Abstract
Heterozygous mutations in the human POU-homeobox TCF2 (vHNF1, HNF1β) gene are associated with maturity-onset diabetes of the young, type 5, and abnormal urogenital tract development. Recently, pancreas atrophies have been reported in several maturity-onset diabetes of the young type 5 patients, suggesting that TCF2 is required not only for adult pancreas function but also for its normal development. Tcf2-deficient mice die before gastrulation because of defective visceral endoderm formation. To investigate the role of this factor in pancreas development, we rescued this early lethality by tetraploid aggregation. We show that TCF2 has an essential function in the first steps of pancreas development, correlated with its expression domain that demarcates the entire pancreatic buds from the earliest stages. Lack of TCF2 results in pancreas agenesis by embryonic day 13.5. At earlier stages, only a dorsal bud rudiment forms transiently and expresses the transcription factors Ipf1 and Hlxb9 but lacks the key transcription factor involved in the acquisition of a pancreatic fate, Ptf1a, as well as all endocrine precursor cells. Regional specification of the gut also is perturbed in Tcf2-/- embryos as manifested by ectopic expression of Shh and lack of Ihh and Ipf1 in the posterior stomach and duodenum. Our results highlight the requirement of Tcf2 for ensuring both accurate expression of key regulator molecules in the stomach–duodenal epithelium and proper acquisition of the pancreatic fate. This study provides further insights into early molecular events controlling pancreas development and may contribute to the development of cell-replacement strategies for diabetes.
Keywords: diabetes MODY5, homeodomain transcription factor, pancreas development, gut regionalization, tetraploid aggregation
In mammals, the pancreas emerges as ventral and dorsal evaginations from the foregut–midgut junction that subsequently fused to form a complex organ. The signaling molecule Sonic Hedgehog (SHH) demarcates a molecular boundary between the prepancreatic endoderm and adjacent stomach and duodenal anlagen and exerts an inhibitory action on pancreas development (1–3). Genetic studies in mice have identified a hierarchical regulatory network involved in pancreas morphogenesis, with significant and sequential differences between ventral and dorsal pancreas. In the mouse, the dorsal bud appears at embryonic day 9.5 (E9.5) concomitantly with the first differentiated glucagon-producing cells. The homeobox gene Ipf1(Pdx1) is expressed before and during this budding, and all pancreatic cell types derive from IPF1+ progenitors (4, 5). However, in Ipf1-deficient mice, pancreas development is arrested after budding (6, 7), implying that other factors promote pancreas specification. Recently, the transcription factor Ptf1a (P48) has been shown to be essential for the acquisition of a pancreatic fate by undifferentiated ventral foregut endoderm, being required for the specification of the ventral pancreas and robust outgrowth of the dorsal bud. In its absence, ventral pancreas progenitors differentiate into duodenal cells by default (8). By contrast, the homeobox gene Hlxb9 is required only dorsally, for specifying the gut epithelium to a pancreatic fate (9, 10). A key regulator of endocrine development is the basic helix–loop–helix protein Neurogenin3 (Ngn3), which is absolutely required to promote islet cell development (11). The Isl1 gene, which encodes a LIM-homeodomain protein, performs two functions in the developing pancreas. It is initially required in the dorsal mesenchyme for proper exocrine differentiation and later in the pancreatic epithelium for islet survival (12). Downstream of them, other transcription factors are essential for proper pancreatic endocrine differentiation such as Nkx2.2 and Pax6 (13–15). However, the initial stages of pancreatic development occur early in mammalian embryogenesis, and molecular mechanisms governing these first steps remain to be elucidated.
In humans, mutations in the POU-homeobox TCF2 gene are associated with the human disease maturity-onset diabetes of the young type 5, a form of dominantly inherited type II diabetes mellitus characterized by pancreatic beta cell dysfunction at the age of 25 years or younger, nondiabetic early onset renal disease, liver dysfunction, and abnormal urogenital tract development (16–18). In addition to these phenotypes, variable levels of pancreas atrophies have recently been associated with different TCF2 mutations (19, 20). Remarkably, we have recently identified a severe pancreas hypoplasia in two fetuses carrying previously undescribed mutations in the TCF2 gene (A. L. Delezoide, C.H., and S.C., unpublished results). These data, together with the observation that vHnf1(Tcf2)-mutant embryos show underdevelopment of the pancreas in zebrafish (21), strongly suggest a critical function of Tcf2 in pancreas development. However, the molecular bases of these pancreatic phenotypes are poorly understood, as are the Tcf2 target genes involved. In mice, the precise implication of Tcf2 during early organogenesis remains essentially unknown, because Tcf2-deficient embryos die before gastrulation due to defective visceral endoderm formation (22, 23).
In this study, we rescued this early lethality by tetraploid aggregation, by using Tcf2-/- embryonic stem (ES) cells. We observed in these rescued Tcf2-null embryos an absence of the ventral pancreatic bud and an extremely reduced and transient dorsal bud that leads to pancreas agenesis by E13.5. Our results uncover the requirement of Tcf2 for the specification of the ventral pancreas and for proper morphogenesis and differentiation of the dorsal pancreas. They further suggest that Tcf2 also is required for both accurate regionalization of the primitive gut through Hedgehog (Hh) signaling and proper acquisition of the pancreatic fate by regulating Ptf1a expression, thus placing this transcription factor at one of the highest positions in the genetic network that controls pancreas development.
