S, Angel J, Lunau M, Wright K, Fex C, Le Saux N, Dales R. Granulocyte inflammatory markers and airway infection during acute exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;163:349–355. doi: 10.1164/ajrccm.163.2.2003122. [DOI] [PubMed] [Google Scholar]
- 19.Bathoorn E, Liesker JJ, Postma DS, Koeter GH, van der Toorn M, van der Heide S, Ross HA, van Oosterhout AJ, Kerstjens HA. Change in inflammation in out-patient COPD patients from stable phase to a subsequent exacerbation. Int J Chron Obstruct Pulmon Dis. 2009;4:101–109. doi: 10.2147/copd.s4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roland M, Bhowmik A, Sapsford RJ, Seemungal TA, Jeffries DJ, Warner TD, Wedzicha JA. Sputum and plasma endothelin-1 levels in exacerbations of chronic obstructive pulmonary disease. Thorax. 2001;56:30–35. doi: 10.1136/thorax.56.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papi A, Bellettato CM, Braccioni F, Romagnoli M, Casolari P, Caramori G, Fabbri LM, Johnston SL. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am J Respir Crit Care Med. 2006;173:1114–1121. doi: 10.1164/rccm.200506-859OC. [DOI] [PubMed] [Google Scholar]
- 22.Bafadhel M, McKenna S, Terry S, Mistry V, Reid C, Haldar P, McCormick M, Haldar K, Kebadze T, Duvoix A, et al. Acute exacerbations of chronic obstructive pulmonary disease: identification of biologic clusters and their biomarkers. Am J Respir Crit Care Med. 2011;184:662–671. doi: 10.1164/rccm.201104-0597OC. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, Qiu YS, Majumdar S, Gamble E, Matin D, Turato G, Fabbri LM, Barnes N, Saetta M, Jeffery PK. Exacerbations of bronchitis: bronchial eosinophilia and gene expression for interleukin-4, interleukin-5, and eosinophil chemoattractants. Am J Respir Crit Care Med. 2001;164:109–116. doi: 10.1164/ajrccm.164.1.2007050. [DOI] [PubMed] [Google Scholar]
- 24.Qiu Y, Zhu J, Bandi V, Atmar RL, Hattotuwa K, Guntupalli KK, Jeffery PK. Biopsy neutrophilia, neutrophil chemokine and receptor gene expression in severe exacerbations of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2003;168:968–975. doi: 10.1164/rccm.200208-794OC. [DOI] [PubMed] [Google Scholar]
- 25.Drost EM, Skwarski KM, Sauleda J, Soler N, Roca J, Agusti A, MacNee W. Oxidative stress and airway inflammation in severe exacerbations of COPD. Thorax. 2005;60:293–300. doi: 10.1136/thx.2004.027946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickens J, Miller B, Edwards L, Silverman E, Lomas D, Tal-Singer R for the Evaluation of COPD Longitudinally to Identify Surrogate Endpoints (ECLIPSE) study investigators. COPD association and repeatibility of blood biomarkers in the eclipse cohort. Respir Res. 2011;12:146. [Google Scholar]
- 27.Agusti A, Edwards LD, Rennard SI, MacNee W, Tal-Singer R, Miller BE, Vestbo J, Lomas DA, Calverley PM, Wouters E, et al. Persistent systemic inflammation is associated with poor clinical outcomes in COPD: a novel phenotype. PLoS ONE. 2012;7:e37483. doi: 10.1371/journal.pone.0037483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Celli BR, Locantore N, Yates J, Tal-Singer R, Miller BE, Bakke P, Calverley P, Coxson H, Crim C, Edwards LD, et al. Inflammatory biomarkers improve clinical prediction of mortality in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;185:1065–1072. doi: 10.1164/rccm.201110-1792OC. [DOI] [PubMed] [Google Scholar]
- 29.Wouters EF, Reynaert NL, Dentener MA, Vernooy JH. Systemic and local inflammation in asthma and chronic obstructive pulmonary disease: is there a connection? Proc Am Thorac Soc. 2009;6:638–647. doi: 10.1513/pats.200907-073DP. [DOI] [PubMed] [Google Scholar]
- 30.Sethi S, Murphy T. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med. 2008;359:2355–2365. doi: 10.1056/NEJMra0800353. [DOI] [PubMed] [Google Scholar]
- 31.Patel I, Seemungal T, Wilks M, Lloyd-Owen S, Donaldson G, Wedzicha J. Relationship between bacterial colonisation and the frequency, character, and severity of COPD exacerbations. Thorax. 2002;57:759–764. doi: 10.1136/thorax.57.9.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banerjee D, Khair OA, Honeybourne D. Impact of sputum bacteria on airway inflammation and health status in clinical stable COPD. Eur Respir J. 2004;23:685–691. doi: 10.1183/09031936.04.00056804. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson T, Patel I, Wilks M, Donaldson G, Wedzicha J. Airway bacterial load and FEV1 decline in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2003;167:1090–1095. doi: 10.1164/rccm.200210-1179OC. [DOI] [PubMed] [Google Scholar]
- 34.Bresser P, Out TA, van Alphen L, Jansen HM, Lutter R. Airway inflammation in nonobstructive and obstructive chronic bronchitis with chronic Haemophilus influenzae airway infection. Comparison with noninfected patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2000;162:947–952. doi: 10.1164/ajrccm.162.3.9908103. [DOI] [PubMed] [Google Scholar]
