Abstract
Reactivation of latent viral reservoirs is on the forefront of HIV-1 eradication research. However, it is unknown if latency reversing agents (LRAs) increase the level of viral transcription from cells producing HIV RNA or harboring transcriptionally-inactive (latent) infection. We therefore developed a microfluidic single-cell-in-droplet (scd)PCR assay to directly measure the number of CD4+ T cells that produce unspliced (us)RNA and multiply spliced (ms)RNA following ex vivo latency reversal with either an histone deacetylase inhibitor (romidepsin) or T cell receptor (TCR) stimulation. Detection of HIV-1 transcriptional activity can also be performed on hundreds of thousands of CD4 + T-cells in a single experiment. The scdPCR method was then applied to CD4+ T cells obtained from HIV-1-infected individuals on antiretroviral therapy. Overall, our results suggest that effects of LRAs on HIV-1 reactivation may be heterogeneous—increasing transcription from active cells in some cases and increasing the number of transcriptionally active cells in others. Genomic DNA and human mRNA isolated from HIV-1 reactivated cells could also be detected and quantified from individual cells. As a result, our assay has the potential to provide needed insight into various reservoir eradication strategies.
Keywords: Human Immunodeficiency Virus (HIV), Single cell quantification, Digital PCR, HIV reservoirs, HIV reactivation, Histone deacetylase inhibitors
Highlights
-
•
A common approach to HIV cure involves reactivating HIV-infected cells.
-
•
Developed single cell assay to directly quantify HIV transcriptionally reactivated cells.
-
•
Single cell effects of latency reversing agents on HIV reactivation are heterogeneous.
-
•
Bulk cell-associated HIV RNA levels are divergent from the number of RNA-producing cells.
-
•
Assay allows for single-cell quantification of genomic DNA and mRNA following target cell identification.
We designed and implemented a microfluidic, single-cell assay to directly measure the number of individual cells from individuals on antiretroviral therapy that produce HIV-1 RNA. The assay also allows for single-cell quantification of human genomic DNA and messenger RNA following identification and isolation of actively HIV-1-infected cells. The ability to directly measure transcriptional activity of HIV-1 in individual cells followed by downstream characterization of human and viral genetic information within these cells has the potential to provide a greater mechanistic understanding of experimental strategies to purge residual HIV-1 reservoirs.
1. Introduction
Despite the success of combination antiretroviral therapy (ART) to reduce disease-related morbidity and mortality in HIV-1 infection, viral reservoirs still persist in the setting of intensive therapy. The main challenge in achieving a cure for HIV-1 is the elimination of these latent viral reservoirs (Chun et al., 1997, Deeks et al., 2016, Finzi et al., 1997, Richman et al., 2009, Siliciano, 2010, Wong et al., 1997). Many HIV-1 eradication strategies are based on the “shock and kill” approach, which involves reactivating infected cells with latency reversing agents (LRAs; e.g.,vorinostat, romidepsin, panobinostat), triggering viral production. Ideally, HIV-1 producing cells are then cleared by immune mediated and direct cytopathic mechanisms, while uninfected cells are protected by ART (Archin et al., 2014, Barton et al., 2016, Bullen et al., 2014, Elliott et al., 2014, Laird et al., 2015, Leth et al., 2016, Rasmussen et al., 2013, Rasmussen et al., 2014). However, viral reactivation alone is insufficient to reduce HIV-1 DNA reservoirs (Rasmussen et al., 2014), and associations between HIV-1 transcriptional activity and the number or percentage of reactivated individual cells are poorly understood (Chun et al., 1997, Eriksson et al., 2013, Ho et al., 2013, Josefsson et al., 2011, Laird et al., 2013, Procopio et al., 2015, Siliciano and Siliciano, 2005, Varadarajan et al., 2012). Even when latency is effectively reversed, it remains unknown if the enhanced HIV transcription comes from an already active pool, or from latently infected cells that reactivate and newly express HIV-1 RNA.
Cellular HIV-1 burden can be characterized by PCR quantification of cell-associated genomic and episomal DNA or cell-associated total, unspliced (us)RNA and multiply spliced (ms)RNA (Eriksson et al., 2013, Kiselinova et al., 2014, Pasternak et al., 2008, Procopio et al., 2015, Strain and Richman, 2013). However, assays involving these methods have traditionally focused on quantifying nucleic acid levels from bulk mononuclear cell lysates rather than from individual cells. As a result, these assays may attribute viral production to the entire pool of infected cells, whereas only a portion of latently infected cells may be able to reactivate and produce replication competent virus (Ho et al., 2013). Techniques to quantify inducible HIV-1 burden with single-cell resolution have recently been developed, including an assay that couples intracellular HIV RNA by probe hybridization with HIV p24 detection in flow cytometric methods (Baxter et al., 2016). However, these assays typically use or require a high degree of ex vivo stimulation in order to detect and quantify transcriptionally or translationally active cells. Other droplet-based single cell systems are either cost prohibitive, currently lack built in technology to combine viral target specific detection in addition to human polyadenylated mRNA, or lack sufficient throughput to survey the millions of cells required to indentify and characterize HIV in the setting of ART (Macosko et al., 2015, Shalek et al., 2014, Trivedi et al., 2015, Zheng et al., 2017). As a result, there is an urgent need for studies that examine the number or percentage of individual reactivated primary human cells that may be susceptible to targeted immune or pharmacologic killing (Josefsson et al., 2011, Strain and Richman, 2013), and how these numbers correlate with the total amount of cell-associated RNA being produced by an infected cell pool. Such studies may provide insight regarding whether LRAs increase the level of viral transcription from cells already activated, or from inactive, latently infected cells that reactivate and newly express HIV-1 RNA.
Here, we describe a methodology that allows for isolation and enumeration of individual latently infected lymphocytes and other tissue-derived cells. Individual CD4+ lymphocytes, macrophages, and brain-derived glial cells were encapsulated into nanoliter-scale reaction droplets via bioprinting and/or microfluidic approaches. Once encapsulated, downstream applications such as intra-droplet lysis and PCR amplification of HIV-1 usRNA and msRNA target sequences were performed. We have successfully applied this method to identify and quantify transcriptionally active individual CD4+ T cells from peripheral blood of HIV-1-infected individuals on ART to determine the ex vivo responses to various reactivating agents. We also have performed proof-of-concept studies demonstrating that genomic DNA and human mRNA can be isolated and quantified from individual encapsulated and lysed human cells. Our results suggest that the number of single cells that undergo HIV-1 reactivation is independent of total cell-associated HIV-1 RNA levels measured by traditional assays, and that the production of usRNA and msRNA in response to LRA may vary between clinical samples as well as within cells from a single individual. Our results highlight the importance of direct single-cell analysis to fully understand the impact of reactivating agents on latently HIV-infected cells.
2. Materials and Methods
2.1. Materials and Reagents
Cell culture reagents include RPMI-1640 + l-glutamine, penicillin/streptomycin 100 ×, Hepes buffer, fetal bovine serum heat inactivated, and 1 × phosphate buffered saline (Corning Cellgro, Manassas, VA). Cell staining for microscopy and flow cytometry consist of MitoTracker® Orange (Life Technologies, Carlsbad, CA), LIVE/DEAD® Fixable Blue Dead Cell Stain Kit (Life Technologies), Anti-HLA-DR APC-Cy7 (BD Biosciences, San Jose, CA), and CD38 PE-Cy7 (BD Biosciences). Cell encapsulation and lysis were performed using commercially available droplet generation oil (BioRad, Hercules, CA) and lysis buffer containing up to 10% final concentrations of decaethylene glycol mono-dodecyl ether (Sigma, St. Louis, MA), Tris-HCl, Ph8 (Fisher Scientific, Pittsburgh, PA). scdPCR mastermix included the OneStep RT ddPCR Kit (BioRad). Bulk genomic extraction for qPCR was completed with an AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany). PCRs were performed on either ABI 7300 Real Time PCR Machine (Applied Biosystems), Roche LightCycler® 480 II (Roche Diagnostics, Basel, Switzerland), or QX100 Droplet Reader (BioRad), and thermocycled using a GeneAmp® PCR System 9700 (Applied Biosystems). Laboratory cell lines included the HIV-infected 8E5/LAV lymphocyte, ACH2 and U1 cell lines (NIH AIDS Reagent Program); adherent macrophages were derived from mononuclear cells (human myeloid U937 cells) using phorbol myristate acetate (Hattori et al., 1983).
2.2. Human Subjects, Sample Collection, Cell Isolation, Culture, and Reactivation
Approval was obtained by the Brigham and Women's Hospital/Partners Healthcare Review Board and the University of California Review Committee on Human Research for the collection and testing of cells from HIV-1-infected individuals. Participants provided written informed consent prior to enrollment. Peripheral blood mononuclear cells (PBMC) were obtained from peripheral blood by density-gradient centrifugation using the Ficoll-Paque (Sigma). CD4 + T-cells were isolated from frozen peripheral blood mononuclear cells (PBMCs) using the EasySep™ Human CD4 + T-cell Enrichment Kit (Stem Cell Technologies) under manufacturer's instructions. Purified CD4 + T-cells were then cultured in RPMI-1640 with 10% FBS, 1% penicillin/streptomycin, 1% HEPES buffer, and supplemented with 1 ng/mL interleukin-2 (R-10). To prevent any potential HIV-1 infection in culture all samples, including HIV-1 negative controls, were treated with 8 nM efavirenz and 10 nM darunavir (ART).