Materials and Methods
Diploid and Tetraploid Chimera. Because chimeric embryos generated with our previously isolated Tcf2-/- and Tcf2+/- ES cells presented neural tube defects that were inherent to the parental cell line (22), we isolated seven previously undescribed ES cell lines (four Tcf2+/- and three Tcf2-/-) from blastocysts obtained after crossing Tcf2-heterozygous mice (129sv background), as described in ref. 24. Tetraploid embryos were generated by electrofusion on a cell fusion instrument (CF-150, BLS Ltd., Budapest; voltage, 80 V; duration, 80 ms; 1 pulse) at the two-cell stage. Tetraploid or diploid chimeric embryos were generated as described in ref. 25. Two four-cell stage CD1 tetraploid embryos or a wild-type (WT) CD1 morula were aggregated with a single loose clump of 15–20 Tcf2-deficient ES cells, cultured in M16 medium (Sigma) up to the blastocyst stage, and implanted into pseudopregnant females. We first confirmed that tetraploid or diploid embryos generated with our Tcf2+/- ES cells were similar to WT or heterozygous Tcf2 embryos, indicating that the phenotype of Tcf2-/- ES cells-derived embryos is specifically due to the lack of TCF2. We used as control embryos in a given litter blastocysts obtained from cultured morulae not aggregated with ES cells and implanted together with ES-cell-aggregated embryos. Because the yield of tetraploid embryos was low, we established optimal conditions to obtain diploid chimera with maximal ES-cell contribution, and we verified that these very highly diploid chimeric embryos displayed the same phenotype as tetraploid embryos. Because we disrupted the Tcf2 gene by inserting the LacZ gene, the relative contribution of WT and mutant cells in ES-cell-derived embryos was evaluated by wholemount X-Gal staining (22). We analyzed here only very highly chimeric and tetraploid embryos, characterized by the presence of exclusively β-gal+ mutant cells in the Tcf2-expressing tissues (defined as Tcf2-/- embryos).
Immunohistochemistry, in Situ Hybridization, and TUNEL. Mouse embryos were fixed in 4% paraformaldehyde and embedded in paraffin. Then, 5-μm sagittal sections were dewaxed, rehydrated, and subjected to microwave antigen retrieval in 10 mM citrate. For immunostaining, we used rabbit anti-Ipf1 (M. German, Hormone Research Institute, San Francisco), mouse anti-glucagon (Sigma), rabbit anti-Hlxb9 (10), mouse anti-Islet-1 (39.4D5 and 40.2D6), mouse anti-Pax6 (Developmental Studies Hybridoma Bank, Iowa City, IA), and rabbit anti-phosphohistone H3 (Upstate Biotechnology, Lake Placid, NY) as primary antibodies, and FITC- and Cyanine3-conjugated (The Jackson Laboratory) as secondary antibodies. For in situ hybridization, we prepared frozen sections from timed embryos, as described in ref. 11. The following cRNA probes were used: Ptf1a (P. Wellauer, Swiss Institute for Experimental Cancer Research, Epalinges, Switzerland); Hnf6 (F. Lemaigre, Université Catholique de Louvain, Brussels); Ngn3 (11); Shh and Ihh (A. P. McMahon, Harvard University, Cambridge, MA); and Ptc (M. Scott, Howard Hughes Medical Institute, Stanford, CA). TUNEL was performed by using the fluorescein cell death detection kit (Roche).
Results and Discussion
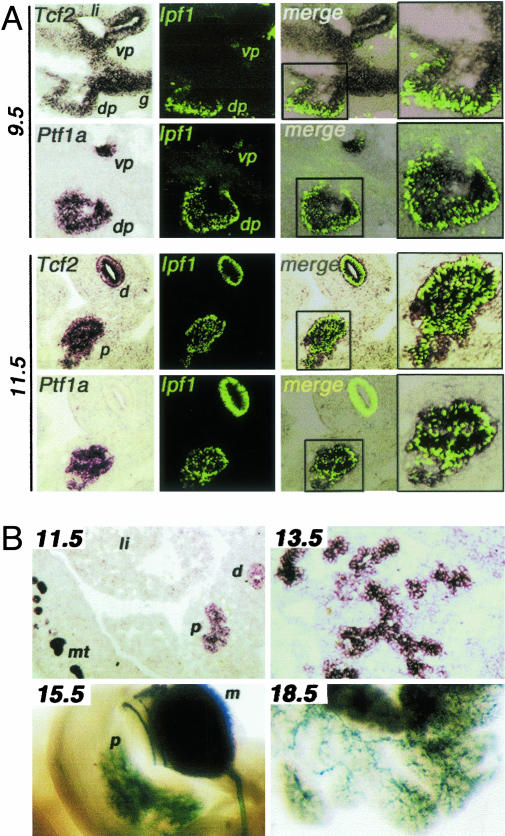
Tcf2 Is Expressed in the Developing Pancreas from Its Early Stages. Tcf2 heterozygous embryos for a null allele with the LacZ gene under the control of regulatory regions of the Tcf2 locus exhibit at E8–E8.5 high β-gal expression in the neural tube and in the entire gut from the foregut–midgut region and by E9.5 in the hepatic, ventral, and dorsal pancreatic primordia (22) (see also Fig. 2 A and C). As the ventral and dorsal pancreatic buds started to form, we observed Tcf2 transcripts at high levels in the entire epithelial cells of the pancreatic buds (Fig. 1A). Interestingly, Tcf2 expression domain included that of Ptf1a and Ipf1, two of the earliest markers of the pancreatic bud (6–8) (Fig. 1 A), as well as early glucagon-expressing cells (data not shown). At E13.5, Tcf2 transcripts were detected in the branched pancreatic epithelium. As the buds grew and fused, Tcf2 appeared more intensely expressed in exocrine ducts, as shown by X-Gal staining of Tcf2+/- embryos at E15.5 and E18.5 (Fig. 1B).