- 35.Marin A, Garcia-Aymerich J, Sauleda J, Belda J, Millares L, Garcia-Nunez M, Serra I, Benet M, Agusti A, Anto JM, et al. on behalf of the PAC-COPD Study Group. Effect of bronchial colonisation on airway and systemic inflammation in stable COPD COPD(In press) [DOI] [PubMed] [Google Scholar]
- 36.Soler N, Ewig S, Torres A, Filella X, Gonzalez J, Zaubet A. Airway inflammation and bronchial microbial patterns in patients with stable chronic obstructive pulmonary disease. Eur Respir J. 1999;14:1015–1022. doi: 10.1183/09031936.99.14510159. [DOI] [PubMed] [Google Scholar]
- 37.Hill AT, Campbell EJ, Hill SL, Bayley DL, Stockley RA. Association between airway bacterial load and markers of airway inflammation in patients with stable chronic bronchitis. Am J Med. 2000;109:288–295. doi: 10.1016/s0002-9343(00)00507-6. [DOI] [PubMed] [Google Scholar]
- 38.Monso E, Ruiz J, Rosell A, Manterola J, Fiz J, Morera J, Ausina V. Bacterial infection in chronic obstructive pulmonary disease: a study of stable and exacerbated outpatients using the protected specimen brush. Am J Respir Crit Care Med. 1995;152:1316–1320. doi: 10.1164/ajrccm.152.4.7551388. [DOI] [PubMed] [Google Scholar]
- 39.Fagon J, Chastre J, Trouillet J, Domart Y, Dombret M, Bornet M, Gibert C. Characterization of distal bronchial microflora during acute exacerbation of chronic bronchitis. Use of the protected specimen brush technique in 54 mechanically ventilated patients. Am Rev Respir Dis. 1990;142:1004–1008. doi: 10.1164/ajrccm/142.5.1004. [DOI] [PubMed] [Google Scholar]
- 40.Soler N, Torres A, Ewig S, Gonzalez J, Celis R, El-Ebiary M, Hernandez C, Rodriguez-Roisin R. Bronchial microbial patterns in severe exacerbations of chronic obstructive pulmonary disease (COPD) requiring mechanical ventilation. Am J Respir Crit Care Med. 1998;157:1498–1505. doi: 10.1164/ajrccm.157.5.9711044. [DOI] [PubMed] [Google Scholar]
- 41.Pela R, Marchesani F, Agostinelli C, Staccioli D, Cecarini L, Basotti C, Sanguinetti C. Airways microbial flora in COPD patients in stable clinical conditions and during exacerbations: a bronchoscopic investigation. Monaldi Arch Chest Dis. 1998;53:262–267. [PubMed] [Google Scholar]
- 42.Sethi S, Maloney J, Grove L, Wrona C, Berenson CS. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173:991–998. doi: 10.1164/rccm.200509-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosell A, Monso E, Soler N, Torres F, Angrill J, Riise G, Zalacain R, Morera J, Torres A. Microbiologic determinants of exacerbations in chronic obstructive pulmonary disease. Arch Intern Med. 2005;165:891–897. doi: 10.1001/archinte.165.8.891. [DOI] [PubMed] [Google Scholar]
- 44.Bandi V, Apicella M, Mason E, Murphy T, Siddiqi A, Atmar R, Greenberg S. Nontypeable Haemophilus influenzae in the lower respiratory tract of patients with chronic bronchitis. Am J Respir Crit Care Med. 2001;164:2114–2119. doi: 10.1164/ajrccm.164.11.2104093. [DOI] [PubMed] [Google Scholar]
- 45.Sethi S, Evans N, Grant B, Murphy T. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med. 2002;347:465–471. doi: 10.1056/NEJMoa012561. [DOI] [PubMed] [Google Scholar]
- 46.Chin C, Manzel L, Lehman E, Humlicek A, Shi L, Starner T, Denning G, Murphy T, Sethi S, Look D. Haemophilus influenzae from COPD patients with exacerbation induce more inflammation than colonizers. Am J Respir Crit Care Med. 2005;172:85–91. doi: 10.1164/rccm.200412-1687OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Musher D, Kubitschek K, Crennan J, Baughn RE. Pneumonia and acute febrile tracheobronchitis due to haemophilus influenzae. Ann Intern Med. 1983;99:444–450. doi: 10.7326/0003-4819-99-4-444. [DOI] [PubMed] [Google Scholar]
- 48.Yi K, Sethi S, Murphy T. Human immune response to non-typable Haemophilus influenzae in chronic bronchitis. J Infect Dis. 1997;176:1247–1252. doi: 10.1086/514119. [DOI] [PubMed] [Google Scholar]
- 49.Bakri F, Brauer A, Sethi S, Murphy T. Systemic and mucosal antibody response to Moraxella catarrhalis after exacerbations of chronic obstructive pulmonary disease. J Infect Dis. 2002;185:632–640. doi: 10.1086/339174. [DOI] [PubMed] [Google Scholar]
- 50.Sethi S, Wrona C, Grant B, Murphy T. Strain-specific immune response to haemophilus influenzae in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2004;169:448053. doi: 10.1164/rccm.200308-1181OC. [DOI] [PubMed] [Google Scholar]