CD4 + T-cell reactivation was performed through either T-cell receptor (TCR) activation or histone deacetylase inhibitor (HDACi) treatment. For a majority of experiments, TCR mediated reactivation was achieved through overnight culture in the presence of 100 ng/mL anti-CD-3 and anti-CD-28 antibodies (Miltenyi Biotech). HDACi treatment included a 4 h pulse with 5 nM romidepsin followed by washing, resuspension in new medium and culture in R-10 + ART for an additional 14 h. Nonstimulated controls were cultured in R-10 + ART. Laboratory-infected U1 promonocyte cells were reactivated with anti-CD-3 and anti-CD-28 antibodies as above or with 10μg/ml of PHA for approximately 18 h. Experiments involving CD4 + T cells were also performed using both 18 h and 3 day activation culture times prior to encapsulation.
2.3. Cell Encapsulation and scdPCR
Cell encapsulation was performed using an in-house set-up consisting of a syringe pump, commercially available microfluidic chips (Biorad), droplet generation oil, cell lysis buffer, and PCR mastermix (all Biorad). Cells were washed and resuspended in PBS at 5 × 105 or 1 × 106 cells/mL, and mixed 1:1 with PCR mastermix (Supplementary Table 1). To minimize extradroplet lysis and RNA contamination, 5% final concentration of lysis buffer was added immediately prior to encapsulation. Droplet generation and cell encapsulation was performed under negative pressure flow conditions using a reverse syringe pump attached to the outflow well of a BioRad ddPCR encapsulation chip with flexible tubing. Droplet generator oil (70 μL) was placed in the oil inlet well of the chip, followed by 20 μL of cell/mastermix suspension in the middle, sample loading well. Immediately prior to negative pressure encapsulation, lysis buffer was added and mixed gently in the sample well. The droplet/oil suspension was then transferred from the output well to a PCR plate well using caution as to not shear droplets. scdPCR thermocycling conditions are listed in Supplementary Table 1. Finally, thermocycled droplets were transferred to a BioRad QX100 Droplet Digital PCR system for quantification. Quantification is performed by first setting a threshold above negative control samples, followed by normalization to the estimated number of encapsulated cells, and then a final background subtraction for any low-level false positives. For cell encapsulation efficiency studies cells were first stained with MitoTracker® Orange under manufacturer's instructions, and 2 × Control Buffer was used in place of mastermix. Encapsulation efficiency was calculated by analyzing multiple fluorescent images using ImageJ software (NIH, Bethesda, MD).
2.4. Bulk PCR and Flow Cytometry
Flow cytometry was used to determine the effects of culture conditions on cell viability and cellular activation. Briefly, cells were washed and resuspended in PBS at 1 × 106 cell/mL, stained with LIVE/DEAD® Fixable Blue Dead Stain Kit at room temperature for 30 min, washed and resuspended in 100 μL PBS, stained with HLA-DR APC-Cy7 and for 20 min at room temperature in the dark, and fixed with 200 μL of 4% paraformadehyde for 30 min. Flow cytometry was performed on a BD™ LSRII flow cytometer using FACSDiva™ software (BD).
Bulk quantitative PCR methods were utilized to determine amounts of cell-associated HIV-1 unspliced and multiply-spliced RNA. RNA and DNA were extracted from cells using the AllPrep DNA/RNA Mini Kit (Qiagen). Unspliced RNA was quantified using reverse transcriptase, quantitative PCR as previously described (Malnati et al., 2008). Multiply-spliced RNA was quantified using droplet digital PCR as described(Kiselinova et al., 2014). Quantified RNA values were then normalized to total cell counts determined through qPCR for CCR5 DNA, expressed as number of copies per 1 × 106 cells as previously described (Malnati et al., 2008).
2.5. Droplet Sorting, Isolation and gDNA/mRNA Quantification
Patient-derived CD4 + T cells or mixtures of 8E5 and HIV-uninfected PBMC were encapsulated. Following scdPCR, droplets were layered onto a thin film of droplet generator oil under an inverted fluorescent microscope on the underside of a 6-well culture plate cover. Individual droplets containing HIV-1 usRNA producing cells were then selected based on positive fluorescence using an ultrafine (32 gauge) flexible, stainless steel needle (Hamilton) attached to a 2 ml lure-lock syringe. For RNA isolation, single droplets were placed in 10 μL of single-cell lysis solution with DNase I (Ambion/ThermoFisher) followed by incubation at room temperature for up to 10 min as per manufacture protocols. For DNA isolation, single droplets were placed in 7.5 μL of PicoPure (Applied Biosystems) DNA extraction solution followed by heat incubation in a thermocycler as per the manufacturer protocols. Human CCR5 DNA was quantified using real-time PCR as previously described (Malnati et al., 2008). Human IPO8 transcripts were quantified using the PrimePCR (Bio-Rad) ddPCR Gene Expression Probe assay using manufacturer protocols following reverse transcription of the RNA droplet/cell lysis extracts suing the RETROscript® Reverse Transcription Kit (Ambion; ThermoFisher Scientific). Fluorescent droplets containing HIV-1 us RNA without cellular material were used as negative controls to very that there was no fluorescent signal carried over into IPO8 ddPCR experiments.
2.6. Data and Statistical Analysis
Fluorescent micrographs were analyzed using ImageJ Software. PCR data was analyzed using either the ABI 7300 SDS Shell, LightCycler® 480 Software 1.5.1, or QuantaSoft 1.3 software. Regression, correlation, Student's t-test, and/or ANOVA (alpha level = 0.05) were performed using GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA) and SPSS vs 21 (IBM).
2.7. Droplet Bioprinting
Droplets were printed using pressurized nitrogen gas nozzle (TechElan, Mountainside, NJ) driven by a pulse/function generator (Hewlett-Packard, Palo Alto, CA) with the following optimized settings: Square pulse waveform with a 25–200 Hz frequency, 60 μs width, 2 V amplitude with no offset, 2m30s time, and 2 PSI pressure.
3. Results
3.1. System Overview: Single-Cell-in-Droplet (scd)PCR
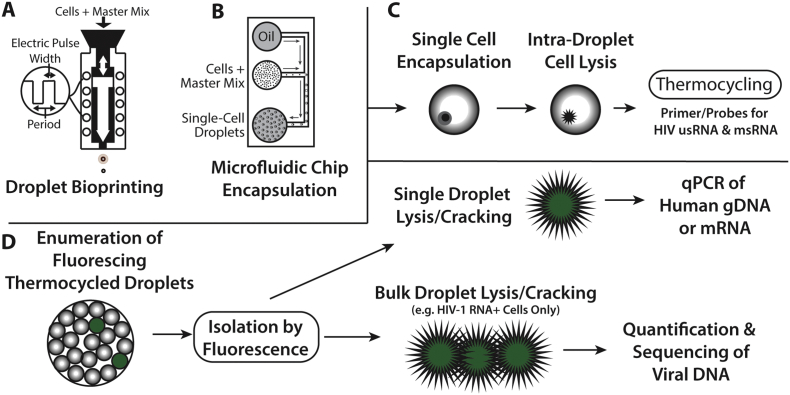
Single-Cell-in-Droplet (scd)PCR is a technique developed to enable intra-droplet cell lysis and PCR amplification of intracellular RNA without interfering with histone/chromatin bound DNA or PCR enzymes as shown in Fig. 1. scdPCR can be used to quantify HIV-1 latent reservoir reactivation frequencies by encapsulating and lysing single cells within the isolated microenvironment of a droplet, with subsequent intra-droplet PCR amplification of target HIV-1 usRNA and msRNA. Following PCR amplification of a specific target of interest, such as HIV-1 cell-associated RNA, enumeration of fluorescing droplets is performed. Droplets can then be isolated based on fluorescence followed by downstream quantification of genomic DNA and human mRNA from bulk or single cells.
Fig. 1.
Schema of the single cell encapsulation, lysis, HIV-1 detection, and rescue of cellular genomic DNA and mRNA. Cells are encapsulated in a master mix including PCR enzymes, primers, probes, and cell lysis agents (Supplementary Table 1) into oil by either brioprinting or microfluidic chip encapsulation (A, B) as described in the methods. Up to 20,000 droplets are then added to each well of a 96 well PCR plate. Cells are lysed within isolated droplet microenvironenments followed by PCR amplification of Tat-Rev spliced or unspliced cell-associated HIV-1 RNA (C). Cells are contained in the hydrophilic droplets which are stabilized by the oil-droplet interface. Droplets containing infected cells can be enumerated using an oil-based commercial flow cytometer or by direct visualization followed by microfluidic sorting of positive droplets (D). HIV-1 RNA positive or negative cells can be isolated using ultrafine needle aspiration, placed into individual microwell tubes or plate wells followed by droplet “cracking” (i.e. lysing droplets to release nucleic acids) and further characterized. In this study, human or viral genomic DNA and human mRNA from droplets containing a single encapsulated cell or from bulk droplets containing HIV-1 RNA positive cells were quantified or sequenced.