Fig. 2.
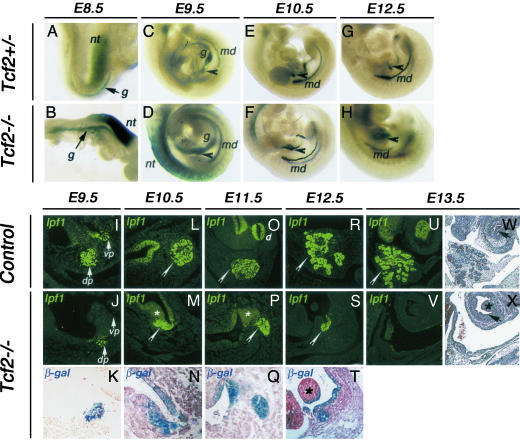
Early defective pancreas development in Tcf2-/- embryos. (A–H) Overall morphology in whole-mount β-gal-stained Tcf2 heterozygous and homozygous mutant embryos between E8.5 and E12.5. Tcf2-/- embryos exhibit, in addition to severe liver hypoplasia and abnormal ureteric branching (L. Lokmane, M. Pares-Fessy, C.H., and S.C., unpublished data), defective pancreas development. β-gal activity is more intensely observed in the neural tube (nt) of E8.5 Tcf2-/- than Tcf2+/- embryos (A and B), as well as in mesonephric ducts (md) and primitive gut (g) at E9.5 (C and D), probably as a result of the presence of two copies of the LacZ gene in mutant embryos and/or a negative autoregulation of Tcf2. The protruding dorsal pancreatic bud externally detected in heterozygous embryos between E9.5 and E12.5 is not observed in homozygous mutant embryos (black arrowhead, C–H). (I–X) Pancreatic bud morphogenesis in sagittal sections of control and Tcf2 homozygous mutant embryos between E9.5 and E13.5. (I–K) IPF1 immunostainings of sagittal sections of whole-mount β-gal-stained embryos reveal ventral and dorsal pancreatic buds in Tcf2+/- embryos, whereas the ventral pancreatic bud is totally absent and only a very reduced dorsal bud is observed in Tcf2-/- embryos at E9.5. vp, ventral pancreas; dp, dorsal pancreas. (L–T) At later stages, the dorsal pancreatic bud (white arrowhead), the duodenum (d), and the posterior stomach are stained by IPF1 in control embryos, but only a remnant pancreatic bud is stained in Tcf2-deficient embryos, which is abnormally close to the posterior stomachal epithelium. Whereas the pancreatic bud exhibits an important growth in control embryos particularly from E12.5, the remnant pancreatic bud in Tcf2-/- embryos regresses by E12.5 and is not further detected at E13.5 (pancreas agenesis) (U–X). (K, N, Q, and T) β-gal staining of the remnant pancreatic bud in sagittal sections of Tcf2 homozygous mutant embryos. β-gal and IPF1-stained sections are counterstained by safranin. β-gal+ mutant cells are detected in the rudiment of the pancreatic bud in Tcf2-/- embryos coexpressing IPF1 and display a broader expression domain including a thickness of the stomachal epithelium between E10.5 and E11.5. Note that the magnification in K, N, Q, and T is higher than in the corresponding IPF1-stained section in J, M, P, and S. (U–X) Stomachal epithelium morphology in E13.5 Tcf2 control and homozygous mutant embryos. Trichromic staining of the IPF1-stained sagittal sections. Arrows indicate the posterior stomachal epithelium, which is surrounded by the stomachal mesenchyme. Whereas the normal posterior stomach exhibits a columnar vacuolized epithelium, the posterior stomach of Tcf2-/- embryos appears squamous and nonvacuolized, as is normally the anterior stomach. In M, P, T, and X, an asterisk indicates thickening of the gastric epithelium.
Fig. 1.
Tcf2 expression in the embryonic pancreas. (A) Demarcation of the entire pancreatic buds at early stages by Tcf2 expression domain. Tcf2 and Ptf1a transcripts are visualized in the ventral and dorsal pancreatic buds in sagittal sections of E9.5 and E11.5 embryos by in situ hybridization (Left) with the corresponding IPF1 immunostaining on the same section (Middle). Merge images at lower and higher magnifications (Right) revealed that Tcf2 expression domain is correlated with Ptf1a expression domain and include Ipf1-expressing cells. Note also that Tcf2 and Ipf1 are coexpressed in the duodenum where Ptf1a is absent. vp, ventral pancreatic bud; dp, dorsal pancreatic bud, li, liver; g, gut; d, duodenum; p, pancreas. (B) Tcf2 expression in the mouse developing pancreas. At E11.5, Tcf2 transcripts are present in the pancreas (p) and duodenum (d) at lower levels than in the mesonephric tubules (mt). li, liver. At E13.5 the pancreatic epithelium is labeled by Tcf2 transcripts. β-gal staining of Tcf2 heterozygous embryos reveals an intense Tcf2 expression in ductal cells at E15.5 and E18.5. m, metanephros.
