Abstract
Background
Cancer stem cell (CSC) epithelial cell adhesion molecule (Ep-CAM) is frequently expressed in colorectal cancer (CRC). However, the clinical significance of Ep-CAM expression in CRC is not clear. This study evaluated whether Ep-CAM provided valuable insight as a molecular biomarker for CRC diagnosis and prognosis and the potential of Ep-CAM as a novel therapeutic target in CRC.
Methods
Publications were selected online using electronic databases. The pooled odds ratios (ORs) or hazard ratios (HRs) with their 95% confidence intervals (95% CIs), and the combined sensitivity, specificity, and area under the curve (AUC) were calculated and summarized.
Results
Eleven eligible articles published in English involving 4561 cases were analyzed in this study. Ep-CAM expression was significantly higher in CRC compared with normal controls, and its overexpression was negatively linked to tumor differentiation, tumor stage, vascular invasion, depth of tumor invasion, lymph node metastasis, distant metastasis, and tumor budding in CRC patients. The loss of Ep-CAM expression positively correlated with these characteristics. Multivariate analysis of loss of Ep-CAM expression correlated with a poor prognosis in disease-free survival (DFS), disease-specific survival (DSS), and overall survival (OS). The pooled sensitivity, specificity and AUC values of Ep-CAM expression in patients with CRC vs. normal controls were 0.93, 0.90, and 0.94, respectively.
Conclusions
The present findings suggest that Ep-CAM expression may be associated with CRC carcinogenesis, while the loss of Ep-CAM expression is correlated with the progression, metastasis, and poor prognosis of CRC. Ep-CAM expression may be a useful biomarker for the clinical diagnosis of CRC.
Abbreviations: CRC, colorectal cancer; CSC, cancer stem cell; Ep-CAM, epithelial cell adhesion molecule; OR, odds ratio; HR, hazard ratio; 95% CI, 95% confidence interval; AUC, area under the curve; DFS, disease-free survival; DSS, disease-specific survival; OS, overall survival; ESA, epithelial-specific antigen; EMT, epithelial-to-mesenchymal transition; CTC, circulating tumor cell; PRISMA, the preferred reporting items for systematic reviews and meta-analyses; IHC, immunohistochemistry
Keywords: Ep-CAM, Expression, CRC, Prognosis, Diagnosis, Biomarker
Highlights
-
•
Cancer stem cell (CSC) epithelial cell adhesion molecule (Ep-CAM) expression may correlate with CRC tumorigenesis.
-
•
Frequent overexpression of Ep-CAM was a favorable factor for CRC progression and metastasis.
-
•
Loss of Ep-CAM expression correlated with the progression, metastasis, and poor prognosis of patients with CRC.
-
•
Ep-CAM expression may be a potential marker for the detection of CRC.
Ep-CAM expression was reported in CRC, but no clear direction for the diagnostic and prognostic effects of Ep-CAM expression was documented in patients with CRC. We performed a systematic meta-analysis of the existing evidence to determine the clinical significance of Ep-CAM expression in CRC. The findings indicated that Ep-CAM expression was associated with CRC risk. Frequent overexpression of Ep-CAM correlated with a decreased risk of CRC progression and metastasis, and loss of Ep-CAM expression played an important role in CRC progression, metastasis and prognosis. The detection of Ep-CAM expression may be a promising biomarker in diagnosing CRC.
1. Introduction
Colorectal cancer (CRC) is the fifth most frequent malignancy and the fifth leading cause of death in all human cancers in China (Chen et al., 2016). Global cancer statistics noted approximately 1.4 million new cases were clinically diagnosed with CRC in 2002 and an estimated 693,900 deaths occurred due to CRC worldwide (Torre et al., 2015). CRC is a major public health problem because approximately 50% of CRC cases develop metastasis and exhibit a poor survival rate despite advances in early detection and treatment (Aranda et al., 2015, Ferlay et al., 2015, Van Cutsem et al., 2010). Therefore, there is a need for a potential biological marker that allows early detection and predicts clinical prognosis of CRC.
A special subpopulation of cancer cells, known as cancer stem cells (CSCs), drive cancer progression. CSCs self-renew, uncontrollably proliferate, differentiate, and form the bulk of the tumor (Majumdar et al., 2012, Chandler and Lagasse, 2010). CSCs also regulate the development, progression, and metastasis of cancer (da Silva-Diz et al., 2016, Sampetrean and Saya, 2013, Nguyen et al., 2012). Multiple studies reported evidence of CSCs in CRC (Patman, 2016, Zeuner et al., 2014). Epithelial cell adhesion molecule (Ep-CAM), also known as epithelial-specific antigen (ESA) or CD326, is a transmembrane glycoprotein cell adhesion molecule that is encoded by the Ep-CAM gene mapped to chromosomal region 4q (Balzar et al., 1999, Linnenbach et al., 1989). This cell adhesion molecule plays a key role in Ca2 +-independent cell-to-cell adhesion (Litvinov et al., 1994). Ep-CAM correlates with cell proliferation, migration, invasion, motility, and signal transduction (Subramanian et al., 2015, Maetzel et al., 2009). Studies suggest that Ep-CAM is associated with epithelial-to-mesenchymal transition (EMT) and enhances tumor-initiating capacity (Gupta et al., 2009, Morel et al., 2008). Ep-CAM is overexpressed in many types of cancers, such as breast cancer, ovarian cancer, and head and neck squamous cell cancer (Moldenhauer et al., 2012). Some studies demonstrated that Ep-CAM overexpression was an unfavorable prognostic marker in breast cancer and gallbladder carcinoma (Varga et al., 2004, Gastl et al., 2000). Ep-CAM is a CSC marker, and it is frequently expressed or overexpressed in CRC (Dalerba et al., 2007, Mosolits et al., 2004). Ep-CAM is defined as a universal molecular marker for circulating tumor cell (CTC) detection, which is termed the “post-Ep-CAM era” (Nicolazzo et al., 2015, Raimondi et al., 2015). Therefore, it is important to investigate Ep-CAM further.