- 51.Murphy T, Brauer A, Grant B, Sethi S. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune reponse. Am J Respir Crit Care Med. 2005;172:195–199. doi: 10.1164/rccm.200412-1747OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, Flores SC, Fontenot AP, Ghedin E, Huang L, et al. Comparison of the respiratory microbiome in healthy non-smokers and smokers. Am J Respir Crit Care Med. 2013;187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han MK, Huang YJ, Lipuma JJ, Boushey HA, Boucher RC, Cookson WO, Curtis JL, Erb-Downward J, Lynch SV, Sethi S, et al. Significance of the microbiome in obstructive lung disease. Thorax. 2012;67:456–463. doi: 10.1136/thoraxjnl-2011-201183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Dore J. Direct analysis of genes encoding 16s rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16s rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46:535–548. doi: 10.1111/j.1348-0421.2002.tb02731.x. [DOI] [PubMed] [Google Scholar]
- 56.Sibley CD, Grinwis ME, Field TR, Eshaghurshan CS, Faria MM, Dowd SE, Parkins MD, Rabin HR, Surette MG. Culture enriched molecular profiling of the cystic fibrosis airway microbiome. PLoS ONE. 2011;6:e22702. doi: 10.1371/journal.pone.0022702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sogin SJ, Sogin ML, Woese CR. Phylogenetic measurement in procaryotes by primary structural characterization. J Mol Evol. 1971;1:173–184. [PubMed] [Google Scholar]
- 59.Woese CR, Fox GE. Phylogentic structure of prokaryotic domain - primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woese CR, Sogin ML, Sutton LA. Procaryotic phylogeny. 1. Concerning relatedness of Aerobacter aerogenes to Escherichia coli. J Mol Evol. 1974;3:293–299. doi: 10.1007/BF01796044. [DOI] [PubMed] [Google Scholar]
- 61.Pace NR, Stahl DA, Lane DJ, Olsen GJ. Analyzing natural microbial populations by rRNA sequences. ASM News. 1985;51:4–12. [Google Scholar]
- 62.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 63.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 64.Huang YJ, Lynch SV. The emerging relationship between the airway microbiota and chronic respiratory disease: clinical implications. Expert Rev Respir Med. 2011;5:809–821. doi: 10.1586/ers.11.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Transl Res. 2012;160:258–266. doi: 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16s rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zoetendal EG, Collier CT, Koike S, Mackie RI, Gaskins HR. Molecular ecological analysis of the gastrointestinal microbiota: a review. J Nutr. 2004;134:465–472. doi: 10.1093/jn/134.2.465. [DOI] [PubMed] [Google Scholar]
- 68.Hall N. Advanced sequencing technologies and their wider impact in microbiology. J Exp Biol. 2007;210:1518–1525. doi: 10.1242/jeb.001370. [DOI] [PubMed] [Google Scholar]
- 69.Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11:3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- 77.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brodie EL, Desantis TZ, Joyner DC, Baek SM, Larsen JT, Andersen GL, Hazen TC, Richardson PM, Herman DJ, Tokunaga TK, et al. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol. 2006;72:6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL. High-density universal 16s rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol. 2007;53:371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- 80.Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, Hwang J, Bushman FD, Collman RG. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS ONE. 2010;5:e15216. doi: 10.1371/journal.pone.0015216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Chen H, Berger KI, Goldring RM, Rom WN, Blaser MJ, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, et al. Disordered microbial communities in asthmatic airways. PLoS ONE. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang YJ, Kim E, Cox MJ, Brodie EL, Brown R, Wiener-Kronish JP, Lynch SV. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS. 2010;14:9–59. doi: 10.1089/omi.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS ONE. 2012;7:e47305. doi: 10.1371/journal.pone.0047305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cabrera-Rubio R, Garcia-Nunez M, Seto L, Anto JM, Moya A, Monso E, Mira A. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J Clin Microbiol. 2012;50:3562–3568. doi: 10.1128/JCM.00767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV, Cooper J, Sin DD, Mohn WW, Hogg JC. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;185:1073–1080. doi: 10.1164/rccm.201111-2075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. The role of the lung microbiome in health and disease: a National Heart, Lung and Blood Institute workshop report. Am J Respir Crit Care Med. 2013;187:1382–1387. doi: 10.1164/rccm.201303-0488WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Larsen J, Steen-Jensen D, Laursen J, Sondergaard J, Musavian H, Butt T, Brix S. Divergent pro-inflammatory profile of human dendritic cells in response to commensal and pathogenic bacteria associated with the airway microbiota. PLoS ONE. 2012;7:e31976. doi: 10.1371/journal.pone.0031976. [DOI] [PMC free article] [PubMed] [Google Scholar]