Thus, in the ventral and dorsal pancreatic anlagen, Tcf2, Ptf1a, and Ipf1 are expressed concurrently, suggesting that TCF2 might control early steps of pancreas differentiation.
Lack of TCF2 Disrupts Early Pancreas Development. Tcf2-deficient mice die before gastrulation due to defective extra-embryonic visceral endoderm formation (22, 23). Therefore, to examine the role of Tcf2 in pancreas development, we generated diploid and tetraploid chimeric mouse embryos by aggregation with β-gal+ Tcf2-deficient ES cells. In tetraploid embryos, 4n cells contribute to extra-embryonic lineages, whereas the resulting fetuses derive exclusively from ES cells. We set up conditions by which very highly chimeric embryos generated by diploid aggregation exhibited the same phenotype as embryos generated by tetraploid aggregation. In this study, we focused on the severe pancreatic phenotype of these two equivalent types of embryos, further defined as Tcf2-/- embryos. In both cases, we confirmed that these embryos essentially were derived from Tcf2-deficient ES cells, as manifested by β-gal staining of Tcf2-expressing tissues (Fig. 2 B, D, F, and H). Tetraploid chimeric embryos generated by aggregation of Tcf2-heterozygous ES cells were phenotypically normal at the stages examined here (data not shown).
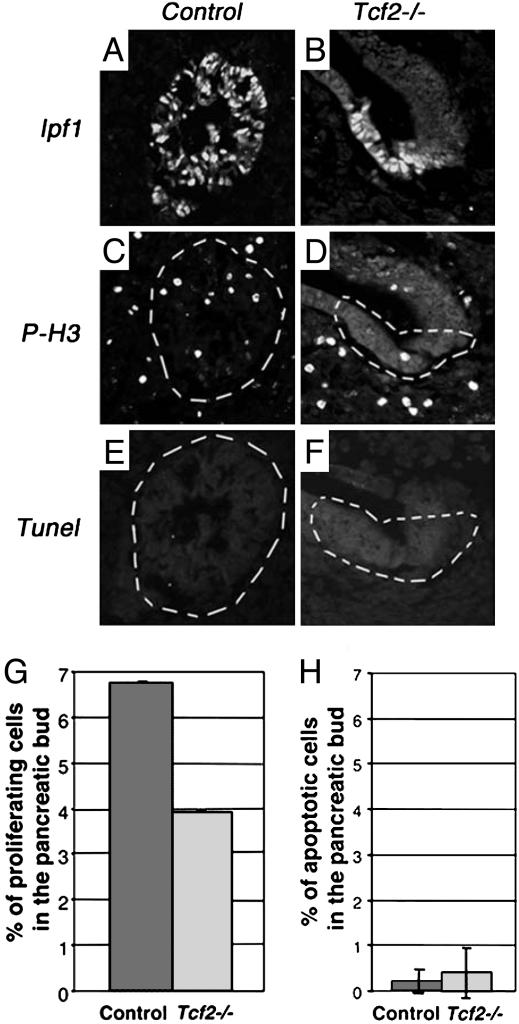
Pancreatic bud formation in Tcf2-/- and control embryos (see Materials and Methods) was analyzed from E9.5 to E13.5, by scoring the expression of Ipf1 (Fig. 2 I, J, L, M, O, P, R, S, U, and V). We verified that all Ipf1+ cells in Tcf2-/- embryos were mutant β-gal+ cells (Fig. 2 K, N, Q, and T). At E9.5, we observed a severely reduced dorsal pancreatic bud (an ≈60% reduction) in Tcf2 mutants, whereas the ventral bud was undetectable (Fig. 2 D and J). The dorsal pancreatic bud rudiment did not grow further, remaining close to the stomach lumen (Fig. 2 M, P, and S), in contrast to control embryos that displayed an important branching phase between E10.5 and E12.5 (Fig. 2 L, O, and R). The pancreatic bud in Tcf2-/- embryos was extremely reduced at E12.5 (Fig. 2S) and became absent at E13.5 (Fig. 2V). In accordance with these observations, by using the mitosis marker phosphorylated histone-H3 (26, 27), we found a lower proliferating rate in the mutant pancreatic bud from E9.5, whereas the number of proliferating cells in the surrounding mesenchyme was not affected (Fig. 3G). TUNEL experiments showed that cells of this remnant dorsal bud did not undergo apoptosis because the percentage of apoptotic cells of Tcf2-/- vs. control pancreatic bud was not significantly different (Fig. 3H), suggesting that these cells were either eliminated by necrosis or recruited to another tissue.
Fig. 3.