Some studies demonstrated inconsistent and controversial conclusions of Ep-CAM expression in CRC patients. For example, Kuhn et al. reported that Ep-CAM expression was not associated with tumor stage or grade (Kuhn et al., 2007). Gosens et al. reported that the loss of Ep-CAM expression was significantly associated with tumor grade and trended towards a correlation with tumor stage (Gosens et al., 2007). Therefore, the present study assessed whether Ep-CAM expression correlated with an increased risk of CRC vs. benign colonic lesions and normal controls. We also analyzed whether Ep-CAM overexpression or the loss of Ep-CAM expression was associated with the prognostic effect and clinicopathological features of CRC. Finally, we evaluated the use of Ep-CAM expression as a biomarker for the early diagnosis of CRC.
2. Materials and Methods
2.1. Literature Search
A comprehensive literature search (PubMed, EMBASE, EBSCO, Web of Science, and Cochrane Library databases) was performed to identify relevant publications on Ep-CAM expression in CRC patients prior to January 16th, 2017. The articles were identified using the following search terms and key words: ‘Ep-CAM’, ‘epithelial cell adhesion molecule’, ‘EpCAM’, ‘CD326’, ‘GA733’, ‘CO17-1A’, ‘EGP’, ‘KS1-4’, ‘ESA’, ‘MOC31’, ‘BerEP4’, and ‘colorectal cancer’ ‘colorectal tumor’, ‘colorectal carcinoma’, ‘colorectal neoplasm’, ‘CRC’ and ‘expression’. The reference lists of the included articles were also screened to obtain other potential studies. This study was performed based on the preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement criteria (Moher et al., 2009) (Table S1).
2.2. Selection Criteria
Eligible studies were selected based on the following inclusion criteria: 1) patients were limited to a diagnosis of CRC; 2) articles were published in English with the full text; 3) Ep-CAM was considered as a positive expression, overexpression or a loss of the expression; 4) articles provided sufficient information to evaluate the correlation of Ep-CAM expression between CRC and nonmalignant controls; 5) articles provided sufficient data to assess the relationship of Ep-CAM expression with the clinicopathological characteristics of patients with CRC; and 6) the pooled hazard ratios (HRs) and their 95% confidence intervals (CIs) were extracted to evaluate the prognostic role when the data from the original papers were available. The most complete paper containing the most information was selected when authors published several articles using duplicated data.
2.3. Data Extraction
The following data were collected from eligible studies: first author's surname, year of publication, country, ethnic population, age, detection method, clinical stage, staining patterns, cut-off values, Ep-CAM expression (overexpression or loss), expression frequency, number of cases and controls, survival data of multivariate analysis, and clinicopathological features. The clinicopathological characteristics included tumor differentiation (poor vs. well/moderate), tumor stage (3–4 vs. 1–2), vascular invasion (yes vs. no), depth of tumor invasion (pT3–4 vs. pT1–2), lymph node metastasis (yes vs. no), distant metastasis (yes vs. no), tumor budding (yes vs. no), and tumor location (colon vs. rectum). Loss of Ep-CAM expression consisted of complete and partial loss.
2.4. Statistical Analysis
Data were analyzed using Stata software (version 12.0, Stata Corporation, College Station, TX, USA). The combined odds ratios (ORs) and their 95% confidence intervals (95% CIs) were calculated to determine the relationship of Ep-CAM expression between CRC and nonmalignant controls. The associations of Ep-CAM overexpression or loss of Ep-CAM expression with the clinicopathological features of CRC were also calculated using pooled ORs with 95% CIs. The overall hazard ratios (HRs) with 95% CIs were used to determine the impact of Ep-CAM expression on the survival of CRC patients, if possible. Between-study heterogeneity was measured using the Cochran's Q statistic (Zintzaras and Ioannidis, 2005). The random-effects model was used to increase the reliability of the results in the present analysis (heterogeneity: P < 0.1). A sensitivity analysis was performed to determine the influence of one study on the results and heterogeneity via omission of a single study when the pooled results with greater than two studies had substantial heterogeneity (P < 0.1) (Higgins et al., 2003, Lau et al., 1997). Potential publication bias was assessed using Egger's test for the results with greater than or equal to ten studies (Egger et al., 1997). The pooled sensitivity, specificity, and the summary receiver operator characteristic (SROC) curve (AUC) values were calculated and constructed according to the bivariate analysis to evaluate the performance of the diagnostic capacity of Ep-CAM expression to CRC in this study (Reitsma et al., 2005, Jones and Athanasiou, 2005).
