Abstract
The luciferase of Lingulodinium polyedrum, a marine bioluminescent dinoflagellate, consists of three similar but not identical domains in a single polypeptide. Each encodes an active luciferase that catalyzes the oxidation of a chlorophyll-derived open tetrapyrrole (dinoflagellate luciferin) to produce blue light. These domains share no sequence similarity with any other in the GenBank database and no structural or motif similarity with any other luciferase. We report here the 1.8-Å crystal structure of the third domain, D3, at pH 8, and a mechanism for its activity regulation by pH. D3 consists of two major structural elements: a β-barrel pocket putatively for substrate binding and catalysis and a regulatory three-helix bundle. N-terminal histidine residues previously shown to regulate activity by pH are at the interface of the helices in the bundle. Molecular dynamics calculations indicate that, in response to changes in pH, these histidines could trigger a large molecular motion of the bundle, thereby exposing the active site to the substrate.
Keywords: dinoflagellate, β-barrel, pH regulation, lingulodidium, histidine
Luminous dinoflagellates are responsible for much of the bioluminescence, sometimes incorrectly called phosphorescence, in ocean surface waters. One of the species, Lingulodinium polyedrum (formerly known as Gonyaulax polyedra), emits blue-light flashes (λmax 475 nm) after mechanical or electrical stimulation (1, 2). Light is emitted from scintillons, specialized organelles formed as outpocketings from the cytoplasm into the acidic vacuole (3, 4).
The scintillons contain the three components required for light emission: luciferase (LCF) (5), its tetrapyrrole substrate, called dinoflagellate luciferin (LH2) (Fig. 1A) (6), and luciferin-binding protein (LBP) (7–9). The L. polyedrum LCF comprises three homologous domains within a single molecule, each with an active site preceded by an N-terminal sequence of ≈100 aa of unknown function (5). The full-length LCF and each of the individual domains are most active at pH 6.3, and there is very little activity at pH 8.0. LBP binds luciferin at pH 8.0 but not at pH 6.3 (10); upon acidification of the scintillon, the luciferin is therefore released for reaction with an activated LCF. Upon mechanical stimulation, an action potential in the vacuolar membrane triggers flashing, postulated to act by opening voltage-gated channels in the scintillon membrane, thus allowing protons from the acidic vacuole to enter and lower the pH inside the scintillons to give a flash (11, 12).
Fig. 1.
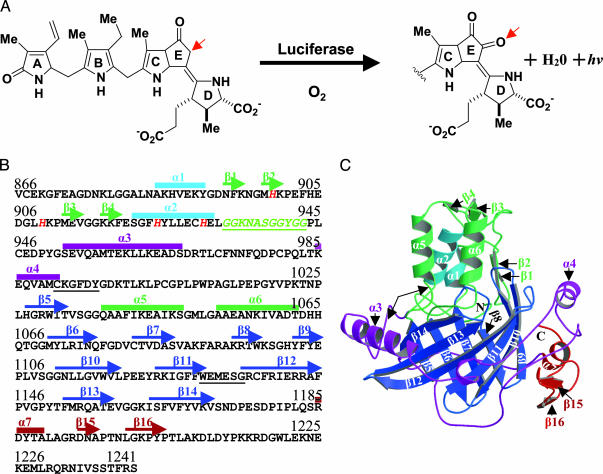
Biochemistry of the reaction and the amino acid sequence and three-dimensional structure of LCF D3. (A) Bioluminescent reaction of dinoflagellate luciferin, a chlorophyll-like open tetrapyrrole. The arrow shows the position of enzymatic oxidation. (B) The sequence of LCF D3 with regions of α-helix (α) and β-strand (β) annotated. The Gly-rich sequence (italicized and underlined) connects the N-terminal subdomain to the β-barrel comprising strands β5to β14. The four N-terminal histidines (899, 909, 924, and 930) are shown in red and italicized. Two underlined six-residue sequences show the beginning and end of the highly conserved region (991–1136) found in LCFs of all seven species examined (23). (C) A ribbon diagram of the crystal structure of D3 with helices and β-strands numbered and color coded as in B. The two N-terminal helices (light blue) and the helix–loop–helix motif (green) are at the top, and the β-barrel (dark blue) is below. Ribbon diagrams were produced by using molscript (42) and raster3d (43).