Decreased cell proliferation in pancreas of Tcf2-/- embryos. (A and B) The pancreatic bud in control and Tcf2-homozygous mutant embryos is defined by Ipf1 expression domain. (C and D) The mitosis marker antiphosphorylated histone H3 (P-H3) antibody detects proliferating cells in the pancreatic bud (circled by white dashed lines). (E and F) TUNEL experiments show cells in apoptosis in control and Tcf2-mutant pancreas (circled by white dashed lines). (G) The percentage of P-H3+ cells among IPF1+ cells of control and Tcf2-/- pancreatic buds reveals an important cell proliferation decrease in Tcf2-mutant pancreas. (H) The percentage of TUNEL+ cells among IPF1+ cells reveals no significant difference between control and Tcf2-/- pancreatic buds. A total of 13 sections from control (n = 5) and 7 sections from Tcf2-/- (n = 4) of E9.5 and E10.5 embryos were evaluated.
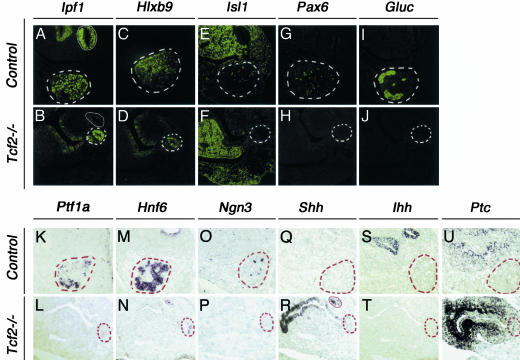
We subsequently examined the expression of early pancreatic markers (28) by immunohistochemical (Fig. 4 A–J) and in situ hybridization (Fig. 4 K–V) analyses in E11.5 sagittal sections. We observed that the Ipf1 dorsally reduced expression domain in mutant embryos also expressed the early pancreatic marker Hlxb9 (Fig. 4 B and D). By contrast, both Ipf1 and Hlxb9 expression were not detected in the presumptive ventral pancreatic bud area. Remarkably, we found no expression of the key transcription factor Ptf1a (8) (Fig. 4L). Moreover, Tcf2-/- mutants displayed a very reduced expression of Hnf6 and no Ngn3 expression in the remnant dorsal pancreatic bud, two factors required for endocrine fate acquisition (11, 29) (Fig. 4 N and P). Consistent with this absence of Ngn3, the earliest known marker of endocrine precursors (11), expressions of Isl1, Pax6, and glucagon were lost (Fig. 4 F, H, and J). Isl1 remained, however, expressed in the mesenchyme (7) (Fig. 4F), a tissue where Tcf2 was not expressed. Thus, endocrine precursors are totally absent in Tcf2-/- pancreatic epithelium.
Fig. 4.
Impaired early pancreatic cell differentiation in Tcf2-/- embryos. (A–J) Immunohistochemical analysis of E11.5 control and Tcf2-/- dorsal pancreatic buds. Anti-Ipf1 (A and B), anti-Hlxb9 (C and D), anti-Isl1 (E and F), anti-Pax6 (G and H), and anti-glucagon (I and J) antibodies were used on sagittal sections of WT and Tcf2-mutant embryos. (A and B) Ipf1 was detected in the remnant dorsal pancreatic bud (marked by dashed lines), but not in the duodenum (circled by a white line), of Tcf2-/- embryos. (C and D) Hlxb9 also is expressed in this bud. (E–J) None of the early endocrine pancreatic markers, Isl1, Pax6, and glucagon, were detected in the dorsal pancreatic epithelium of Tcf2 mutants. (E and F) Isl1, although absent in pancreatic epithelial cell clusters, is still expressed in the mesenchyme. (K–V) In situ hybridization analysis of control and Tcf2-/- E11.5 dorsal pancreatic buds. In contrast to Ipf1, Ptf1a and Ngn3 are not expressed in the remnant dorsal pancreatic bud of Tcf2 mutants (K, L, O, and P), whereas Hnf6 is severely reduced (M and N). Whereas Ihh expression is abolished in Tcf2-/- mutants (S and T), Shh is highly expressed in the anterior stomach with an expanded expression domain in posterior stomach and duodenum (Q and R) and induced Ptc expression (U and V).
Taken together, our results show that Tcf2 controls initial specification of the ventral pancreas and is required for proper proliferation and differentiation of the dorsal pancreas.
Lack of TCF2 Perturbs Regionalization of the Primitive Gut. Because Hh signaling was shown to exert an inhibitory action on pancreatic development (1, 30), we further investigated whether the impairment of pancreatic development in Tcf2-/- embryos could result from a modified expression of either Sonic hedgehog (Shh) or Indian hedgehog (Ihh). Whereas Ihh was expressed at E12 in the caudal epithelium of the stomach and duodenum in WT embryos (Fig. 4S), no Ihh transcripts were detected in Tcf2-/- embryos (Fig. 4T). We propose that Ihh could be a direct target gene of Tcf2, because we found that Ihh is also absent in Tcf2-/- embryoid bodies (31). By contrast, Shh, normally not expressed in the caudal stomach and in the pancreatic bud (Fig. 4Q), was highly ectopically expressed in the stomachal epithelium and duodenum with a rostrocaudal gradient of expression, but remained excluded from the mutant pancreatic bud (Fig. 4R). As expected, the Hh receptor Patched (Ptc) was intensively expressed along the Shh expression domain with the corresponding rostro-caudal expression gradient (Fig. 4V), suggesting that the Hh pathway is active. Interestingly, in E13.5 Tcf2-/- embryos, the posterior stomach exhibited characteristics of the anterior stomach and appeared essentially squamous and non-vacuolized, instead of exhibiting a columnar epithelium with mucin-negative vacuoles as posterior stomach of control embryos (Fig. 2 W and X). A similar anteriorly directed transformation of the posterior stomach resulting from ectopic Shh expression also has been observed in mice carrying mutations of activin receptors ActRIIA and ActRIIB (32). However, Tcf2-/- embryos present a more severe pancreatic phenotype than that of ActRIIA+/- ActRIIB-/- double-mutant embryos, suggesting an additional role of TCF2 in early pancreas development.