3. Results
3.1. Characteristics of the Included Studies
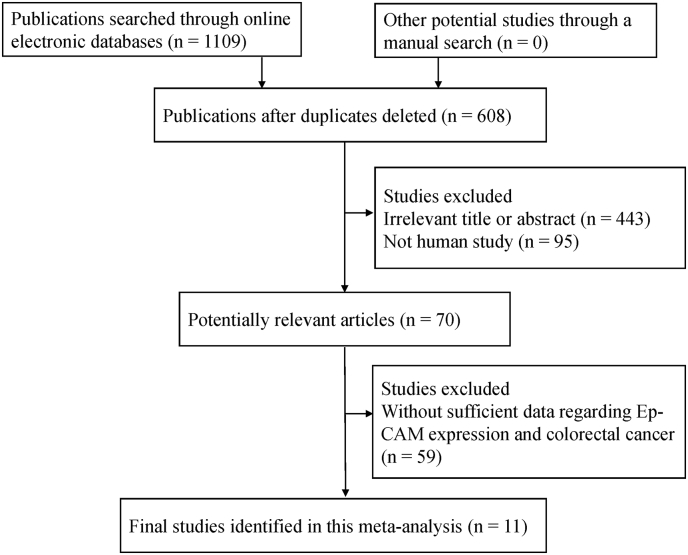
Fig. 1 shows the detailed selection procedure used for the potential literature. The abovementioned inclusion criteria resulted in 11 eligible studies published in English from 2005 to 2016 (Kim et al., 2016, Chai et al., 2015, Zhou et al., 2015, Kim et al., 2014, Goossens-Beumer et al., 2014, Lugli et al., 2010, Paret et al., 2007, Kuhn et al., 2007, Gosens et al., 2007, Went et al., 2006, Karanikiotis et al., 2005) for the meta-analysis, which included 4103 CRC patients and 458 controls. Five studies of 331 patients with CRC and 458 controls analyzed the relationship of Ep-CAM expression between CRC and controls (Chai et al., 2015, Zhou et al., 2015, Paret et al., 2007, Kuhn et al., 2007, Karanikiotis et al., 2005). Six studies analyzed the correlation of Ep-CAM overexpression or loss with the clinicopathological characteristics of 3772 patients with CRC (Kim et al., 2016, Kim et al., 2014, Goossens-Beumer et al., 2014, Lugli et al., 2010, Gosens et al., 2007, Went et al., 2006). Two studies of 1031 CRC patients reported multivariate survival analyses of Ep-CAM expression (Kim et al., 2016, Goossens-Beumer et al., 2014). Ep-CAM expression was assessed on tissues using immunohistochemistry (IHC). Tables 1 and S2 present the main characteristics of all eligible studies.
Fig. 1.
Flow chart of the selection procedure.
Table 1.
Basic characteristics of the publications included in this study.
| First author | Country | Ethnicity | Age | Method | Stage | Staining patterns |
Cut off scores (positive) | Sample | Status | CRC | Benign | Normal | Clinicopathological features | MA-survival |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N (E + %) |
N (E + %) | N (E + %) |
||||||||||||
| Karanikiotis et al. (2005) | Greece | Caucasians | NA | # | NA | NA | NA | Blood | E | 27 (96.3) | 20 (0) | No | NA | |
| Went et al. (2006) | Iran | Caucasians | 69.7 | IHC | NA | M/C | 70% | Tissue | High | 1186 (97.7) | Yes | NA | ||
| Kuhn et al. (2007) | Germany | Caucasians | NA | IHC | NA | M | 0% | Tissue | E | 104 (94.2) | 6 (100) | 92 (40.2) | No | NA |
| Paret et al. (2007) | Germany | Caucasians | NA | IHC | NA | NA | 25% | Tissue | E | 100 (94) | 100 (20) | No | NA | |
| Gosens et al. (2007) | The Netherlands | Caucasians | NA | IHC | 1–4 | M | NA | Tissue | High | 109 (64.2) | Yes | NA | ||
| Lugli et al. (2010) | Switzerland | Caucasians | NA | IHC | NA | M | 100% | Tissue | High | 1278 (89.6) | Yes | NA | ||
| Kim et al. (2014) | Korea | Asians | NA | IHC | 1–4 | M/C | 90% | Tissue | High | 168 (85.7) | Yes | NA | ||
| Goossens-Beumer et al. (2014) | The Netherlands | Caucasians | NA | IHC | 1–4 | M | 89% | Tissue | High | 305 (49.8) | Yes | Yes | ||
| Zhou et al. (2015) | China | Asians | NA | IHC | 1–4 | C | 0% | Tissue | E | 50 (92) | 50 (6) | 100 (5) | No | NA |
| Chai et al. (2015) | China | Asians | NA | IHC | 1–3 | C | 15% | Tissue | E | 50 (92) | 90 (5.6) | No | NA | |
| Kim et al. (2016) | Korea | Asians | NA | IHC | 1–4 | M/C | 95% | Tissue | High | 726 (93) | Yes | Yes |
NA: not applicable; E: expression; MA: multivariate analysis; “*” stands for a novel chromatographic method (molecular strip) for the detection of PCR-amplified product; IHC: immunohistochemistry; M: membrane; C: cytoplasm.
3.2. Association of Ep-CAM Expression Between CRC and Controls
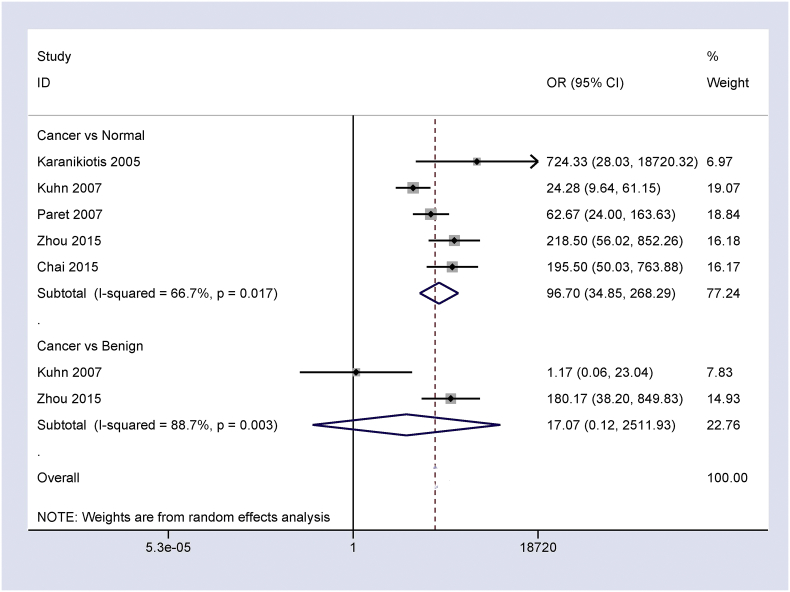
The overall OR from five studies of 331 patients with CRC and 402 normal controls demonstrated that the frequency of Ep-CAM expression in CRC was notably higher than in normal controls (OR = 96.70, 95% CI = 34.85–268.29, P < 0.001) (Fig. 2).