3.2. Single-cell Encapsulation
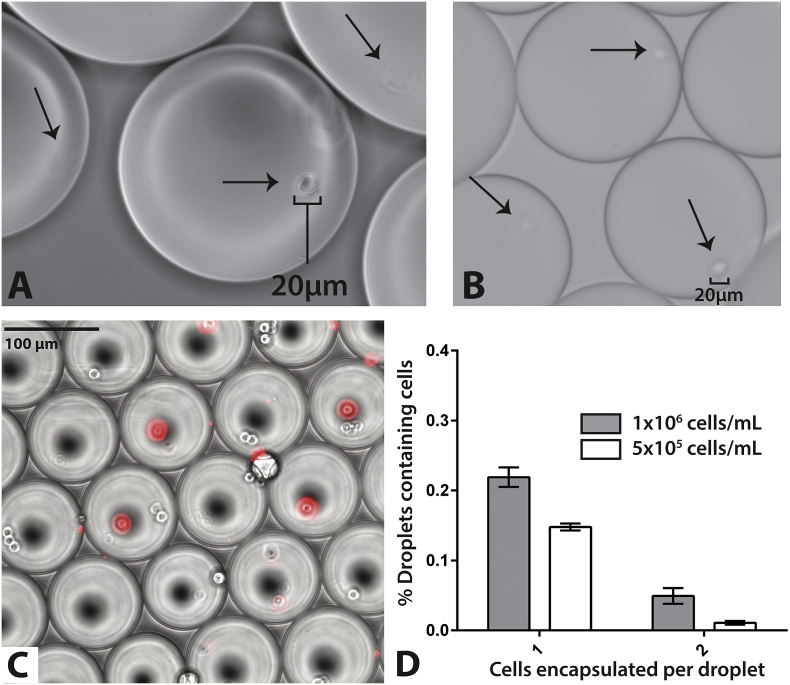
We packaged primary human CD4 + T lymphocytes, macrophages derived from the U937 mononuclear cell line and brain derived U118-MG gliobastoma cells into droplet digital PCR (ddPCR) mastermix droplets using both bioprinting and microfluidic technologies as shown in Fig. 1. Bioprinting techniques were based on our previously published nanoscale droplet methods to isolate and pattern single cells from heterogeneous cell suspensions (Ceyhan et al., 2012, Moon et al., 2011). In brief, cell/mastermix suspensions are simultaneously forced through a micro-nozzle by pressurized inert gas. An input concentration of 105 cells/mL yielded the most efficient single-cell encapsulation, with 32% of bioprinted droplets containing a single cell and 5.7% containing 2 cells, as determined by light micoscopy. We identified similar packaging efficiencies with glioblastoma cells, adherent macrophages and CD4 + T cells (Fig. 2). Bioprinting allows for relatively tight control over droplet size, but involves the use of specialized equipment and trained personnel. As a result, we incorporated a commercially available microfluidic chip for single-cell encapsulation (ddPCR cartridge originally designed for the QX100/200 platform). These chips generate 0.9 to 1.1 nanoliter droplets that can be used with the QX100/200 droplet reader (a flow cytometer involving oil rather than sheath fluid in order to preserve droplet integrity during enumeration) and were used for the remainder of experiments described below. We encapsulated single-cells by applying negative pressure to the output well of microfluidic chips, combining mastermix with surfactant laden silicon-based oil. This method negated the need for the costly commercial equipment required for efficient, high-throughput encapsulation. The compositions of droplet lysis and PCR mastermixes are shown in Supplemental Table 1.
Fig. 2.
Single cell packaging of various human cell lines. (A) Adherent macrophages and (B) brain-derived glioma cells packaged into microdoplets using bioprinting are shown under light microscopy following bioprinting. Arrows show individual cells encapsulated within fluid droplets suspended in droplet generation oil. Approximately 32% of droplets containing single cells. Few droplets contained 2 or more cells (< 6%) and packaging efficiency was similar for macrophages, glioma cells and CD4 + T lymphoctes. (C) Human CD4 + T cells stained with MitroTracker orange encapsulated using microfluidic chips stained with MitoTracker in an overlay of fluorescent signal (Red) on light microscopic images of droplets. MitoTracker staining allowed for the differentiation of encapsulated cells from air bubbles created during microfluidic chip encapsulation which are of a similar size and appearance with light microscopy. (D) Packaging using microfluidic yielded 22% of droplets containing a single cell, and fewer than 5% of droplets containing 2 or more cells at a concentration of 1 × 106 cells/mL input concentration. Error bars represent standard error from 10 replicate experiments.
To determine optimal encapsulation parameters, uninfected laboratory A3.01 lymphocyte cell lines and CD4 + T cells isolated from HIV-uninfected donors were stained with MitoTracker Orange, and packaged at various cell densities using the QX100 microfluidic chips (Fig. 2). A concentration of 1 × 106 cells/mL yielded encapsulation with approximately 22% of droplets containing a single cell, fewer than 5% of droplets containing 2 or more cells, and < 1% of cells containing more than three cells (Fig. 2). This patterns approximates the theoretical Poisson distribution in cell encapsulation as we have previously described (Moon et al., 2011). Cells within droplets were easily visible via standard fluorescent microscopy; approximately 50% of input cells are successfully encapsulated. Up to 20,000 droplets were created in each well of the BioRad chips, which were then transferred to a single well of a 96-well PCR plate, allowing for > 500,000 cells to be surveyed in a single PCR run; 160,000 droplets are able to be generated in < 5 min. As the number of RNA-expressing cells in a patient sample are expected to be very low, this assay was designed to allow repetitive sampling of hundreds of individual PCR wells for each sample or sample condition.
3.3. Encapsulated Cell Lysis and Amplification of Cell-associated HIV-1 RNA
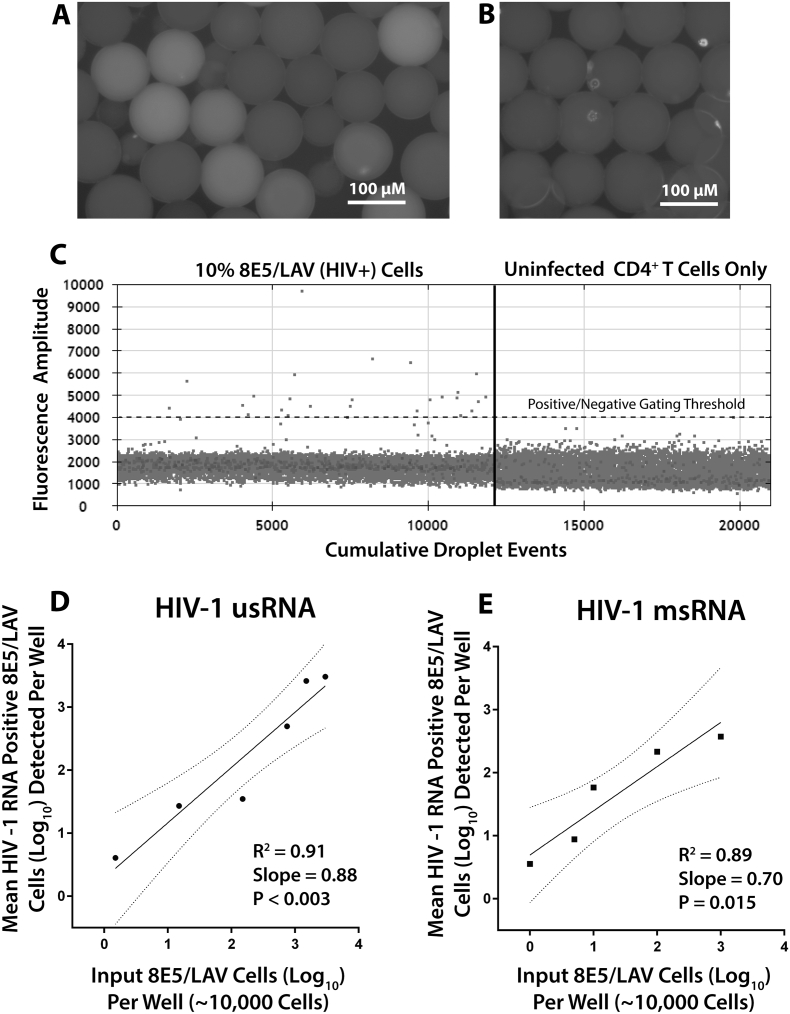
We developed the assay to detect cell-associated HIV-1 RNA while leaving genomic viral DNA intact for downstream characterization and to minimize confounding signal from cell-associated HIV-1 DNA. Following droplet generation, an initial heat step was incorporated to allow reverse-transcription of target RNA and to aid in cell lysis, followed by exposure to higher temperatures (95 °C) to aid in cell lysis prior to repetition of ddPCR cycles as outlined in the methods section. A final high heat step (98 °C for 10 min) was incorporated to promote droplet hardening. Hardened droplets are relatively resilient to sheer stress, and we have demonstrated stability for up to 7 days at 4 °C without significant loss of mechanical integrity. Initially, mixtures of various concentrations of HIV-1-infected 8E5/LAV and uninfected A3.01 lymphocyte cell lines were encapsulated followed by lysis and thermocycling for PCR amplification of cell-associated HIV-1 usRNA and msRNA as described in Supplementary Table 1. The 8E5/LAV line contains a single copy of integrated HIV-1 LAV DNA with a mutation rendering replication incompetent virus. However, the cells are able to efficiently produce HIV RNA (Wilburn et al., 2016). Nearly all 8E5 cells produce some level of cell-associated HIV-1 RNA during routine culture without the presence of ART. Successful amplification resulted in visibly fluorescent droplets (Fig. 3A) containing 8E5/LAV cells but no increased signal in droplets containing uninfected CD4 + T cells (Fig. 3B). Droplets can also be enumerated using the QX100 reader as shown in Fig. 3C; the results demonstrate that positive and negative fluorescent droplets can be readily enumerated using commercially available equipment. Gating between positive and negative cells was performed using the QX100 software, with the cutoff line positioned above the values of the negative droplets. This gating strategy is analogous to a 2-dimensional gating strategy used in flow cytometry. Uninfected CD4 + T cell were encapsulated and assayed in parallel experiments to aid with gating and to provide a measure of background noise; any droplet counts above the cut-off value from negative control experiments were subtracted from the experimental wells. Of note, PCR probe agglomerates within encapsulated cells were observed, causing the bright spots on the images, and were confirmed by dual staining with probe and MitoTracker. However, these agglomerates did not cause interference with the ddPCR Droplet Reader, and the reader software autocorrects for droplet size. Once packaging efficiency was determined, experiments were performed without the use of MitoTracker orange.