We also observed a thickening of the posterior stomach epithelium by E10.5 (Fig. 2 M and P). Subsequently, part of this multilayered stomachal epithelium appeared to be delaminated by E12.5 and remained as clusters of tissue in the lumen of a distended stomach (Fig. 2 T and X). This phenomenon might be caused by the loss of Ihh expression, because a multilayer epithelium is also observed in the colon of Ihh-/- embryos (33).
These results show that loss of Tcf2 function results in a perturbed anteroposterior regionalization of the primitive gut, through a deregulation of Hh signaling.
TCF2 Is a Critical Regulator in the Transcriptional Network That Governs Pancreas Morphogenesis. Our data strongly suggest a critical role for Tcf2 in the orchestratred network of transcription factors and secretory molecules (34) controlling the expansion of endodermal progenitors and their differentiation into pancreatic primordia.
Remarkably, Tcf2-deficient embryos exhibit a very close phenotype to that caused by Ptf1a deficiency, in regard to the absence of a ventral pancreas and a reduced dorsal pancreas. Intriguingly, we identified a TCF1/2-DNA consensus-binding site (ATTAATGTTTAAC) in the Ptf1a promoter at -5,092 bp from the initiation site, within a domain highly conserved between mouse and human, which specifically binds TCF1/2 proteins (data not shown). This finding suggests that TCF2 could regulate Ptf1a expression directly. However, the phenotype of Tcf2-/- embryos is more severe than that of Ptf1a-/- embryos, because in the absence of Ptf1a expression the dorsal pancreas is maintained and relatively developed, with endocrine cells still present (8, 35), indicating that other regulatory factors may contribute to Tcf2 mutant phenotype. In this context, Ipf1 and Ngn3 were previously identified as direct target genes of TCF2 (36, 37). Yet, Ipf1 was still detectable in the Tcf2-/- pancreatic bud rudiment (Fig. 2), implying that TCF2 is not required for Ipf1 initial induction. By contrast, the expression of Ngn3, a gene whose expression was transiently reduced in Hnf6-/- embryos (29, 38), was completely abolished in Tcf2-/- embryos. Because Hnf6 expression also was severely reduced in Tcf2-/- dorsal pancreatic bud, activation of Ngn3 may require the concurrent action of Tcf2 and Hnf6. Recent studies have reported a transient reduction in TCF2 levels in the pancreatic duct cells in Hnf6-deficient embryos from E13.5 to E15.5, leading to the suggestion that Hnf6 is upstream of Tcf2 (38). Nevertheless, the more severe pancreatic phenotype of Tcf2-mutant embryos, compared with that of Hnf6-/- embryos (29), indicates that this transcriptional hierarchy most likely is not involved at earlier stages of pancreas development.
Regulatory circuits also are involved in regional specification of the gut endoderm. One important aspect of this regionalization is the restricted expression of Ipf1 and Shh, essential to permit pancreas development (12). Several studies have suggested that Shh and Ipf1 mutually repress their expression through a regulatory loop in the gut endoderm (1, 21, 30). In correlation with this finding, we found in Tcf2-/- embryos an expanded domain of Shh expression in the posterior stomach and duodenum, whereas the Ipf1 expression domain remained restricted to the rudimentary dorsal pancreatic bud but was absent in posterior stomach and duodenum (Fig. 4). Thus, Tcf2 appears to regulate regional specification of the gut endoderm through the Shh-Ipf1 network.
Taken together, these findings allow us to propose a model highlighting the critical role played by Tcf2 in the control of pancreas development, in relation with the regionalization of the primitive gut (Fig. 5). We propose that in the absence of TCF2, Ptf1a expression is not induced, leading to defective specification of the ventral pancreas and a reduced dorsal pancreas, which is subsequently not maintained because of an altered regionalization of the gut through deregulation of Hh signaling.
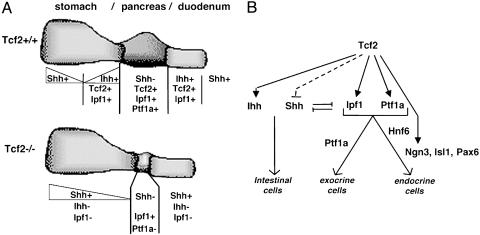
Fig. 5.