Fig. 2.
Forest plot showing the relationship between Ep-CAM expression and CRC in cancer vs. controls, CRC vs. normal controls: OR = 96.70, 95% CI = 34.85–268.29, P < 0.001; and CRC vs. benign colonic lesions: OR = 17.07, 95% CI = 0.12–2511.93, P = 0.265.
Two studies of 154 CRC patients and 56 benign colonic lesions revealed no significant difference between CRC and benign colonic lesions in Ep-CAM expression (OR = 17.07, 95% CI = 0.12–2511.93, P = 0.265) (Fig. 2).
3.3. Association of Ep-CAM Overexpression with Tumor Differentiation and Clinical Stage of CRC
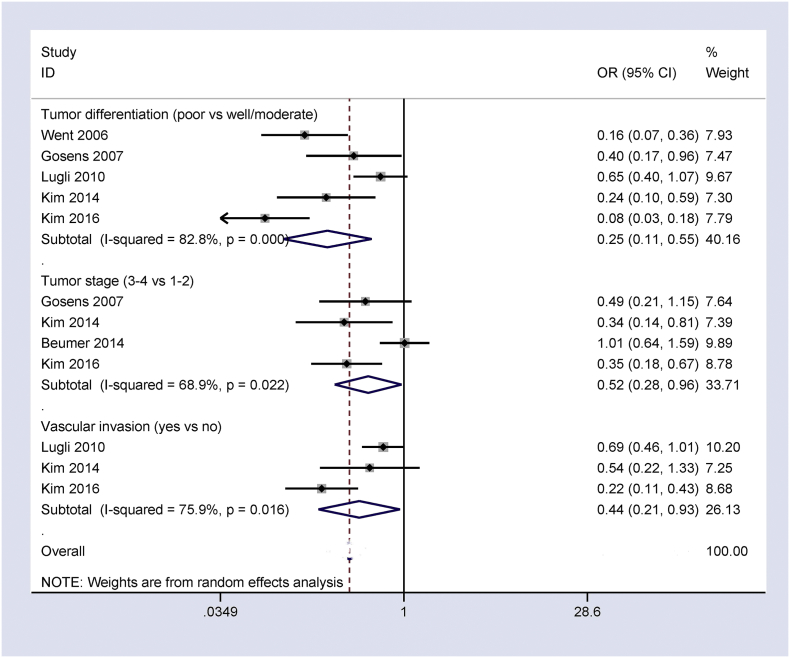
Ep-CAM overexpression negatively correlated with tumor differentiation and clinical stage (OR = 0.25, 95% CI = 0.11–0.55, P = 0.001; OR = 0.52, 95% CI = 0.28–0.96, P = 0.036; respectively) (Fig. 3) in our analysis of the results from five studies of 3433 CRC patients and four studies with 1308 CRC patients, respectively.
Fig. 3.
Forest plot showing the relationship of Ep-CAM overexpression with some clinicopathological features of CRC, including tumor differentiation: OR = 0.25, 95% CI = 0.11–0.55, P = 0.001; clinical stage: OR = 0.52, 95% CI = 0.28–0.96, P = 0.036; and vascular invasion: OR = 0.44, 95% CI = 0.21–0.93, P = 0.032.
3.4. Association of Ep-CAM Overexpression with Vascular Invasion and Depth of Tumor Invasion of CRC
The results from three studies of 2147 patients with CRC demonstrated a negative correlation between Ep-CAM overexpression and vascular invasion (OR = 0.44, 95% CI = 0.21–0.93, P = 0.032) (Fig. 3).
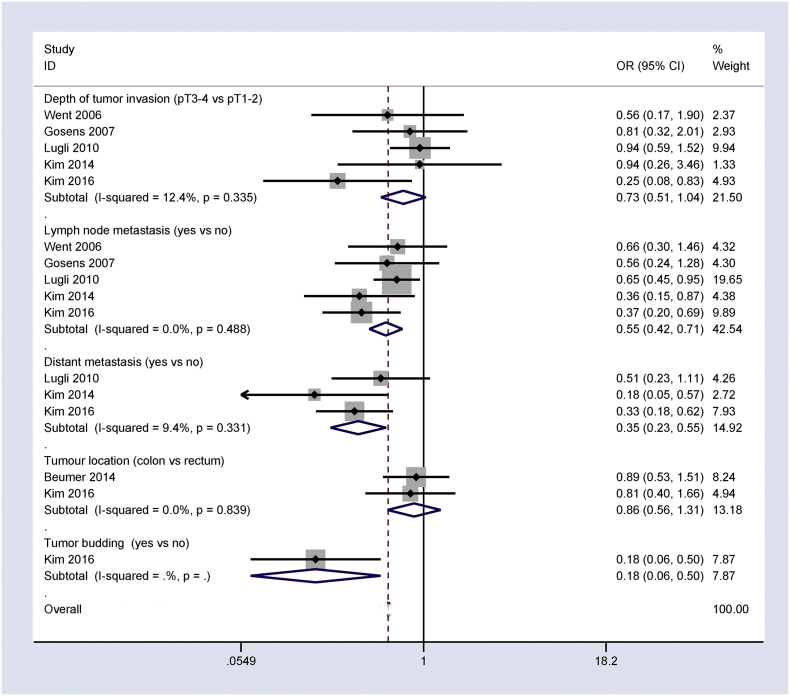
The pooled OR from five studies of 3429 patients with CRC revealed a trend towards a negative association between Ep-CAM overexpression and depth of tumor invasion (OR = 0.73, 95% CI = 0.51–1.04, P = 0.078) (Fig. 4).