Whereas each of the three domains has a pH–activity profile similar to that of the full-length molecule, domains truncated by ≈60 amino acid residues at the N terminus have significantly higher pH 8.0/6.3 activity ratios. Previous studies have implicated four intramolecularly conserved histidines in the N-terminal region of each domain in this effect, because their mutation to alanine greatly increases the relative activity at pH 8.0 (13). It was thus concluded that these histidines, although not needed for catalysis, play a role in the regulation of LCF activity.
We report here the crystal structure of domain 3 (D3) of L. polyedrum LCF at pH 8. In this structure, a β-barrel pocket, putatively for substrate binding and catalysis, can be identified by comparison with the structures of human muscle fatty-acid-binding protein (M-FABP)–fatty acid complexes and insect bilin-binding protein (BBP)–biliverdin complex (14, 15). The access to the interior of the pocket is restricted by a three-helix bundle, but protonation of the four histidines implicated in pH regulation is postulated to alter the conformation of the bundle and provide entry to an enlarged pocket.
Materials and Methods
Protein Expression and Purification. Expression and crystallization of D3 have been described in ref. 16. For expression of the selenomethionine derivative (17), BL21(DE3) cells transformed with a construct bearing D3 in pQE30 (Qiagen, Valencia, CA) were cultured overnight at 37°C in 25 ml of LB medium. Pelleted cells were resuspended in 25 ml of M9 minimal medium, aliquoted in four 4-liter flasks containing 1 liter of minimal medium supplemented with 8 ml of 50% glucose, 1 ml of thiamin (5 mg/ml), 1 ml of 1 M MgSO4, and 0.1 ml of 1 M CaCl2, and grown at 37°C. At OD600 = 0.4, the temperature was reduced to 22°C, and 10 ml of an amino acid mixture, consisting of 10 mg/ml each of lysine hydrochloride, threonine, and phenylalanine and 5 mg/ml each of leucine, isoleucine, and valine (Sigma), was added, along with 2 ml of 25 mg/ml dl-selenomethionine (Acros, Fisher Scientific). The cells were grown overnight at 22°C with 0.5 mM isopropyl β-d-thiogalactoside, pelleted by centrifugation, and stored at -80°C until they were used.
Histidine-tagged D3 was affinity-purified by using nickel-chelating matrix. The cell pellet from a 1-liter culture was resuspended and disrupted by passing it twice through a French press at 1,200 psi (1 psi = 6.89 kPa). After centrifugation, the supernatant was filtered through a 0.45-μm syringe filter, applied to a nickel metal chelate column (Pharmacia), run at 4°C with 50 mM sodium phosphate buffer (pH 8.0) containing 300 mM NaCl and 10 mM imidazole chloride, and eluted with 250 mM imidazole. The peak protein fractions were pooled and desalted immediately on a GF-25 (Pharmacia) column into the tobacco etch virus (TEV) cleavage buffer [50 mM Tris·HCl buffer (pH 8.0)/10 mM NaCl/1 mM DTT]. Histidine-tagged TEV protease (Invitrogen) was added to a final concentration of 0.1 mg/ml and incubated overnight at 4°C. The reaction mixture was diluted and desalted in metal chelate-binding buffer and run through a nickel metal chelate column to remove the tagged protein and TEV protease. The protein in the flowthrough was concentrated by using an Amicon YM-10 membrane and a Millipore 10K filter device and, finally, passed over a GF75 (Pharmacia) size-exclusion column to remove any aggregates. Dynamic light scattering was used to determine that the protein for crystallization was monodisperse.