Proposed model for Tcf2 function in the development of the pancreas and gut endoderm. (A) Expression domains of the transcription factors Tcf2, Ipf1, and Ptf1a, as well as the signaling molecules Shh and Ihh, in the fore-midgut area of Tcf2+/+ and Tcf2-/- embryos. Tcf2 deficiency leads to an extremely reduced pancreatic bud expressing Ipf1 but missing Ptf1a expression, associated with a perturbed gut regionalization reflected by an expansion of Shh expression domain and an absence of Ihh and Ipf1 expression in stomach and duodenum. +, expressed gene; -, nonexpressed gene. (B) Proposed model of the regulatory network that governs differentiation of pancreatic cells. The diagram illustrates the epistatic relations of genes required for pancreas differentiation, leading to endocrine and exocrine cells. Tcf2 is required early in pancreas development, activating Ptf1a, and regulating the Ipf1-Shh network, with Shh repression vs. Ihh activation. TCF2 also activates Ngn3 in endocrine precursor cells, in agreement with the hypothesis that TCF2-positive cells are the precursors of NGN3 positive cells (38).
This study provides further insight into the early molecular events controlling pancreas development in mice and the function of the transcription factor TCF2 in this process. Our observation of pancreas hypoplasia in two fetuses carrying novel TCF2 mutations (A. L. Delezoide, C.H, and S.C, unpublished data) suggests that decreased levels of TCF2 also perturb normal pancreas growth and function in humans. The role played by Tcf2 in pancreas development thus appears to be conserved during evolution. Then, understanding how Tcf2 together with other regulatory molecules direct early pancreas development in mice may help to elaborate cell-replacement strategies for diabetes mellitus.
Acknowledgments
We thank B. Thorens (Institute of Physiology of Lausanne, Lausanne, Switzerland), M. German, P. Wellauer, F. Lemaigre, A. P. McMahon, M. Scott, and S. Schneider-Maunoury (Unité Mixte de Recherche 7622, Centre National de la Recherche Scientifique, Université Pierre et Marie Curie) for reagents, and J. F. Colas, J. L. Duband, and S. Schneider-Maunoury for comments on the manuscript. This work was supported by Association pour la Recherche sur le Cancer Contracts 5824/3231, Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, and Université Pierre et Marie Curie. C.H. is a recipient of Ph.D. student fellowships from Ministère de la Recherche et de la Technologie and Association pour la Recherche sur le Cancer.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: En, embryonic day n; ES, embryonic stem; Hh, hedgehog; vHnf1, variant Hepatocyte nuclear factor 1; Shh, Sonic Hedgehog; Ihh, Indian Hedgehog; Ngn3, Neurogenin3; Isl1, Islet-1; Hnf6, Hepatocyte nuclear factor 6.
References
- 1.Apelqvist, A., Ahlgren, U. & Edlund, H. (1997) Curr. Biol. 7, 801-804. [DOI] [PubMed] [Google Scholar]
- 2.Hebrok, M., Kim, S. K. & Melton, D. A. (1998) Genes Dev. 12, 1705-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim, S. K., Hebrok, M. & Melton, D. A. (1997) Development (Cambridge, U.K.) 124, 4243-4252. [DOI] [PubMed] [Google Scholar]
- 4.Ohlsson, H., Karlsson, K. & Edlund, T. (1993) EMBO J. 12, 4251-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu, G., Dubauskaite, J. & Melton, D. A. (2002) Development (Cambridge, U.K.) 129, 2447-2457. [DOI] [PubMed] [Google Scholar]
- 6.Offield, M. F., Jetton, T. L., Labosky, P. A., Ray, M., Stein, R. W., Magnuson, M. A., Hogan, B. L. & Wright, C. V. (1996) Development (Cambridge, U.K.) 122, 983-995. [DOI] [PubMed] [Google Scholar]
- 7.Ahlgren, U., Jonsson, J. & Edlund, H. (1996) Development (Cambridge, U.K.) 122, 1409-1416. [DOI] [PubMed] [Google Scholar]
- 8.Kawaguchi, Y., Cooper, B., Gannon, M., Ray, M., MacDonald, R. J. & Wright, C. V. (2002) Nat. Genet. 32, 128-134. [DOI] [PubMed] [Google Scholar]
- 9.Li, H., Arber, S., Jessell, T. M. & Edlund, H. (1999) Nat. Genet. 23, 67-70. [DOI] [PubMed] [Google Scholar]
- 10.Harrison, K. A., Thaler, J., Pfaff, S. L., Gu, H. & Kehrl, J. H. (1999) Nat. Genet. 23, 71-75. [DOI] [PubMed] [Google Scholar]
- 11.Gradwohl, G., Dierich, A., LeMeur, M. & Guillemot, F. (2000) Proc. Natl. Acad. Sci. USA 97, 1607-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahlgren, U., Pfaff, S. L., Jessell, T. M., Edlund, T. & Edlund, H. (1997) Nature 385, 257-260. [DOI] [PubMed] [Google Scholar]