Fig. 4.
Forest plot showing the association of Ep-CAM overexpression with other clinicopathological features of CRC, including depth of tumor invasion: OR = 0.73, 95% CI = 0.51–1.04, P = 0.078; lymph node metastasis: OR = 0.55, 95% CI = 0.42–0.71, P < 0.001; distant metastasis: OR = 0.35, 95% CI = 0.23–0.55, P < 0.001; tumor budding: OR = 0.18, 95% CI = 0.06–0.50, P = 0.001; and tumor location: OR = 0.86, 95% CI = 0.56–1.31, P = 0.486.
3.5. Association of Ep-CAM Overexpression with Lymph Node Metastasis and Distant Metastasis of CRC
Analysis of the pooled data of five studies of lymph node metastasis with 3393 CRC patients and three studies of distant metastasis with 1348 CRC patients demonstrated that Ep-CAM overexpression negatively correlated with lymph node metastasis and distant metastasis (OR = 0.55, 95% CI = 0.42–0.71, P < 0.001; OR = 0.35, 95% CI = 0.23–0.55, P < 0.001; respectively) (Fig. 4).
3.6. Association of Ep-CAM Overexpression with Tumor Budding and Tumor Location of CRC
The results of a study of 726 CRC patients revealed a negative relationship between Ep-CAM overexpression and tumor budding (OR = 0.18, 95% CI = 0.06–0.50, P = 0.001) (Fig. 4).
The pooled OR from two studies of 1031 CRC patients revealed that Ep-CAM overexpression was similar in colon and rectal cancer (OR = 0.86, 95% CI = 0.56–1.31, P = 0.486) (Fig. 4).
3.7. Association of the Loss of Ep-CAM Expression with the Clinicopathological Features of CRC
Ep-CAM overexpression negatively correlated with the clinicopathological characteristics of CRC patients, including tumor differentiation, tumor stage, vascular invasion, depth of tumor invasion, lymph node metastasis, distant metastasis, and tumor budding. Therefore, we further evaluated whether the loss of Ep-CAM expression positively correlated with these clinicopathological features.
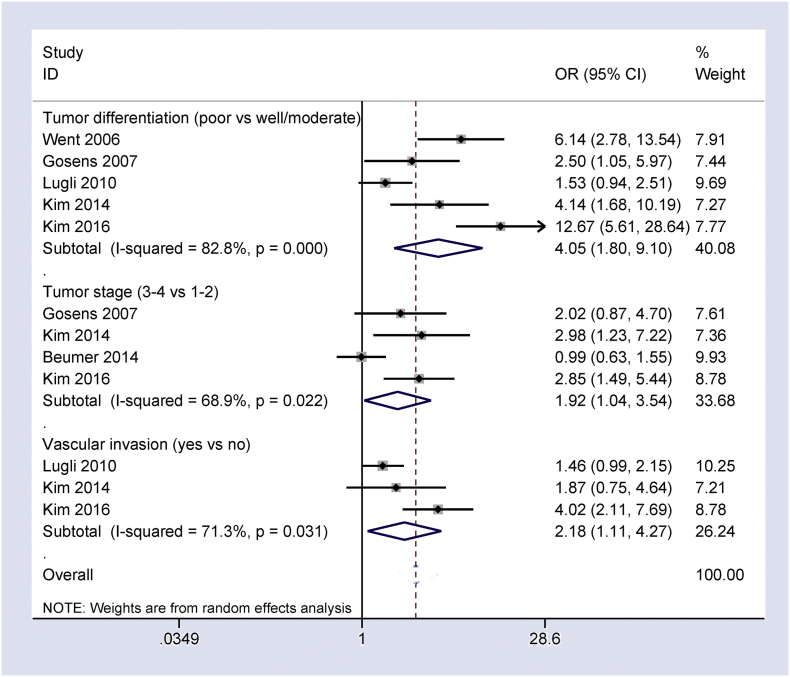
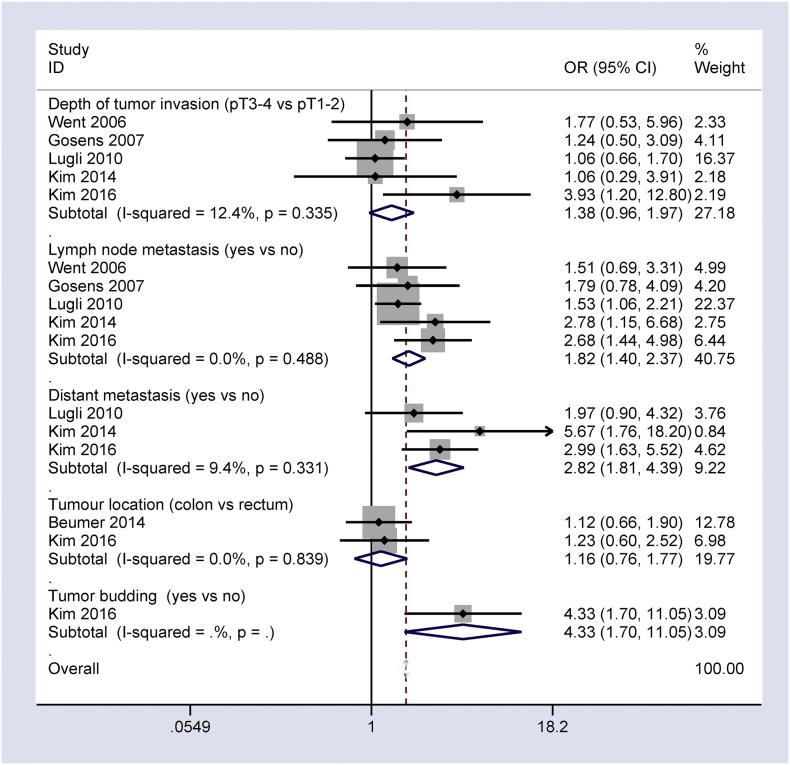
Fig. 5, Fig. 6 show that the loss of Ep-CAM expression positively associated with tumor differentiation, tumor stage, vascular invasion, depth of tumor invasion, lymph node metastasis, distant metastasis, and tumor budding in CRC patients (OR = 4.05, 95% CI = 1.80–9.10, P = 0.001; OR = 1.92, 95% CI = 1.04–3.54, P = 0.036; OR = 2.18, 95% CI = 1.11–4.27, P = 0.024; OR = 1.38, 95% CI = 0.96–1.97, P = 0.078; OR = 1.82, 95% CI = 1.40–2.37, P < 0.001; OR = 2.82, 95% CI = 1.81–4.39, P < 0.001; OR = 4.33, 95% CI = 1.70–11.05, P = 0.001, respectively). No correlation was found between the loss of Ep-CAM expression and tumor location (OR = 1.16, 95% CI = 0.76–1.77, P = 0.486).