Crystallography. Crystals of the selenomethionine derivative of the D3 LCF were prepared by vapor diffusion of a protein solution containing 10 mg/ml selenomethionine D3 in 10% glycerol (vol/vol)/25 mM NaCl/20 mM Tris·HCl (pH 8.0) against a reservoir solution containing 18–20% (wt/vol) polyethylene glycol 2,000 methyl ether and 100 mM (2-hydroxyethyl)piperazine-N-(3-propanesulfonic acid) (pH 8.0). Drops were formed by mixing equal amounts (3 μl) of protein and reservoir solution. Crystals appeared within 3 days and grew to a final size of 0.8 × 0.1 × 0.05 mm8. The crystals are in space group P212121 with a = 58.86, b = 63.98, and c = 95.76 Å and contain one molecule per asymmetric unit with a solvent content of 46%. Crystals were cryopreserved in a solution created by adding 20% (vol/vol) glycerol to the reservoir solution, mounted in 1 mM cryoloops (Hampton), and frozen directly in the nitrogen cold stream (133 K). Multiwavelength anomalous dispersion data were measured at three wavelengths near the Se K edge by using an ADSC (Poway, CA) Quantum-210 charge-coupled device detector at beamline F2 of MacCHESS (Cornell High Energy Synchrotron Source). denzo/scalepack (18) were used for data processing and solve (19) was used to find six of eight potential selenomethionine-substitution sites and calculate experimental phases to 1.8-Å resolution. The experimental phases were used in conjunction with resolve (20) to automatically fit 290 of 375 possible residues. Several gaps and turns were fitted manually by using turbo-frodo (21), and the initial model was refined with cns (22) against all data in the remote-wavelength data set from 50 to 1.8 Å. Methionine residues were modeled as selenomethionine. The density is continuous from residue 868 to residue 1218 (numbering corresponds to the full-length LCF). Water molecules were assigned to 3σ peaks in the observed structure factor (Fo) minus calculated structure factor (Fc) difference map and were required to have at least 1σ 2Fo - Fc density and form a hydrogen bond with the protein or other water molecules.
Molecular Dynamics. Calculations were performed by using sybyl 7.0 (Tripos Associates, St. Louis) with histidine residues 899, 909, 924, and 930 replaced by alanines and hydrogen atoms added. Gasteiger–Hückel charges were calculated with a single layer of water molecules added, and 100 cycles of Powell minimization were performed. By using the tripos force field and a dielectric constant of 1.0, the model was equilibrated for 1,000 cycles (1 ps per cycle) of molecular dynamics at 50, 100, and 200 K and then subjected to 10,000 cycles at 300 K.
Results
Overall Structure. The crystal structure of L. polyedrum D3 LCF was solved by multiwavelength anomalous dispersion phasing from a single selenomethionyl crystal (Table 1). The model obtained contains residues 868-1218 and 133 water molecules. The complete sequence of D3 comprises residues 866-1241, where the numbers refer to the full-length LCF. The 23-aa C terminus was disordered and not visible in the electron density, even though its presence was confirmed by mass spectrometry. The D3 sequence is given in Fig. 1B with regions of secondary structure noted; there are a total of seven α-helices (α1 to α7) and 16 β-strands (β1 to β16), organized into two prominent subdomains: a regulatory region consisting primarily of a three-helix bundle at the top (green and light blue) and a β-barrel below (Fig. 1C) (dark blue) comprised of 10 antiparallel β-strands arranged in a continuous +1 topology labeled β5 to β14. The conformations of all nonglycine residues were found to lie within allowed regions of the Ramachandran plot.
Table 1. X-ray diffraction data.
| Se remote | Se peak | Se inflection | |
|---|---|---|---|
| Data collection statistics | |||
| Wavelength, Å | 0.9949 | 0.9791 | 0.9793 |
| Resolution, Å | 30–1.8 | 30–1.8 | 30–1.8 |
| Completeness, %* | 96.1(77.2) | 97.8(86.4) | 97.3(83.4) |
| Rsym, %† | 4.7(23.8) | 6.4(25.9) | 6.3(26.5) |
| 〈I/σI〉 | 20.4(3.0) | 21.1(3.4) | 19.6(3.0) |
| Total observations | 124,738 | 147,306 | 146,745 |
| Unique reflections | 35,921 | 36,957 | 36,802 |
| Average redundancy | 3.5 | 4.0 | 4.0 |
| Overall FOM (solve) | 0.45 | ||
| After solvent flip (resolve) | 0.59 | ||
| Refinement statistics | |||
| R factor‡ | 0.199 | ||
| Rfree,§ (8% test set) | 0.230 | ||
| Geometry | |||
| rmsd bonds, Å | 0.01 | ||
| rmsd angles, ° | 1.5 | ||
| rmsd dihedrals, ° | 24.9 | ||
| Average B factors, Å2 | 23.3 |
FOM, figure of merit; rmsd, rms deviation.