Fig. 3.
scdPCR detection and quantification of HIV-1-infected 8E5/LAV cells. Fluorescent microscopic images of droplets containing 8E5/LAV cells (A) and uninfected CD4 + T cells (B) after intradroplet lysis and target quantification for HIV-1 usRNA. Droplets containing 8E5/LAV cells demonstrate fluorescence from amplification of FAM probe that can be discerned from droplets without cells or those that contain uninfected lymphocytes. Positive fluorescent droplets can readily be quantified using the QX100 oil-based droplet reader; results from a mixture of 10% 8E5 and 90% uninfected lymphocytes is shown (C). Infected cells have fluorescent amplitudes above baseline as determined by droplets containing uninfected lymphocytes. Linear regression of the number of input 8E5/LAV cells to the mean number of infected cells containing HIV-1 usRNA (D) and msRNA (E) detected per reaction well using scdPCR. Linearity was preserved down to 1 infected cell per 10,000 total input cells in a single PCR well. Given the ability to perform repetitive sampling in multiple wells for each sample, the assay has the sensitivity to detect as low as 10 cells per 106 total input cells using a 96 well plate with known encapsulation efficiencies. Dotted lines represent the 95% confidence intervals from curve-fit regression.
Mixtures of infected and uninfected laboratory cells. Ten to 200,000 HIV (LAV) infected 8E5/LAV cells were suspended along with 1 × 106 HIV-1-uninfected CD4 + T cells from donors in a final concentration of 1 × 106 cells/mL. Linear regression analyses comparing input and expected positive cell frequencies for each reaction well are shown in Fig. 3D (usRNA) and Fig. 3E (msRNA). Quantification of usRNA and msRNA producing droplets was then performed on approximately 50 to 100,000 positive cells spiked in uninfected lymphocytes across multiple wells. Over and entire experiment involving scdPCR was able to consistently detect 10 cells expressing HIV-1 usRNA and msRNA per 1 × 106 total cells. This infected cell detection sensitivity was close and within error to that predicted from data averaging performance within individual wells (2 to 5 infected cell per 106 total input cells) as shown in Fig. 3, taking into account minor variations in encapsulation efficiencies.
3.4. Digital Droplet PCR Amplification of HIV-1 usRNA and msRNA
We performed experiments to determine if the usRNA or msRNA primer/probes and single-cell conditions would also detect HIV-1 DNA, which could confound interpretation of cell-associated RNA target amplification. The usRNA primer and probe sequences are the same as those used for detection of cell-associated HIV-1 DNA (Malnati et al., 2008). Unspliced RNA from encapsulated 8E5 cells was packaged into droplets consisting of the same mastermix as described above. Following thermocycling, fluorescent droplets were readily detected using the QX100 droplet reader and fluorescent microscopy. Approximately 2xlog10 fewer positive droplets were observed when reverse-transcriptase was omitted, suggesting that the vast majority of the positive signal in our assay arises from RNA rather than genomic DNA. The msRNA primers are designed for amplification of the Tat-Rev spliced sequence spanning near full-length HIV-1 envelope, and are designed to have little to no amplification of intact DNA sequences (Pasternak et al., 2008). We did not detect DNA using the msRNA primers and probes despite high levels of input HIV-1 DNA.
3.5. Safety of Thermocycled Droplets Containing HIV-1-infected Cells
In order to determine the safety of using thermocycled droplets for downstream manipulations outside of a BL2 + containment setting, we packaged droplets with ACH2 cells containing replication competent HIV-1 proviral DNA; these cells are capable of producing infectious virus upon stimulation. We combined HIV uninfected A3.01 cells (negative control), with ACH2 cells, which were then subject to either thermocycling alone or thermocycling after droplet encapsulation, followed by incorporation into an infectivity assay using TZM-black adherent cells or into a viral co-culture assay. High levels of β-galactosidase, a surrogate for HIV-1 entry into TZM cells, were observed in experiments incorporating unmanipulated ACH2 cells and the positive control wells inoculated directly with infectious HIV-1 JRCSF (RLU > 400,000; Supplementary Fig. 1). However, no signal was detected in wells incorporating encapsulated or unencapsulated ACH2 cells following scdPCR thermocycling. Encapsulated, thermocycled ACH2 cells did not lead to increasing HIV-1 RNA in supernatants over 14 days of co-culture, with RNA levels being similar to DNA in supernatants derived from dead cells (Supplementary Fig. 1). As a result, themocycled droplets were determined to have negligible infectious risk.
3.6. Single-cell Responses to HIV-1-reactivation and Mitogen Stimulation of Cell Lines Using scdPCR
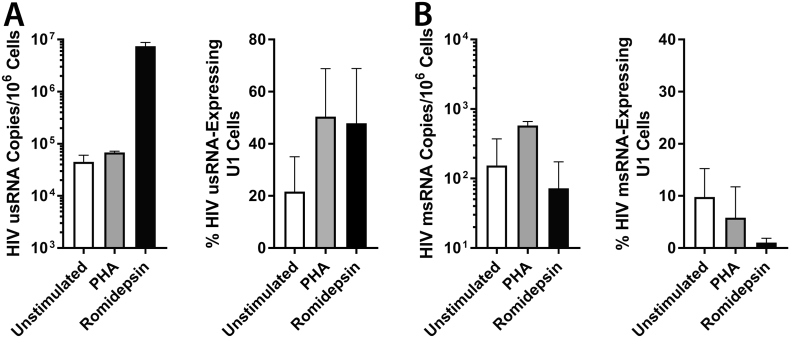
In order to test the performance of scdPCR to detect increases in HIV transcriptional activity following latency reversal or global cell activation, we incubated the HIV-1-infected U 937 (U1) promonocyte line (Folks et al., 1987) following a 4 h pulse of the HDAC inhibitor, romidepsin (RMD) and overnight culture in RMD-free growth medium, or for 18 h in the presence of growth medium alone or with the mitogen, phytohaemagglutinin (PHA). Cells were then washed, encapsulated and sdcPCR performed using both HIV-1 usRNA and msRNA primers and probes. U1 cells were used in lieu of 8E5-LAV cells as fewer U1 cells express detectable HIV-1 RNA without activation, and better represents viral latency. Romidepsin exposure led to a large increase in bulk HIV-1 usRNA, and both RMD and PHA increased the percentage of cells expressing usRNA (Fig. 4A). Changes in HIV-1 msRNA in the setting of various stimuli were more variable, with decreased bulk RNA levels and percentages of cells expressing msRNA observed following romidepsin exposure (Fig. 4B).
Fig. 4.
Single cell and bulk HIV-1 usRNA and msRNA responses in HIV-1-infected U1 cells to various stimuli. (A) Bulk and single cell HIV-2 usRNA U1 responses to mitogen (PHA) or HDAC inhibitor stimulation are shown in the left and right panels respectively. Romidepsin increased usRNA in bulk cell extracts as well as the percentage of cells expressing RNA. In contrast, RMD decreased bulk msRNA production and the percentage of U1 cells expressing msRNA (B). Error bars represent standard error for two parallel experiments (conventional qPCR) or up to 16 individual experimental wells (scdPCR).
3.7. Effects of T-cell Stimulation and HIV-1 Reactivation on Individual Viral Transcriptionally Active Cells From Individuals on ART
Next, to determine the ex vivo single cell responses to T-cell receptor stimulation and HDACi reactivation, CD4 + T-cells were isolated from cryopreserved peripheral blood mononuclear cells (PBMC) obtained from individuals on combination ART with various HIV-1 clinical settings including those with undetectable viral load measurements or low-level blips or viremic events (Table 1). These participants were included in order to test the assay on a range of clinical scenarios that may be expected in various interventional studies. Following overnight resting in culture medium without exogenous cytokines or drugs, isolated CD4 + T cells were activated through the T-cell receptor (TCR) using αCD3/αCD28 antibodies or treated with pulse dosed HDACi (romidepsin; RMD) in the setting of at least two antiretroviral drugs in order to reverse HIV-1 latency, and induce viral transcription and usRNA and msRNA production without de novo infection of HIV-1 uninfected cells. Cells were then split equally for: (i) encapsulation and scdPCR at a concentration of 106 cells/mL, and (ii) bulk cell lysis and usRNA and msRNA quantification using traditional real-time PCR (usRNA) or ddPCR (msRNA) based on previously reported methods (Kiselinova et al., 2014, Malnati et al., 2008, Pasternak et al., 2008).
Table 1.
Participant demographics, clinical status, CD4 + T cell counts, HIV-1 cell associated DNA, and viral load at the time of sampling.
| Participant | Age | Years of ART at time-point tested | Other clinical diagnosis | Last HIV load (RNA copies/mL) | HIV DNA (DNA copies/106 CD4 + T cells) | CD4 + T cell counts (cells/μl) |
|---|---|---|---|---|---|---|
| A | 44 | 0.25 | None | < 50 | 3231 | 921 |
| B | 67 | > 10 | None | < 50 | 898 | |
| C | 54 | 4 | None | < 50 | 957 | |
| D | 56 | See notea | 4.7 years following allogeneic HSCT | < 50 | 5528 | 618 |
| E | 27 | See noteb | 6.4 years following allogeneic HSCT | 1530 | 16,157 | |
| F | 52 | > 5 | > 1 year post autologous HSCT for Hodgkin lymphoma | 246 | 13,865 | 1063 |
| G | 46 | 2.6 | 7.4 months following completion of chemotherapy for B cell lymphoma | 85 | 422 | 221 |
| H | 37 | 9 | None | < 40 | – | 418 |
| I | 61 | 21 | None | < 40 | – | 371 |
ND = HIV RNA not detected.