Fig. 5.
Forest plot indicating the correlation of loss of Ep-CAM expression with some clinicopathological features of CRC, including tumor differentiation: OR = 4.05, 95% CI = 1.80–9.10, P = 0.001; clinical stage: OR = 1.92, 95% CI = 1.04–3.54, P = 0.036; and vascular invasion: OR = 2.18, 95% CI = 1.11–4.27, P = 0.024.
Fig. 6.
Forest plot indicating the correlation of loss of Ep-CAM expression with other clinicopathological features of CRC, including depth of tumor invasion: OR = 1.38, 95% CI = 0.96–1.97, P = 0.078; lymph node metastasis: OR = 1.82, 95% CI = 1.40–2.37, P < 0.001; distant metastasis: OR = 2.82, 95% CI = 1.81–4.39, P < 0.001; tumor budding: OR = OR = 4.33, 95% CI = 1.70–11.05, P = 0.001; and tumor location: OR = 1.16, 95% CI = 0.76–1.77, P = 0.486.
3.8. Sensitivity Analysis
Heterogeneity was observed in the comparison of CRC and normal controls, in tumor differentiation, clinical stage, and vascular invasion (all Ps < 0.1). We removed the study (Kuhn et al., 2007) and recalculated the overall OR (OR = 137.14, 95% CI = 59.32–317.08, P < 0.001) in CRC vs. normal controls, which dramatically reduced the heterogeneity (P = 0.243). We successively removed two studies (Lugli et al., 2010) and (Kim et al., 2016) from our correlation analyses between the loss of Ep-CAM expression and tumor differentiation, and the pooled OR was 4.09 (95% CI = 2.42–6.92, P < 0.001), with no heterogeneity (P = 0.319). The combined OR between loss of Ep-CAM expression and tumor stage was 2.62 (95% CI: 1.68–4.08, P < 0.001) based on the omission of the study (Goossens-Beumer et al., 2014), with no evidence of heterogeneity (P = 0.776). One study(Kim et al., 2016) was removed from the correlation analyses of the loss of Ep-CAM expression with vascular invasion, and the pooled OR did not change substantially (OR = 1.52, 95% CI = 1.06–2.17, P = 0.023), but heterogeneity was significantly reduced (P = 0.624). The above analyses demonstrated that our results were stable and credible.
3.9. Prognostic Impact of Ep-CAM Expression in CRC Patients Using Multivariate Analysis
Only two studies reported a multivariate survival analysis of Ep-CAM expression in CRC patients (Kim et al., 2016, Goossens-Beumer et al., 2014). The loss of Ep-CAM expression correlated with a poor prognosis in disease-specific survival (DSS) and a trend towards a poor prognosis between loss of Ep-CAM expression and a 5-year overall survival (OS) in 305 CRC patients (Goossens-Beumer et al., 2014). Multivariate analysis of 725 CRC patients demonstrated that the loss of Ep-CAM expression was an unfavorable prognostic factor in disease-free survival (DFS) (HR = 1.57, 95% CI = 1.04–2.39, P = 0.03) (Kim et al., 2016).
3.10. Diagnostic Capacity of Ep-CAM Expression in CRC vs. Normal Controls
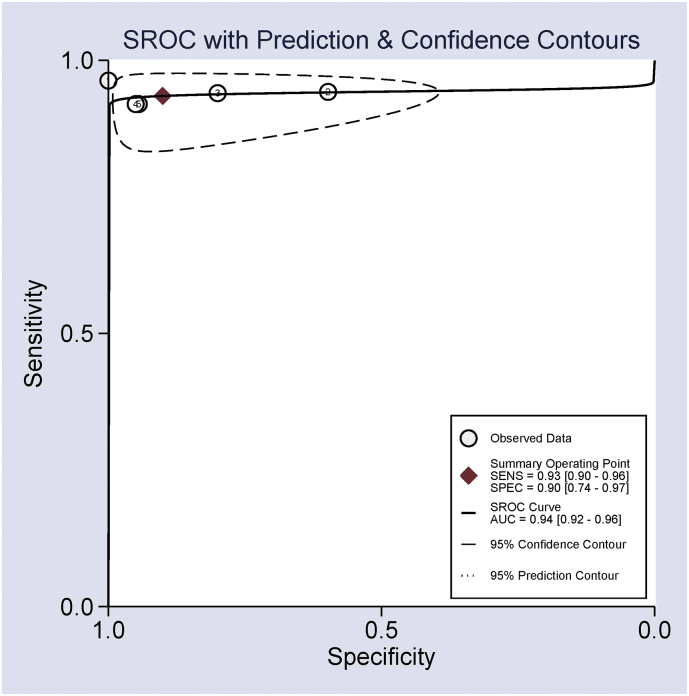
We also evaluated the diagnostic value of Ep-CAM expression as a potential biomarker in the discrimination of CRC patients and normal controls. The pooled sensitivity, specificity and AUC values of Ep-CAM expression were 0.93 (95% CI = 0.90–0.96), 0.90 (95% CI = 0.74–0.97), and 0.94 (95% CI = 0.92–0.96) (Fig. 7), respectively, which suggests that Ep-CAM expression exhibited good diagnostic performance in CRC.