Completeness represents all data from 30–1.8 Å (1.9–1.8 Å)
Rsym = Σhkl|I – 〈I〉|/ΣhklI, where I is the observed intensity and |I| is the average intensity obtained from multiple observations of symmetry-related reflections
R = Σhkl|Fo – Fc|/Σhkl|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively
Rfree = Σtest set‖Fo|–|Fc‖/Σtest set|Fo|
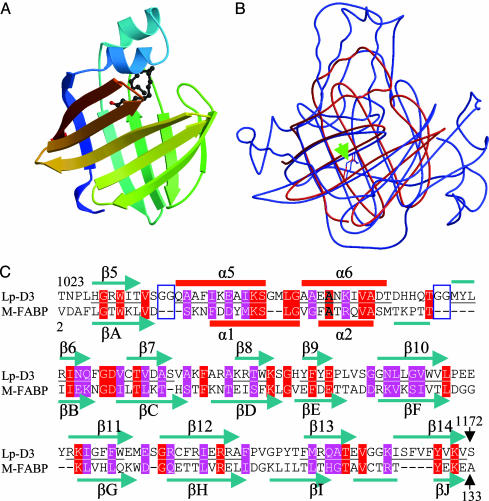
The Core of the Structure Is a 10-Stranded β-Barrel. A blast search with the sequence of D3 indicated a close similarity to only six other dinoflagellate LCFs (23). However, when the coordinates of D3 were compared to published structural data by using dali (24), a structural homology was found with M-FABP (protein database code 1HMT; Fig. 2A), a 10-stranded β-barrel, also referred to as the β-clam (14, 25). These 10-stranded β-barrels are grouped with lipocalins, which are 8-stranded β-barrels, known for binding hydrophobic molecules (26). By virtue of its enzymatic activity, which other members lack, the D3 structure represents a previously undescribed class of the lipocalin family. In addition, D3 also shares a low but appreciable structural homology with the cellular retinoic-acid-binding proteins and insect Pieris brassicae BBP, which have FABP-like structure containing 8-stranded β-barrels.
Fig. 2.
Structural similarity of D3 to FABP. (A) A ribbon diagram of M-FABP with a bound stearate (14). (B) Superposition of LCF D3 (blue) and M-FABP (red). The D3 and FABP helix–loop–helices differ, whereas the β-barrels are closely aligned. An arrow shows the location of residue 1142. (C) Alignment of primary and secondary sequences of D3 and M-FABP within the β-barrel region. Red and purple residues are identical or similar, respectively, in the two sequences. Two Gly-Gly sequences (boxes) are present in D3 but absent in M-FABP. β-Strands 5–14 in D3 correspond to strands A–J in M-FABP.
Relationship of the Three-Helix Bundle with the β-Barrel. In D3, the structure formed from the N-terminal 67 aa comprises two helices (α1 and α2) and two short two-stranded antiparallel β-hairpins (β1 and β2 and β3 and β4) (Fig. 1 B and C) and is stabilized by a small hydrophobic core and a highly organized hydrogen-bonding structure. It is tethered to the rest of D3 by the glycine-rich sequence, GGKNASGGYGG (Fig. 1B underlined and Fig. 1C arrows at left).
Following the N-terminal structure is a long linker (residues 944-1026, thus including α3 and α4), which partially surrounds the β-barrel. The chain continues into the β-barrel (residues 1027–1170), interrupted only between β5 and β6 by a helix–loop–helix motif formed by α5 and α6 located at the top of the β-barrel. This motif interacts with the N-terminal helix (α2), forming a loosely associated three-helix bundle that blocks access to a cavity within the β-barrel, the putative ligand-binding site. Helices α5 and α6 are arranged antiparallel to each other and are not conformationally related to the helix–loop–helix calcium-binding or helix–turn–helix DNA-binding motifs. After the β-barrel, residues 1171–1218 are ordered and pack against the outer surface of the barrel and continue to an aspartic acid residue (D1218), which forms a salt bridge with an arginine (R1098), beyond which the last 23 aa in the peptide chain are not visible in the electron density, although they are present in the crystal.