Restarted ART within a week of viral rebound following analytical treatment interruption; participant had been on ART for approximately 18 months following re-initiation.
Restarted stable ART within 4 weeks of viral rebound following analytical treatment interruption; participant had been on ART for approximately 15 months following re-initiation.
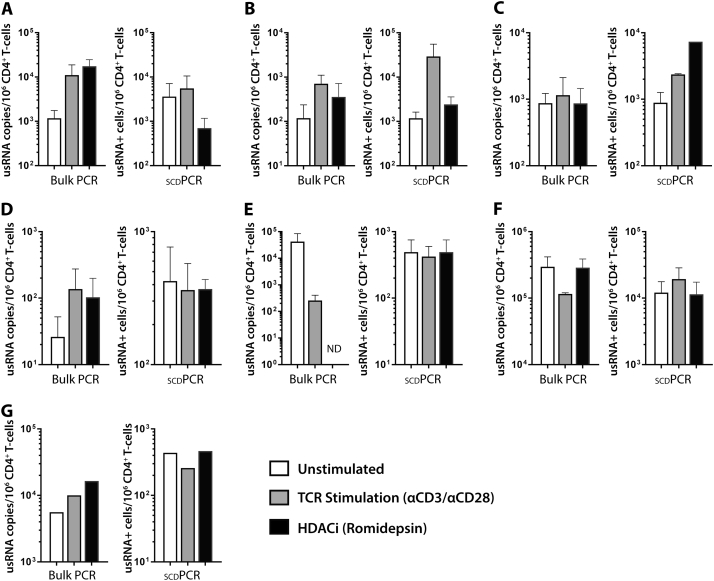
The number of cells expressing HIV-1 usRNA and msRNA with and without TCR simulation or HDACi use, as well as the absolute cell-associated usRNA levels quantified by traditional quantitative PCR from bulk extracts, are shown in Fig. 5. Overall, usRNA copies/106 CD4 + T cells measured by traditional assay on bulk lysates increased after TCR stimulation by αCD3/αCD28 antibodies in all participants with undetectable plasma HIV load measurements at the time of blood collection (Fig. 5; participants A, B, C, and D). Three of these four individual (A, B, and D) also experienced an increase in usRNA copies 24 h after romidepsin pulse. Despite these expected increases in bulk usRNA levels, changes in the number of cells producing usRNA as measured by scdPCR after stimulation were variable, and at times, discordant with results from traditional RNA measures. For example, despite a > 1 log10 increase in bulk RNA following HDAC inhibition for participant A, the number of cells expressing usRNA decreased. Traditional qPCR results were more variable in participants with low-level detectable viremia on ART at the time of sample collection (E, F, G). Nonetheless, dichotomies between qPCR and sdcPCR assay results were notable in each of these samples. A high degree of reproducibility between independent experiments was observed based on the size of error bars, and all scdPCR values were greater than the assay limit of detection. A total of 500,000 to 1,000,000 CD4 + T cells were input into the usRNA and msRNA assays for each activation condition. Given the high degree of variation in HIV-RNA frequencies and levels between individuals, there were no significant differences in responses to various stimuli across all participants.
Fig. 5.
Comparison of single cell HIV-1 usRNA results by scdPCR with traditional quantitative PCR measures of us RNA from participants on ART. usRNA copies/106 CD4 + T cells by conventional assay on bulk lysates increased after 24 h of TCR stimulation by αCD3/αCD28 antibodies in all participants with undetectable plasma HIV load measurements at the time of blood collection (A, B, C, D). Three of these four individual (A, B, and D) also experienced an increase in usRNA copies ~ 18 h after romidepsin pulse. Despite these expected increases in bulk usRNA levels, changes in the number of cells producing usRNA as measured by scdPCR after stimulation were variable, and at times, discordant with results from traditional RNA measures. For example, despite a > 1 log10 increase in bulk RNA following HDAC inhibition for participant A, the number of cells expressing usRNA decreased. Conventional qPCR results were more variable in participants with low-level detectable viremia on ART at the time of sample collection (E, F, G). Assays were repeated two to four times with the exception of participant G (limited sample), and error bars represent standard error of the mean between independent experiments. Reproducibility between independent experiments was observed, and all scdPCR values were greater than the both the detection limit of the assay (1 log10 RNA expressing CD4 + T cells) and limit of linearity (2 log10 cells).
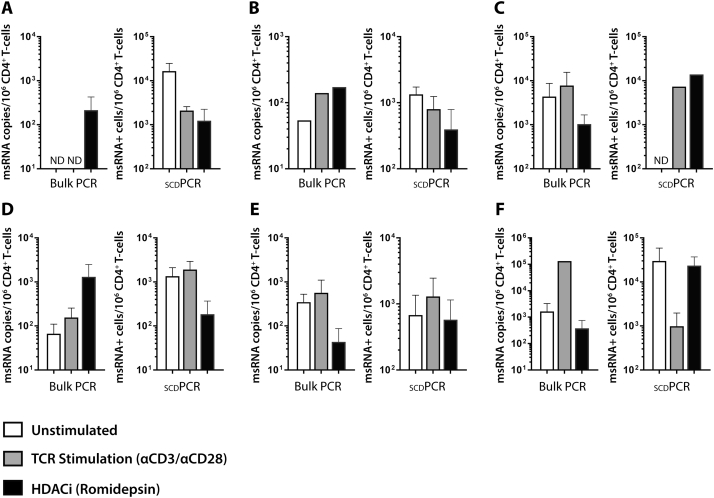
Traditional qPCR and scdPCR measures of msRNA in participants on ART are shown in Fig. 6. With the exception of participant A, in whom msRNA could not be detected in unstimulated or TCR stimulated wells, msRNA copies/106 CD4 + T cells by traditional assay on bulk lysates increased following TCR stimulation in all participant samples tested (A though F). The responses to romidepsin pulse stimulation were more variable, with increases noted in participants A, B, and D. The numbers of cells expressing HIV msRNA were different from the responses seen by traditional assays. In some instances, increases in the amount of bulk msRNA after stimulation or no stimulation increased while the numbers of msRNA expressing cells decreased, and vice versa. Given the high degree of variation in HIV-RNA frequencies and levels between individuals, there were no significant differences in responses to various stimuli across all participants. However, each experiment was reproducible when performed two to four times using different cell aliquots on different experimental days.
Fig. 6.
Comparison of single cell HIV-1 tat-rev msRNA results by scdPCR with traditional quantitative PCR measures of CD4 + T cell-associated msRNA from participants on ART. With the exception of participant A, in whom msRNA could not be detected in unstimulated or TCR stimulated wells, msRNA copies/106 CD4 + T cells by traditional assay on bulk lysates increased following TCR stimulation in all participant samples tested (A though F). As with traditional usRNA measures on bulk cell lysates, the responses to romidepsin pulse stimulation were more variable, with increased noted in participants A, B, and D. The numbers of cells expressing HIV msRNA were different from the responses seen by traditional assays. In some cases, increases in the amount of bulk msRNA after stimulation or no stimulation increased while the numbers of msRNA expressing cells decreased, and vice versa. Assays were repeated twice in independent experiments, and error bars represent standard error between independent experiments. With the exception of a lack of detectable msRNA expressing cells in samples from participant C, all scdPCR values were greater than both the detection limit of the assay (1 log10 RNA expressing CD4 + T cells) and limit of linearity (2 log10 cells), and results were reproducible between independent experiments. (ND = no HIV RNA detected).
In order to determine whether or not cell death during stimulation and culture may have led to differences between ex vivo treatment groups, viability decreased < 6% for all samples following TCR stimulation with αCD3/αCD28 antibodies, but RDP exposure decreased cell viability by up to 16.7% in participants A and D (Supplementary Table 2). We also obtained flow data on the fold increase in the early activation marker following stimulation in cells from participants B and C in order to demonstrate that TCR stimulation led to global cell activation (i.e. positive activation control). Overall, exposure to αCD3/αCD28 antibodies led to a 7.2 and 117-fold increases in the amount of CD69 expression in participants B and C, respectively, whereas RMD led to 0 and 1.8 fold increases.
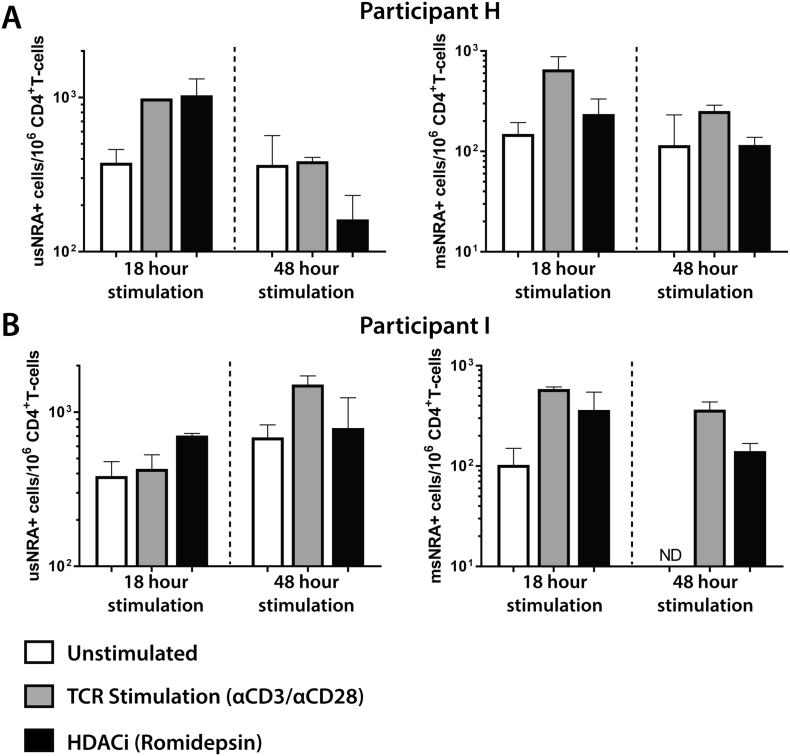
We also performed scdPCR for HIV-1 usRNA and msRNA in two individuals on suppressive ART with plasma viral load measurements < 50 RNA copies/mL following reactivation for up to 3 days in order to determine single cell changes in transcriptional activity over time. As shown in Fig. 7, we noted variability between the two patients with respect to patterns of reactivation and between various time points (18 h and 3 days). Again, results were reproducible over two separate experiments. The number of HIV-1 usRNA expressing CD4 + T cells from participant H increased after both TCR and RMD stimulation after 18 h whereas usRNA levels decreased with romidepsin exposure following 48 h in culture. TCR stimulation resulted in increased HIV-1 msRNA expressing cell numbers at both time points, with little change in the RMD experiments compared with unstimulated controls.