Fig. 7.
Summary receiver operating characteristics (SROC) estimation of Ep-CAM expression in patients with CRC vs. normal controls, sensitivity = 0.93 (95% CI = 0.90–0.96), specificity = 0.90 (95% CI = 0.74–0.97), and AUC = 0.94 (95% CI = 0.92–0.96).
4. Discussion
Cancer stem cells (CSCs) are a special subpopulation of cells within a tumor. Ep-CAM overexpression is a CSC marker of tumor cells that closely correlates with tumor progression, including colorectal cancer cell lines (Liao et al., 2015). Ep-CAM overexpression is a poor prognostic factor in some cancers, such as breast and gallbladder carcinomas (Schmidt et al., 2010, Varga et al., 2004), but its overexpression is a favorable prognostic factor in ovarian cancer (Battista et al., 2014). The present study found that Ep-CAM was commonly expressed in CRC, and its expression was significantly higher in CRC than in normal controls (Chai et al., 2015, Zhou et al., 2015, Paret et al., 2007, Kuhn et al., 2007, Karanikiotis et al., 2005). Ep-CAM was slightly less frequently expressed in CRC than in benign lesions (Kuhn et al., 2007), but its expression level was notably higher in CRC than in benign lesions (Zhou et al., 2015). The current results revealed that the frequency of Ep-CAM expression was significantly more common in CRC compared with normal controls, which suggests that Ep-CAM expression is associated with the carcinogenesis of CRC. Ep-CAM expression stimulates cell differentiation and cell proliferation via up-regulation of the proto-oncogene c-myc, which causes carcinogenic effects (Munz et al., 2004). However, no correlation in Ep-CAM expression was found between CRC and benign colonic lesions because the sample size was small. No association between Ep-CAM expression and DNA mismatch repair protein expression was reported in 218 microsatellite instability (MSI-high) CRCs (Kim et al., 2016).
Ep-CAM overexpression exhibited a high frequency in the present study (88.68%). We analyzed the relationship of Ep-CAM overexpression with the clinicopathological characteristics of CRC patients, including tumor differentiation, tumor stage, vascular invasion, depth of tumor invasion, lymph node metastasis, distant metastasis, and tumor budding. Our findings demonstrated that Ep-CAM overexpression negatively correlated with these clinicopathological characteristics, which indicates that Ep-CAM overexpression was a favorable factor in CRC progression, metastasis, and relapse.
We further investigated whether the loss of Ep-CAM expression was an unfavorable factor in relation to these features. The loss of Ep-CAM expression was significantly associated with tumor differentiation (Kim et al., 2016, Kim et al., 2014, Gosens et al., 2007, Went et al., 2006), and another study demonstrated a positive trend towards an association between the loss of Ep-CAM expression and tumor differentiation (Lugli et al., 2010). An association between the loss of Ep-CAM expression and tumor stage was found in two studies (Kim et al., 2016, Kim et al., 2014), but the other studies demonstrated no association (Goossens-Beumer et al., 2014, Gosens et al., 2007). Loss of Ep-CAM expression correlated with vascular invasion (Kim et al., 2016, Lugli et al., 2010), but there was no correlation with vascular invasion in a small population (168 patients) (Kim et al., 2014). No significant relationship was observed between loss of Ep-CAM expression and depth of tumor invasion (Kim et al., 2014, Lugli et al., 2010, Gosens et al., 2007, Went et al., 2006), but the remaining one study reported a significant correlation (Kim et al., 2016). Three studies demonstrated a correlation between loss of Ep-CAM expression and lymph node metastasis (Kim et al., 2016, Kim et al., 2014, Lugli et al., 2010), but two studies failed to demonstrate any correlation (Gosens et al., 2007, Went et al., 2006). Two studies demonstrated that the loss of Ep-CAM expression significantly correlated with distant metastasis (Kim et al., 2016, Kim et al., 2014), and another study demonstrated a trend towards the relationship between loss of Ep-CAM expression and distant metastasis (Lugli et al., 2010). A study of 726 CRC patients reported a close correlation between loss of Ep-CAM expression and tumor budding (Kim et al., 2016). Therefore, we integrated the eligible studies into a large population, and the results suggested a close association of the loss of Ep-CAM expression with tumor differentiation, tumor stage, vascular invasion, lymph node metastasis, distant metastasis, and tumor budding. We also found that loss of Ep-CAM expression exhibited a positive trend with depth of tumor invasion (OR = 1.38, 95% CI = 0.96–1.97, P = 0.078). These analyses indicate that loss of Ep-CAM expression plays a crucial role in CRC progression, metastasis, and relapse.