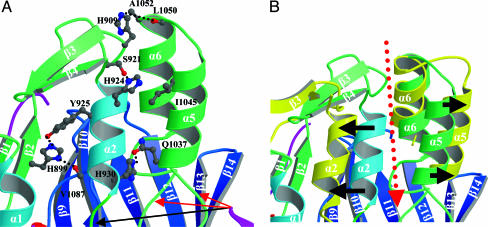
Positions of Four Histidines Involved in pH Regulation. In the D3 structure, the four N-terminal histidines are positioned (Fig. 3A) so that they stabilize the helix–loop–helix at pH 8 and cause a change in its conformation at pH 6.3. H899 acts as a core residue in the N-terminal subdomain, forming hydrogen bonds with Y925 and another to the main-chain carbonyl of V1087. This valine is probably an important residue, because it is located on a short turn of the β-barrel and might serve to anchor the N-terminal subdomain to the β-barrel. H909 is in van der Waals contact with A1052 and forms a hydrogen bond with the main-chain carbonyl of L1050, both of which lie in the loop of the helix–loop–helix motif. H924 forms a hydrogen bond with S921 and is also in van der Waals contact with I1045 of that motif. Last, H930 forms a hydrogen bond with Q1037 of the motif and is resting in a hydrophobic pocket created by A1088, A1038, and M1070.
Fig. 3.
Enlarged views of the flexible regulatory three-helix bundle rotated 90° counterclockwise from Fig. 1C. (A) Structure at pH 8 showing the helix bundle and the four histidines involved in pH regulation. Three arrows point to Gly-Gly residues in D3; shorter red arrows are the pair absent in the FABP sequence. For clarity, residues A1088, A1038, and M1070 are not shown. (B) Structure after molecular dynamics with four histidines replaced by alanines, showing displaced positions of α2, α5, and α6A (yellow-brown) superposed on the starting structure. The black arrows indicate movement of the helices away from each other to open a solvent channel (red arrow) to the active site.
Discussion
Comparison with Other LCFs. L. polyedrum LCF is not homologous in either its sequence or structural motifs to any of the other structurally known types of LCF, namely bacterial (27), firefly (28), or coelenterate LCFs (29, 30). This finding is not surprising, given that, aside from the fact that all involve the oxidation by molecular oxygen of a luciferin substrate (all of which are very different) to form a peroxide intermediate, the reactions are unrelated biochemically (31).
Comparing D3 with M-FABP. The β-barrel (residues 1027–1170) of D3 is most closely related to the human M-FABP (14) with 2.1-Å rms deviation for the main-chain residues comprising the β-barrel (Fig. 2B). Amino acid sequence alignment between the D3 and FABP β-barrels reveals a 19% identity and 38% similarity (Fig. 2C). Whereas both structures contain a helix–loop–helix motif that connects the first two β-strands of the β-barrel (βA and βB in FABP; β5 and β6 in D3) and forms a lid over it, their sequences and conformations are quite different. In the D3 structure, which is inactive at pH 8, these helices are in closer alignment with the axis of the barrel and block access to the interior of the barrel (Fig. 3A). Also, a pair of Gly-Gly sequences, which are absent in the FABP structure, lie at the N and C junctions of the helix–loop–helix to strands β5 and β6 of the β-barrel (two shorter arrows, Fig. 3A and Fig. 2C) (boxed). Gly-Gly sequences offer flexibility to mediate interactions between domains (32, 33), suggesting that the helix–loop–helix is far more mobile in D3 than in M-FABP. In fact, the residues of the N-terminal domain and the helix–loop–helix have higher overall temperature (B) factors (39 Å2) when compared with the rest of the protein (23 Å2). It is also interesting that surrounding the D3 barrel is an extensive set of coils, which may serve as interfaces mediating intramolecular interactions with the two other LCF domains (D1, D2) and/or LBP, or to stabilize the barrel structure.