Fig. 7.
Longitudinal scdPCR results for CD4 + T cells from individuals on suppressive ART following reactivation for 18 h and 3 days. The numbers of HIV-1 usRNA and msRNA expressing CD4 + T cells for participant H (A) and I (B) are shown. TCR stimulation or romidepsin exposure led to increased both unspliced and multiply spliced RNA positive cells following 18 h of stimulation for both participants and after 48 h for participant I. However, 48 h stimulation led to variable responses in cells from participant H. Both individuals were on suppressive ART with plasma RNA levels < 50 copies/mL at the time of sampling. Assays were repeated twice, and error bars represent standard error of the mean between independent experiments.
3.8. Downstream HIV-1 DNA Rescue, Amplification, and Sequencing From Thermocycled Droplets
We performed proof of concept experiments to demonstrate that genomic HIV-1 DNA can be rescued from cells encapsulated and lysed within droplets following scdPCR. First, in order to test if viral or human genomic DNA degrades with repetitive thermocycling, 10 and 100 copies of both HIV-1 and human genomic DNA obtained from infected 8E5/LAV cells were subjected to thermocycling mimicking the conditions of scdPCR. Following thermocycling, real-time PCR was performed on a secondary HIV-1 and human DNA target (HIV integrase and human CCR5, respectively, and as previously described (Cillo et al., 2014, Malnati et al., 2008). Cycle threshold (Ct) values between prior thermocycled and direct input DNA were the same for each paired condition (Supplementary Fig. 2), suggesting cellular DNA is not substantially degraded during scdPCR, at least in the sequence areas targeted by the real-time PCR primers and probes. Additionally, infected 8E5/LAV mixed with uninfected A03.01 cells at ratios of 100% 8E5, 60:30 8E5:A03.01, and 100% A03.01, were encapsulated followed by scdPCR. Thermocycled droplets from each reaction well were washed of residual oil phase, and DNA was extracted from all droplets within a well using established phenol-chloroform lysis and silica gel-based column extraction protocols. HIV-1 integrase real-time PCR was performed on the extracted DNA as described above. HIV-1 integrase gene was detected exclusively in lysates from droplets containing 8E5/LAV cells at any input concentration, but not from DNA extracted from droplets containing only uninfected A03.01 cells. To demonstrate that we could amplify and sequence portions of the HIV-1 genome following scdPCR in aggregate droplets within a single scdPCR reaction well, we rescued HIV-1 DNA from droplets containing either 8E5/LAV or uninfected A3.01 cells. We successfully amplified hyper variable region of HIV-1 envelope [C2-V4 (Sharkey et al., 2011)] (Supplementary Fig. 3) followed by population sequencing of the target gene. Of note, scdPCR experiments of both 8E5 laboratory cell lines and participant samples without reverse-transcriptase, and magnesium in place of manganese in the mastermix yielded approximately 2 log10 fewer positive fluorescent droplets, suggesting that a large majority of signal in our assay arises from HIV RNA rather than HIV DNA.
3.9. Isolation and Quantification of Human Genomic DNA and mRNA From Single HIV-1 Transcriptionally Active Patient-derived Cells
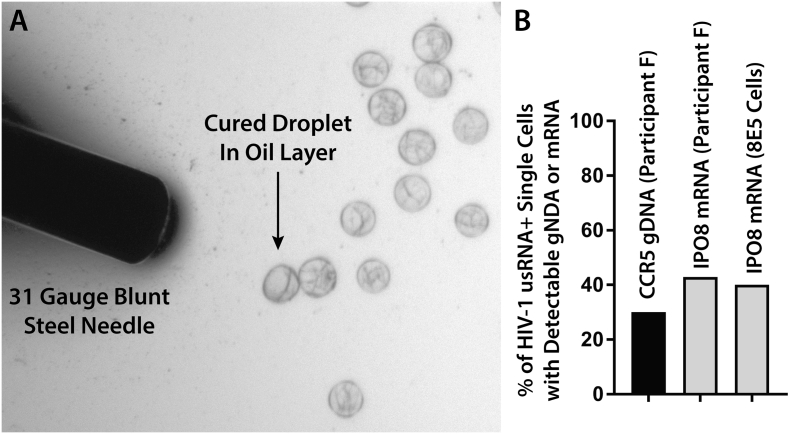
Finally, we performed proof-of-concept studies to demonstrate that single droplets containing one HIV-1 infected and transcrtipionally active cell determined by scdPCR can be isolated followed by rescue and characterization of human genomic DNA or mRNA. Patient-derived CD4 + T cells (Participant F) or a 10% mixture of 8E5 and HIV-uninfected PBMC were encapsulated and HIV-1 usRNA scdPCR performed as above. Following thermocycling, droplets were layered onto a thin film of droplet generator oil under an inverted fluorescent microscope. Individual droplets containing HIV-1 usRNA producing cells were then selected based on positive fluorescence using an ultrafine (32 gauge) flexible, stainless steel needle attached to a 2 ml lure-lock syringe and placed directly into commercially available single-cell lysis buffers (Fig. 8). For RNA isolation, single droplets were placed in 10 μL of single-cell lysis solution with Dnase I (Ambion/ThermoFisher) followed by incubation at room temperature for up to 10 min. For DNA isolation, single droplets from the participant sample were placed in 7.5 μL of PicoPure (Applied Biosystems) DNA extraction solution followed by heat incubation as per the manufacturer protocols. Following lysis, we were able to detect a conserved region of the human CCR5 gene from 3 out of 10 droplets isolated based on HIV-1 usRNA positive produciton from participant F using a qPCR method as described (Malnati et al., 2008). Ct values from each positive reactions corresponded with the 1–5 copy number range based on a standard curve (the qPCR has linearity down to 10 copies/input well) which roughly correlated with the expected two copies of the diploid gene (Fig. 8). No CCR5 was detected in negative template control wells. We were also able to detect the human IPO8 mRNA transcript in 3 out of 7 droplets tested from Participant F and 2 of 5 droplets containing HIV-1 transcriptionally active 8E5 cells using the PrimePCR (Bio-Rad) ddPCR Gene Expression Probe assay following reverse transcription of the RNA droplet/cell lysis extracts (Fig. 8). The mean per-cell IPO8 copy numbers were 2.6 and 21 for participant and 8E5 cells, respectively.
Fig. 8.
Isolation of single scdPCR droplets based on HIV-1 usRNA fluorescence and subsequent human gDNA and mRNA characterization. Fluorescent droplets containing single HIV-1 transcriptionally active 8E5 or participant-derived CD4 + T cell lysates obtained by scdPCR were isolated and placed in single-cell DNA or RNA lysis buffers using an ultrafine gauge blunt needle (A). The percentages of individual droplet/cells with detectable CCR5 genomic DNA using real-time PCR or IPO8 mRNA using traditional ddPCR are shown in (B).
4. Discussion
Assays that allow for the identification and characterization of single HIV-1 infected cells are a top priority in the field of HIV persistence (Strain and Richman, 2013). Here, we report the design and implementation of a single-cell assay for the identification and quantification of individual, transcriptionally active cells (i.e., those that produce HIV-1 usRNA or msRNA) and their responses to various ex vivo stimuli such as antibody engagement of the T cell receptor and pulsed HDACi exposure. We designed the assay to incorporate commercially available PCR reagents, microfluidic droplet generating chips, and available equipment to allow for the rapid implementation in a larger number research groups without the need for customized microfluidic devices. Using this assay, we found that the scdPCR assay is able to reproducibly determine the numbers of transcriptionally active cells from individuals on ART. Furthermore, we found that human genomic DNA and mRNA can be quantified from individual cells using the sdcPCR assay. Overall, our results suggest that effects of LRAs on HIV-1 reactivation may be heterogeneous—increasing transcription from active cells in some cases and increasing the number of transcriptionally active cells in others.
scdPCR assay can provide a measure of the numbers of HIV-1 msRNA and usRNA productive cells that can be directly correlated to traditional measures of cell-associated RNA on bulk cell lysates. Of note, we observed differences between msRNA and usRNA responses to TCR or HDACi stimulation by both our scdPCR method and by traditional qPCR methods. We were also able to identify and quantify transcriptionally active cells using scdPCR in all participant samples above the limit of detection and within the linear range of the assay. Furthermore, scdPCR has the sensitivity to reliably detect various HIV RNA transcripts even without maximal exogenous stimulation. As a result, the assay has the potential to explore more physiologic conditions that exist in participants undergoing HIV curative therapies.