- 13.Sussel, L., Kalamaras, J., Hartigan-O'Connor, D. J., Meneses, J. J., Pedersen, R. A., Rubenstein, J. L. & German, M. S. (1998) Development (Cambridge, U.K.) 125, 2213-2221. [DOI] [PubMed] [Google Scholar]
- 14.St-Onge, L., Sosa-Pineda, B., Chowdhury, K., Mansouri, A. & Gruss, P. (1997) Nature 387, 406-409. [DOI] [PubMed] [Google Scholar]
- 15.Sander, M., Neubuser, A., Kalamaras, J., Ee, H. C., Martin, G. R. & German, M. S. (1997) Genes Dev. 11, 1662-1673. [DOI] [PubMed] [Google Scholar]
- 16.Nishigori, H., Yamada, S., Kohama, T., Tomura, H., Sho, K., Horikawa, Y., Bell, G. I., Takeuchi, T. & Takeda, J. (1998) Diabetes 47, 1354-1355. [DOI] [PubMed] [Google Scholar]
- 17.Lindner, T. H., Njolstad, P. R., Horikawa, Y., Bostad, L., Bell, G. I. & Sovik, O. (1999) Hum. Mol. Genet. 8, 2001-2008. [DOI] [PubMed] [Google Scholar]
- 18.Bingham, C., Ellard, S., Allen, L., Bulman, M., Shepherd, M., Frayling, T., Berry, P. J., Clark, P. M., Lindner, T., Bell, G. I., et al. (2000) Kidney Int. 57, 898-907. [DOI] [PubMed] [Google Scholar]
- 19.Bellanne-Chantelot, C., Chauveau, D., Gautier, J., Dubois-Laforgue, D., Clauin, S., Beaufils, S., Wilhelm, J. M., Boitard, C., Noel, L. H., Velho, G. & Timsit, J. (2004) Ann. Intern. Med. 140, 510-517. [DOI] [PubMed] [Google Scholar]
- 20.Barbacci, E., Chalkiadaki, A., Masdeu, C., Haumaitre, C., Lokmane, L., Loirat, C., Cloarec, S., Talianidis, I., Bellanne-Chantelot, C. & Cereghini, S. (2004) Hum. Mol. Genet. 13, 3139-3149. [DOI] [PubMed] [Google Scholar]
- 21.Sun, Z. & Hopkins, N. (2001) Genes Dev. 15, 3217-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbacci, E., Reber, M., Ott, M., Breillat, C., Huetz, F. & Cereghini, S. (1999) Development (Cambridge, U.K.) 126, 4795-4805. [DOI] [PubMed] [Google Scholar]
- 23.Coffinier, C., Thepot, D., Babinet, C., Yaniv, M. & Barra, J. (1999) Development (Cambridge, U.K.) 126, 4785-4794. [DOI] [PubMed] [Google Scholar]
- 24.Hogan, B., Beddington, R. S., Costantini, F. & Lacy, E. (1994) Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 25.Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W. & Roder, J. C. (1993) Proc. Natl. Acad. Sci. USA 90, 8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmiesing, J. A., Gregson, H. C., Zhou, S. & Yokomori, K. (2000) Mol. Cell. Biol. 20, 6996-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bort, R., Martinez-Barbera, J. P., Beddington, R. S. & Zaret, K. S. (2004) Development (Cambridge, U.K.) 131, 797-806. [DOI] [PubMed] [Google Scholar]
- 28.Wilson, M. E., Scheel, D. & German, M. S. (2003) Mech. Dev. 120, 65-80. [DOI] [PubMed] [Google Scholar]
- 29.Jacquemin, P., Durviaux, S. M., Jensen, J., Godfraind, C., Gradwohl, G., Guillemot, F., Madsen, O. D., Carmeliet, P., Dewerchin, M., Collen, D., et al. (2000) Mol. Cell. Biol. 20, 4445-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hebrok, M., Kim, S. K., St Jacques, B., McMahon, A. P. & Melton, D. A. (2000) Development (Cambridge, U.K.) 127, 4905-4913. [DOI] [PubMed] [Google Scholar]
- 31.Haumaitre, C., Reber, M. & Cereghini, S. (2003) J. Biol. Chem. 278, 40933-40942. [DOI] [PubMed] [Google Scholar]
- 32.Kim, S. K., Hebrok, M., Li, E., Oh, S. P., Schrewe, H., Harmon, E. B., Lee, J. S. & Melton, D. A. (2000) Genes Dev. 14, 1866-1871. [PMC free article] [PubMed] [Google Scholar]
- 33.Van den Brink, G. R., Bleuming, S. A., Hardwick, J. C., Schepman, B. L., Offerhaus, G. J., Keller, J. J., Nielsen, C., Gaffield, W., van Deventer, S. J., Roberts, D. J. & Peppelenbosch, M. P. (2004) Nat. Genet. 36, 277-282. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, M. & Melton, D. (2003) Curr. Opin. Genet. Dev. 13, 401-407. [DOI] [PubMed] [Google Scholar]
- 35.Krapp, A., Knofler, M., Ledermann, B., Burki, K., Berney, C., Zoerkler, N., Hagenbuchle, O. & Wellauer, P. K. (1998) Genes Dev. 12, 3752-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerrish, K., Cissell, M. A. & Stein, R. (2001) J. Biol. Chem. 276, 47775-47784. [DOI] [PubMed] [Google Scholar]
- 37.Lee, J. C., Smith, S. B., Watada, H., Lin, J., Scheel, D., Wang, J., Mirmira, R. G. & German, M. S. (2001) Diabetes 50, 928-936. [DOI] [PubMed] [Google Scholar]
- 38.Maestro, M. A., Boj, S., Luco, R. F., Pierreux, C. E., Cabedo, J., Servitja, J. M., German, M. S., Rousseau, G. G., Lemaigre, F. P. & Ferrer, J. (2003) Hum. Mol. Genet. 12, 3307-3314. [DOI] [PubMed] [Google Scholar]