Comparing D3 with Other Proteins Binding Open Tetrapyrroles. Dinoflagellate LCF and LBP are the only known proteins interacting with tetrapyrroles derived from chlorophylls. However, several other proteins use heme-type open tetrapyrroles as either substrates or ligands. The crystal structures of several of these proteins have been solved with or without the ligand (15, 34–38). Among them, only insect BBP has an 8-stranded β-barrel, similar to the 10-stranded β-barrel in D3 (15). In BBP, the B and C pyrrole rings of bilin are bound inside the barrel, with the carboxyl groups of the A and D rings exposed on the surface to solvent. Although BBP is an eight-stranded lipocalin, it has a significantly more hydrophobic cavity than does D3, indicating that the binding mode of bilin is different from that of luciferin.
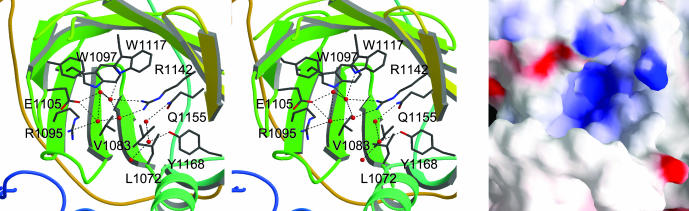
Putative Active Site of LCF. In the D3 structure at pH 8, the cavity within the β-barrel is occupied by seven water molecules, which participate in a hydrogen-bonding network with many of the charged residues on one side of the cavity, including H1065, R1095, E1105, K1125, R1142, Q1155, and Y1168 (Fig. 4). Residues W1097 and W1117 are oriented with the indole nitrogen of the side chain pointing into the cavity and engaged in hydrogen bonds with water. Residues L1072, I1074, V1083, A1085, F1090, and F1103 form a hydrophobic patch on the other wall of the cavity. The presence of polar residues in the interior of the barrel is suggestive of an active site, as it is energetically unfavorable to bury several charged residues.
Fig. 4.
Details of the proposed active site. (Left) Stereoview of putative active site in the β-barrel, rotated 90° counterclockwise from Fig. 1C and tipped forward from the plane of the paper by 90°. (Right) An electrostatic potential surface of the active site with blue (+15 kT) and red (-15 kT) from the same view as Left.
R1142 is the only residue in the putative active site of D3 that is conserved in all FABP family members, as well as in all seven dinoflagellate LCFs (23). The role of this residue in the FABPs is to form water-mediated or direct hydrogen bonds to the carboxylate of the bound fatty acid ligand. Likewise, R1142 of D3 may bind the carboxylate of the luciferin (Fig. 4).
Proposed Mechanism for pH Regulation of LCF Activity. In all FABP members, the ligand-binding site is contained within the β-barrel (volume ≈1,100 Å3), with openings to the outside determined by the position of the helix–loop–helix at the top of the β-barrel (14). Whereas there is a significant pocket within the structure of the D3 barrel at pH 8 (volume ≈174 Å3), there is neither enough room to accommodate the tetrapyrrole [volume ≈564 Å3, as estimated by grasp (39)], nor an opening from the outside. A significant conformational change must thus occur to provide access to and space for a ligand in the active site.
The fact that replacement of four histidines by alanines restores full activity at pH 8 suggests the involvement of the unprotonated histidine residues in a network of hydrogen bonds that blocks substrate access to the active site. The x-ray structure supports this hypothesis. The four histidines make contacts with and stabilize the three-helix bundle that blocks entry to the putative active site (Fig. 3A). Disruption of such interaction between the helices within the bundle by protonation (as in LCF at pH 6.3), or by replacement of the four His residues by Ala, would be expected to cause the helices to move and open access to the catalytic site, as confirmed by molecular dynamics calculations. Starting with the x-ray coordinates but with all four His residues replaced by Ala, H2 and H5 are separated by 10 Å before simulation but by 21 Å after simulation, thus opening the solvent channel (Fig. 3B). The rest of the protein remained stable and deviated <1 Å from the starting model.
Three Gly-Gly sequences, one within the N-terminal helix (G933-G934) and two in the helix–loop–helix motif (G1035–1036 and G1068–1069), could serve as hinges about which the chains rotate (Fig. 3A). Most significantly, a channel forms between the N-terminal helix α2 and the helix–loop–helix, providing solvent access to the active site (Fig. 3B). During the simulation, the active site pocket expands to 693 Å3 (including the solvent channel), which is large enough to accommodate the luciferin substrate (40). In some respects this unusual mechanism of pH-modulation of activity is analogous to that described in the case of low-density lipoprotein receptors, which undergo a tertiary conformational change mediated by histidine in response to low pH (41).