Overall, we observed inter-individual variation in the single-cell response to either global TCR stimulation or ex vivo reactivation of HIV-1 following HDACi exposure. For example, we observed an expected increase in usRNA after TCR and RMD stimulation from bulk extracts, but observed a marked decrease in the number of single cells expressing RNA after RMD exposure in participant A. RMD increased bulk msRNA while both TCR stimulation and RMD exposure led to a decrease in the number of HIV transcriptionally active cells in cells from the same individual. It is possible that cell death from drug toxicity led to a decrease in the total number of RNA positive cells, but we observed a < 5% decrease in live cells following stimulation for participants A and B. Furthermore, we observed increases in the number of usRNA transcribing cells in other individuals in the setting of RMD exposure, despite decreasing total bulk usRNA. Despite these divergent findings, results were consistent when repeated on different days using different cell aliquots. Furthermore, we noticed differences between changes in HIV-1 usRNA and msRNA levels from bulk extracts and the percentage of CD4 + T cell expressing RNA in several other participants.
Several individuals did not have a marked rise in traditional measures of HIV RNA following TCR stimulation. In order to minimize changes in packaging efficiencies, we opted to use free αCD3/αCD28 antibodies rather than the more potent antibody coated beads to perform these stimulations. Free antibodies take longer to stimulate T cells and our activation protocol was restricted to 12–24 h prior to encapsulation, which may have been insufficient to fully activate the CD4 + T cell pools.
Prior studies have shown that increases in msRNA coding for regulatory proteins, such as Tat/Rev. RNA as incorporated in our assay, are seen earlier in the HIV life-cycle and precede increases in usRNA transcripts, which are packaged into virions and are correlated with the lytic phase of infection (Bagnarelli et al., 1996, Pasternak et al., 2013, Pomerantz et al., 1990). It is therefore possible that decreases in the percentage of msRNA producing CD4 + T cells after RMD stimulation observed in our study represent a shift away from early cycles of viral reactivation towards the later, usRNA enriched stage. However, maximal msRNA levels have been observed up to 12 h after stimulation in other studies (Procopio et al., 2015) and our assay was designed to incorporate a similar simulation time. We also observed decreases in bulk usRNA and the numbers of usRNA-expressing cells by traditional and scdPCR methods. Interestingly, we observed stable or increased numbers of msRNA positive cells following RMD exposure at both time-points in our experiments involving 18 h and 3 day stimulations of cells from two ART-suppressed individuals. As a result, shifts from msRNA to usRNA may not completely explain the decreases in the percentage of RNA producing cells observed in this study. Although it is possible that the decreasing frequencies of usRNA or msRNA producing cells observed after TCR or HDACi stimulation may be due, at least in part, to cell death after stimulation, reductions in cell viability following TCR stimulation was minimal, and < 17% in the setting of RMD exposure. Despite the minimal reductions in cell viability, our results may have been influenced by differential loss of HIV-infected cells following viral reactivation.
We included individuals with various clinical settings, including those on long-term ART who initiated treatment during chronic infection and two participants that had no detectable HIV reservoirs following allogeneic HSCT, but experienced viral rebound following ART cessation that mimicked acute infection (Henrich et al., 2014, Henrich et al., 2013). These patients were included as to test the scdPCR assay in the diverse populations that may be incorporated into HIV eradication studies. As above, participant A experienced a decrease in HIV-1 usRNA following ex vivo romidepsin exposure, and was only on 3 months of ART prior to sampling. Given similar cell viabilities with or without stimulation, factors other than cell death likely led to this interesting observation. Results from a single individual are clearly not sufficient to make any conclusions regarding the responses to HDAC inhibitor exposure, but it possible that the observed differences in response to stimulation between participants may be due to factors such as the burden of HIV-1-infection in various T cell immune subsets, CD4 + T cell activation and proliferation state or exhaustion phenotype at the time of sampling (Chomont et al., 2009). It is also possible that technical differences between the traditional and single-cell-in droplet PCR methods led to these varied and sometimes, dichotomous observations. Further investigation in larger, controlled cohorts is needed, and comparisons of scdPCR with recently described single-cell quantification methods involving intracellular HIV-1 RNA in-situ hybridization or HIV antigen detection (Baxter et al., 2016) are warranted.
The scdPCR method carries several limitations. For example, despite its ability to encapsulate single cells, cells may be lost during packaging and subsequent droplet manipulation (e.g., sample transfer prior to thermocycling). In addition, a small percentage of droplets contain greater than a single cell as detailed above which may bias subsequent downstream characterization of individual cells. Nonetheless, we observed a high degree of reproducibility between independent experiments, and levels measured in our patient population were within the linear range of the assay. The assay is also limited by the lack of capacity to provide a direct measure of the percentage of individual cells to produce replication competent virus, such as with the quantitative viral outgrowth assay (qVOA) (Chun et al., 1997, Eriksson et al., 2013, Ho et al., 2013, Laird et al., 2013, Siliciano and Siliciano, 2005). A large proportion of intracellular HIV-1 DNA and RNA do not code for replication competent virus (Ho et al., 2013), and our assay is not designed to enumerate individual cells that may harbor the true replication competent reservoir. Nonetheless, it is of utmost importance to determine the changes in HIV transcriptional activity of infected cells in response to various stimuli. Cells were rested overnight prior to stimulation, which may also have led to increases in HIV transcriptional activity without exogenous stimuli. However, the assay allows the capacity for cells to be encapsulated rapidly following thawing. We were also able to identify transcriptionally active cells from patients without exogenous stimulation, thereby allowing application of scdPCR to samples obtained from in vivo LRA administration.
In this manuscript, we have performed proof-of-concept studies that demonstrate transcriptionally active, HIV-1- infected and transcriptionally active cells may be isolated, allowing for downstream quantification of genomic DNA or mRNA. Integrating the scdPCR assay with previously described fluorescent activated droplet sorting(Baret et al., 2009) may provide a platform in which infected, reactivated HIV-1 + cells can be sorted, isolated, and characterized in a larger number of samples. Our assay may also provide several advantages over previously published or commercialized single-cell platforms. For example, the Fluidigm system allows for deep transcriptional characterization of single cells, but is limited by the relatively low input cells (e.g. 96 to 384) and relatively high cost of processing a single cell. Other platforms such as 10X Genomics microfluidic encapsulation platform and the published DropSeq protocol allow for input of larger number of cells (e.g. thousands), but may lack the throughput to cost-effectively screen the millions of cells required to characterize HIV-1 infection in ART-suppressed individuals (Macosko et al., 2015, Shalek et al., 2014, Trivedi et al., 2015, Zheng et al., 2017). These platforms were also originally designed for the detection of polyadenylated mRNA rather than for specific viral targets, as in scdPCR.
In summary, the quantification of the number of directly observed, individual cells expressing RNA, has the potential to provide insight into how individuals will respond to various HIV reactivation strategies. Our results demonstrate that there may be important inter-patient differences between the number of transcriptionally reactivated cells and the total amount of HIV RNA produced. As a result, our methods may play an important role in determining whether potential increases in cell-associated HIV-1 RNA following reservoir reactivation result from a small number of highly active cells or from a larger, less active pool.
Conflicts of Interest
D.R.K. is a consultant to and has received honoraria from Bionor, Gilead, GlaxoSmithKline, InnoVirVax, Janssen, Merck, and ViiV; he has received grant support from Gilead and Merck, and speaking honoraria from Gilead and Merck. Dr. U Demirci is a founder of, and has an equity interest in: (i) DxNow Inc., a company that is developing microfluidic and imaging technologies, (ii) Koek Biotech, a company that is developing microfluidic IVF technologies for clinical solutions, and (iii) LEVITAS Inc., a company that develops biotechnology tools for genomic analysis in cancer. U.D.’s interests were viewed and managed in accordance with the conflict of interest policies.
Author Contributions
TJH, UD conceptualized the study and obtained funding; RWY and TJH designed and performed experiments, analyzed data and wrote the manuscript; EH, KSH, WN, MOO, RA, MK, KSL, HS, SW, LEH, FI, EG, CT, designed and/or performed experiments and analyzed data; VY, KSL and HB performed experiments; YPR and SGD coordinated participant enrollment and helped obtain samples; DRK helped with study conceptualization and manuscript preparation.
Acknowledgements and Funding
The authors would like to extend their appreciation to Magesh Sadasivam to contribute to the initial stage of this study. This work was supported by the National Institutes of Health/National Institute of Allergy and Infectious Disease R21AI113117-01 (T.J.H. and U.D.); R21AI110277-01 (T.J.H. and U.D.); K23AI098480-01A1 (T.J.H.); a Foundation for AIDS Research (amfAR) ARCHE grant (T.J.H.) and the amFAR Institute for HIV Cure Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. S.W. would like to acknowledge the support from the Ministry of Science and Technology of the People's Republic of China (2016YFC1101302).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.05.006.
Contributor Information
Utkan Demirci, Email: utkan@stanford.edu.
Timothy J. Henrich, Email: timothy.henrich@ucsf.edu.
Appendix A. Supplementary Data
Supplementary material.
References
- Archin N.M., Bateson R., Tripathy M., Crooks A.M., Yang K.H., Dahl N.P., Kearney M.F., Anderson E.M., Coffin J.M., Strain M.C. HIV-1 expression within resting CD4 T-cells following multiple doses of vorinostat. J. Infect. Dis. 2014;210(5):728–735. doi: 10.1093/infdis/jiu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnarelli P., Valenza A., Menzo S., Sampaolesi R., Varaldo P.E., Butini L., Montroni M., Perno C.F., Aquaro S., Mathez D. Dynamics and modulation of human immunodeficiency virus type 1 transcripts in vitro and in vivo. J. Virol. 1996;70:7603–7613. doi: 10.1128/jvi.70.11.7603-7613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baret J.C., Miller O.J., Taly V., Ryckelynck M., El-Harrak A., Frenz L., Rick C., Samuels M.L., Hutchison J.B., Agresti J.J. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab Chip. 2009;9:1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- Barton K., Hiener B., Winckelmann A., Rasmussen T.A., Shao W., Byth K., Lanfear R., Solomon A., McMahon J., Harrington S. Broad activation of latent HIV-1 in vivo. Nat. Commun. 2016;7:12731. doi: 10.1038/ncomms12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A.E., Niessl J., Fromentin R., Richard J., Porichis F., Charlebois R., Massanella M., Brassard N., Alsahafi N., Delgado G.G. Single-cell characterization of viral translation-competent reservoirs in HIV-infected individuals. Cell Host Microbe. 2016;14(20):368–380. doi: 10.1016/j.chom.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen C.K., Laird G.M., Durand C.M., Siliciano J.D., Siliciano R.F. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 2014;20:425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceyhan E., Xu F., Gurkan U.A., Emre A.E., Turali E.S., El Assal R., Acikgenc A., Wu C.A., Demirci U. Prediction and control of number of cells in microdroplets by stochastic modeling. Lab Chip. 2012;12:4884–4893. doi: 10.1039/c2lc40523g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomont N., El-Far M., Ancuta P., Trautmann L., Procopio F.A., Yassine-Diab B., Boucher G., Boulassel M.R., Ghattas G., Brenchley J.M. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T.W., Carruth L., Finzi D., Shen X., DiGiuseppe J.A., Taylor H., Hermankova M., Chadwick K., Margolick J., Quinn T.C. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Cillo A.R., Vagratian D., Bedison M.A., Anderson E.M., Kearney M.F., Fyne E., Koontz D., Coffin J.M., Piatak M., Jr., Mellors J.W. Improved single-copy assays for quantification of persistent HIV-1 viremia in patients on suppressive antiretroviral therapy. J. Clin. Microbiol. 2014;52:3944–3951. doi: 10.1128/JCM.02060-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks S.G., Lewin S.R., Ross A.L., Ananworanich J., Benkirane M., Cannon P., Chomont N., Douek D., Lifson J.D., Lo Y.R. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nat. Med. 2016;22:839–850. doi: 10.1038/nm.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J.H., Wightman F., Solomon A., Ghneim K., Ahlers J., Cameron M.J., Smith M.Z., Spelman T., McMahon J., Velayudham P. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog. 2014;10:e1004473. doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S., Graf E.H., Dahl V., Strain M.C., Yukl S.A., Lysenko E.S., Bosch R.J., Lai J., Chioma S., Emad F. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D., Hermankova M., Pierson T., Carruth L.M., Buck C., Chaisson R.E., Quinn T.C., Chadwick K., Margolick J., Brookmeyer R. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- Folks T.M., Justement J., Kinter A., Dinarello C.A., Fauci A.S. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science. 1987;238:800–802. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- Hattori T., Pack M., Bougnoux P., Chang Z.L., Hoffman T. Interferon-induced differentiation of U937 cells. Comparison with other agents that promote differentiation of human myeloid or monocytelike cell lines. J. Clin. Invest. 1983;72:237–244. doi: 10.1172/JCI110962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich T.J., Hu Z., Li J.Z., Sciaranghella G., Busch M.P., Keating S.M., Gallien S., Lin N.H., Giguel F.F., Lavoie L. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J. Infect. Dis. 2013;207:1694–1702. doi: 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich T.J., Hanhauser E., Marty F.M., Sirignano M.N., Keating S., Lee T.H., Robles Y.P., Davis B.T., Li J.Z., Heisey A. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann. Intern. Med. 2014;161(5):319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y.-C., Shan L., Hosmane N.N., Wang J., Laskey S.B., Rosenbloom D.S., Lai J., Blankson J.N., Siliciano J.D., Siliciano R.F. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–541. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson L., King M.S., Makitalo B., Brannstrom J., Shao W., Maldarelli F., Kearney M.F., Hu W.S., Chen J., Gaines H. Majority of CD4 + T cells from peripheral blood of HIV-1-infected individuals contain only one HIV DNA molecule. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11199–11204. doi: 10.1073/pnas.1107729108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselinova M., Pasternak A.O., De Spiegelaere W., Vogelaers D., Berkhout B., Vandekerckhove L. Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS One. 2014;9:e85999. doi: 10.1371/journal.pone.0085999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird G.M., Eisele E.E., Rabi S.A., Lai J., Chioma S., Blankson J.N., Siliciano J.D., Siliciano R.F. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird G.M., Bullen C.K., Rosenbloom D.I., Martin A.R., Hill A.L., Durand C.M., Siliciano J.D., Siliciano R.F. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J. Clin. Invest. 2015;125(5):1901–1912. doi: 10.1172/JCI80142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leth S., Schleimann M.H., Nissen S.K., Hojen J.F., Olesen R., Graversen M.E., Jorgensen S., Kjaer A.S., Denton P.W., Mork A. Combined effect of Vacc-4x, recombinant human granulocyte macrophage colony-stimulating factor vaccination, and romidepsin on the HIV-1 reservoir (REDUC): a single-arm, phase 1B/2A trial. Lancet HIV. 2016;3:e463–e472. doi: 10.1016/S2352-3018(16)30055-8. [DOI] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnati M.S., Scarlatti G., Gatto F., Salvatori F., Cassina G., Rutigliano T., Volpi R., Lusso P. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat. Protoc. 2008;3:1240–1248. doi: 10.1038/nprot.2008.108. [DOI] [PubMed] [Google Scholar]
- Moon S., Ceyhan E., Gurkan U.A., Demirci U. Statistical modeling of single target cell encapsulation. PLoS One. 2011;6:e21580. doi: 10.1371/journal.pone.0021580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Adema K.W., Bakker M., Jurriaans S., Berkhout B., Cornelissen M., Lukashov V.V. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J. Clin. Microbiol. 2008;46:2206–2211. doi: 10.1128/JCM.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Lukashov V.V., Berkhout B. Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology. 2013;10:41. doi: 10.1186/1742-4690-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz R.J., Trono D., Feinberg M.B., Baltimore D. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model for latency. Cell. 1990;61:1271–1276. doi: 10.1016/0092-8674(90)90691-7. [DOI] [PubMed] [Google Scholar]
- Procopio F.A., Fromentin R., Kulpa D.A., Brehm J.H., Bebin A.G., Strain M.C., Richman D.D., O'Doherty U., Palmer S., Hecht F.M. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine. 2015;2:874–883. doi: 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T.A., Schmeltz Sogaard O., Brinkmann C., Wightman F., Lewin S.R., Melchjorsen J., Dinarello C., Ostergaard L., Tolstrup M. Comparison of HDAC inhibitors in clinical development: effect on HIV production in latently infected cells and T-cell activation. Hum. Vaccin. Immunother. 2013;9:993–1001. doi: 10.4161/hv.23800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T.A., Tolstrup M., Brinkmann C.R., Olesen R., Erikstrup C., Solomon A., Winckelmann A., Palmer S., Dinarello C., Buzon M. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV. 2014;1(1):e13–e21. doi: 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- Richman D.D., Margolis D.M., Delaney M., Greene W.C., Hazuda D., Pomerantz R.J. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- Shalek A.K., Satija R., Shuga J., Trombetta J.J., Gennert D., Lu D., Chen P., Gertner R.S., Gaublomme J.T., Yosef N. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey M., Babic D.Z., Greenough T., Gulick R., Kuritzkes D.R., Stevenson M. Episomal viral cDNAs identify a reservoir that fuels viral rebound after treatment interruption and that contributes to treatment failure. PLoS Pathog. 2011;7:e1001303. doi: 10.1371/journal.ppat.1001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano R.F. What do we need to do to cure HIV infection. Top HIV Med. 2010;18:104–108. [PubMed] [Google Scholar]
- Siliciano J.D., Siliciano R.F. Enhanced culture assay for detection and quantitation of latently infected, resting CD4 + T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol. Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- Strain M.C., Richman D.D. New assays for monitoring residual HIV burden in effectively treated individuals. Curr. Opin. HIV AIDS. 2013;8:106–110. doi: 10.1097/COH.0b013e32835d811b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi S., Neeman T., Jackson R.J., Ranasinghe R., Jack C., Ranasinghe C. Identification of biomarkers to measure HIV-specific mucosal and systemic CD8(+) T-cell immunity using single cell Fluidigm 48.48 Dynamic arrays. Vaccine. 2015;33:7315–7327. doi: 10.1016/j.vaccine.2015.10.085. [DOI] [PubMed] [Google Scholar]
- Varadarajan N., Kwon D.S., Law K.M., Ogunniyi A.O., Anahtar M.N., Richter J.M., Walker B.D., Love J.C. Rapid, efficient functional characterization and recovery of HIV-specific human CD8 + T cells using microengraving. Proc. Natl. Acad. Sci. U. S. A. 2012;109:3885–3890. doi: 10.1073/pnas.1111205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilburn K.M., Mwandumba H.C., Jambo K.C., Boliar S., Solouki S., Russell D.G., Gludish D.W. Heterogeneous loss of HIV transcription and proviral DNA from 8E5/LAV lymphoblastic leukemia cells revealed by RNA FISH:FLOW analyses. Retrovirology. 2016;13:55. doi: 10.1186/s12977-016-0289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.K., Hezareh M., Gunthard H.F., Havlir D.V., Ignacio C.C., Spina C.A., Richman D.D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.