In perspective, it is important to recall that D3 is only a part of the full-length LCF, and the LCF itself is only one component of what must be a much larger, multimeric complex contained within the scintillons, including LBP, presumably positioned favorably for the rapid transfer of LH2 to LCF. Aside from membrane proteins, such as putative proton channels, scintillons contain only these two proteins (8), which can be seen to be already associated in the cytoplasm before their envelopment by the vacuolar membrane (3, 9). How LCF and LBP are arranged at the structural level in scintillons to accomplish the feat of flashing remains to be determined.
Acknowledgments
We thank Walter Pangborn and the staff of the Cornell High Energy Synchrotron Source (CHESS) for help during data collection, George DeTitta and Joe Luft for the use of their crystallization robot, and Dr. Thérèse Wilson for discussions. This work was supported in part by National Science Foundation Grant MCB-0343407 and Office of Naval Research Grants N00014-03-1-0173 (to J.W.H.) and N00014-02-1-0188 (to L.W.S.).
Abbreviations: LCF, luciferase; D3, domain 3 of LCF; LBP, luciferin-binding protein; LH2, luciferin; M-FABP, muscle fatty-acid-binding protein; BBP, bilin-binding protein.
Data deposition: The molecular coordinates for the D3 structure have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1VPR).
References
- 1.Hastings, J. W. (1986) in Light Emission by Plants and Bacteria, eds. Govindjee, J. A. & Fork, D. (Academic, New York), pp. 363-398.
- 2.Hastings, J. W. (2001) in Cell Physiology, ed. Sperelakis, N. (Academic, New York), pp. 1115-1131.
- 3.Nicolas, M. T., Nicolas, G., Johnson, C. H., Bassot, J. M. & Hastings, J. W. (1987) J. Cell. Biol. 105, 723-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fogel, M., Schmitter, R. E. & Hastings, J. W. (1972) J. Cell Sci. 11, 305-317. [DOI] [PubMed] [Google Scholar]
- 5.Li, L., Hong, R. & Hastings, J. W. (1997) Proc. Natl. Acad. Sci. USA 94, 8954-8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura, H., Kishi, Y., Shimomura, O., Morse, D. & Hastings, J. W. (1989) J. Am. Chem. Soc. 111, 7607-7611. [Google Scholar]
- 7.Morse, D., Pappenheimer, A. M., Jr., & Hastings, J. W. (1989) J. Biol. Chem. 264, 11822-11826. [PubMed] [Google Scholar]
- 8.Desjardins, M. & Morse, D. (1993) Biochem. Cell Biol. 71, 176-182. [DOI] [PubMed] [Google Scholar]
- 9.Nicolas, M. T., Morse, D., Bassot, J. M. & Hastings, J. W. (1991) Protoplasma 160, 159-166. [Google Scholar]
- 10.Fogel, M. & Hastings, J. W. (1971) Arch. Biochem. Biophys. 142, 310-321. [DOI] [PubMed] [Google Scholar]
- 11.Eckert, R. & Sibaoka, T. (1968) J. Gen. Physiol. 52, 258-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fogel, M. & Hastings, J. W. (1972) Proc. Natl. Acad. Sci. USA 69, 690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, L., Liu, L., Hong, R., Robertson, D. & Hastings, J. W. (2001) Biochemistry 40, 1844-1849. [DOI] [PubMed] [Google Scholar]
- 14.Young, A. C., Scapin, G., Kromminga, A., Patel, S. B., Veerkamp, J. H. & Sacchettini, J. C. (1994) Structure 2, 523-534. [DOI] [PubMed] [Google Scholar]
- 15.Huber, R., Schneider, M., Mayr, I., Muller, R., Deutzmann, R., Suter, F., Zuber, H., Falk, H. & Kayser, H. (1987) J. Mol. Biol. 198, 499-513. [DOI] [PubMed] [Google Scholar]
- 16.Liu, L., Im, H., Cegielski, M., LeMagueres, P., Schultz, L. W., Krause, K. L. & Hastings, J. W. (2003) Acta Crystallogr. D 59, 761-764. [DOI] [PubMed] [Google Scholar]
- 17.Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993) J. Mol. Biol. 229, 105-124. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 19.Terwilliger, T. C. (2002) Acta Crystallogr. D 58, 1937-1940. [DOI] [PubMed] [Google Scholar]
- 20.Terwilliger, T. C. (2000) Acta Crystallogr. D 56, 965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cambillau, C., Roussel, A., Inisan, A. & Knoops-Mouthay, K. (1997) TURBO-FRODO (Centre National de la Recherche Scientifique/Université Aix-Marseille II, Marseille, France).
- 22.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 23.Liu, L., Wilson, T. & Hastings, J. W. (2004) Proc. Natl. Acad. Sci. USA 101, 16555-16560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm, L. & Sander, C. (1995) Trends Biochem. Sci. 20, 478-480. [DOI] [PubMed] [Google Scholar]
- 25.Sacchettini, J. C., Gordon, J. I. & Banaszak, L. J. (1989) J. Mol. Biol. 208, 327-339. [DOI] [PubMed] [Google Scholar]
- 26.Flower, D. R., North, A. C. & Sansom, C. E. (2000) Biochim. Biophys. Acta 1482, 9-24. [DOI] [PubMed] [Google Scholar]
- 27.Fisher, A. J., Thompson, T. B., Thoden, J. B., Baldwin, T. O. & Rayment, I. (1996) J. Biol. Chem. 271, 21956-21968. [DOI] [PubMed] [Google Scholar]
- 28.Conti, E., Franks, N. P. & Brick, P. (1996) Structure 4, 287-298. [DOI] [PubMed] [Google Scholar]
- 29.Head, J. F., Inouye, S., Teranishi, K. & Simomora, O. (2000) Nature 405, 372-376. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Z. J., Vysotski, E. S., Chen, C. J., Rose, J. P., Lee, J. & Wang, B. C. (2000) Protein Sci. 9, 2085-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson, T. & Hastings, J. W. (1998) Annu. Rev. Cell Dev. Biol. 14, 197-230. [DOI] [PubMed] [Google Scholar]
- 32.Grant, G. A., Xu, X. L. & Hu, Z. Q. (2000) Biochemistry 39, 7316-7319. [DOI] [PubMed] [Google Scholar]
- 33.Vila, R., Ponte, I., Jimenez, M. A., Rico, M. & Suau, P. (2002) Protein Sci. 11, 214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuller, D. J., Wilks, A., de Montellano, P. R. O. & Poulos, T. L. (1999) Nat. Struct. Biol. 6, 860-867. [DOI] [PubMed] [Google Scholar]
- 35.Mathews, M. A. A., Schubert, H. L., Whitby, F. G., Alexander, K. J., Schadick, K., Bergonia, H. A., Phillips, J. D. & Hill, C. P. (2001) EMBO J. 20, 5832-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitby, F. G., Phillips, J. D., Hill, C. P., McCoubrey, W. & Maines, M. D. (2002) J. Mol. Biol. 319, 1199-1210. [DOI] [PubMed] [Google Scholar]
- 37.Kikuchi, A., Park, S. Y., Miyatake, H., Sun, D. Y., Sato, M., Yoshida, T. & Shiro, Y. (2001) Nat. Struct. Biol. 8, 221-225. [DOI] [PubMed] [Google Scholar]
- 38.Pereira, P. J. B., Macedo-Ribeiro, S., Parraga, A., Perez-Luque, R., Cunningham, O., Darcy, K., Mantle, T. J. & Coll, M. (2001) Nat. Struct. Biol. 8, 215-220. [DOI] [PubMed] [Google Scholar]
- 39.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281-296. [DOI] [PubMed] [Google Scholar]
- 40.Kleywegt, G. J. & Jones, T. A. (1994) Acta Crystallogr. D 50, 178-185. [DOI] [PubMed] [Google Scholar]
- 41.Beglova, N., Jeon, H., Fisher, C. & Blacklow, S. C. (2004) Mol. Cell 16, 281-292. [DOI] [PubMed] [Google Scholar]
- 42.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 43.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505-524. [DOI] [PubMed] [Google Scholar]