Loss of Ep-CAM expression with multivariate analysis was a poor prognostic factor in disease-specific survival (DSS) (Goossens-Beumer et al., 2014), 5-year overall survival (OS) (Goossens-Beumer et al., 2014), and disease-free survival (DFS) (Kim et al., 2016), which suggests that loss of Ep-CAM expression may be a potential prognostic biomarker.
Some studies demonstrated the use of CSCs as a marker for the detection and diagnosis of cancer (Takaishi et al., 2008, Yang et al., 2008, Nagrath et al., 2007). We evaluated whether Ep-CAM expression was a useful diagnostic biomarker for CRC. A comparison of CRC and normal controls demonstrated combined sensitivity, specificity and AUC values of Ep-CAM expression of 0.93, 0.90, and 0.94, respectively, which was very good (all values ≥ 0.9). These results suggest that Ep-CAM expression exhibits good diagnostic capacity as a promising molecular biomarker for CRC diagnosis. The importance of Ep-CAM was validated in an Ep-CAM-based assay, the CellSearch® System, which is the only Food and Drug Administration (FDA)-approved assay for the enrichment and detection of circulating tumor cells (CTCs) (Cohen et al., 2008). We also found that the detection of Ep-CAM expression in the blood exhibited a sensitivity of 96.3% and specificity of 100% (Karanikiotis et al., 2005), which indicates that the detection rate of Ep-CAM-expressing circulating tumor cells may be higher in blood samples and serve as a potential noninvasive marker. Additional large-scale and well-designed prospective studies are essential to further confirm the diagnostic effect of Ep-CAM expression in clinical applications, especially the use of blood samples.
There was evidence of heterogeneity in CRC vs. normal controls in tumor differentiation, clinical stage, and vascular invasion (all P < 0.1). We removed one study (Kuhn et al., 2007) in cancer vs. normal controls, two studies (Lugli et al., 2010) and (Kim et al., 2016) for tumor differentiation, the study (Goossens-Beumer et al., 2014) for tumor stage, and the study (Kim et al., 2016) for vascular invasion. The pooled OR remained significant with no evidence of heterogeneity, which indicated the stability and reliability of our results. The reasons for the heterogeneity were not clear, perhaps because of the use of inappropriate or different conditions in immunohistochemistry (IHC) methods, which may have led to the observed bias.
5. Limitations
This meta-analysis has some limitations. First, our study primarily included only Asian and Caucasian populations, and data on other populations, such as African populations, were lacking. Second, the analysis included only available articles published in English. Publications in other languages were excluded due to incomprehensible information, which may result in selection bias. Third, only two studies with small populations were examined to assess the correlation between CRC and benign lesions. Finally, only one study involving blood samples was analyzed in the present meta-analysis, and more studies with large sample sizes are essential to further validate the diagnostic value of Ep-CAM expression.
6. Conclusions
Our findings suggest that Ep-CAM expression is notably higher in CRC patients than in normal controls. Loss of Ep-CAM expression correlated with tumor differentiation, tumor stage, vascular invasion, depth of tumor invasion, lymph node metastasis, distant metastasis, and tumor budding in CRC. Loss of Ep-CAM expression may be a poor prognostic factor of CRC in DFS, DSS and OS. The use of Ep-CAM expression may be a potential biomarker for the diagnosis of CRC patients in the clinical setting. However, more clinical prospective studies using large populations with CRC are necessary to further confirm the diagnostic and prognostic effects.
The following are the supplementary data related to this article.
PRISMA checklist.
Baseline characteristics of Ep-CAM overexpression or loss with the clinicopathological features of patients with CRC.
Authors' Contributions
Susu Han, Fenggang Hou and Wen Li contributed to the conception and design of this research. Susu Han, Shaoqi Zong and Qi Shi contributed to the drafting of the article and final approval of the submitted version. Susu Han, Shaoqi Zong, Qi Shi, Hongjia Li, Shanshan Liu, Wei Yang, Wen Li, and Fenggang Hou contributed to data analyses and interpretation and completion of the figures and tables. All authors read and approved the final manuscript.
Competing Financial Interests
All authors declare they have no competing financial interests.
Ethical Review from Patients
Our study was not primary research involving human samples, but rather a secondary analysis of human subject data published in the public domain.
Acknowledgments and Funding
This research was supported by grants from the Natural Science Foundation of China (81473624). The sponsor had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Contributor Information
Wen Li, Email: 13917367207@163.com.
Fenggang Hou, Email: fghou555@126.com.
References
- Aranda E., Aparicio J., Alonso V., Garcia-Albeniz X., Garcia-Alfonso P., Salazar R., Valladares M., Vera R., Vieitez J.M., Garcia-Carbonero R. SEOM clinical guidelines for diagnosis and treatment of metastatic colorectal cancer 2015. Clin. Transl. Oncol. 2015;17:972–981. doi: 10.1007/s12094-015-1434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzar M., Winter M.J., De Boer C.J., Litvinov S.V. The biology of the 17-1A antigen (Ep-CAM) J. Mol. Med. (Berl.) 1999;77:699–712. doi: 10.1007/s001099900038. [DOI] [PubMed] [Google Scholar]
- Battista M.J., Cotarelo C., Jakobi S., Steetskamp J., Makris G., Sicking I., Weyer V., Schmidt M. Overexpression of epithelial cell adhesion molecule protein is associated with favorable prognosis in an unselected cohort of ovarian cancer patients. J. Cancer Res. Clin. Oncol. 2014;140:1097–1102. doi: 10.1007/s00432-014-1672-9. [DOI] [PubMed] [Google Scholar]
- Chai X.B., Song R.F., Xu F. Expression changes in epithelial cell adhesion molecule during colorectal cancer tumorigenesis. Genet. Mol. Res. 2015;14:7624–7629. doi: 10.4238/2015.July.13.6. [DOI] [PubMed] [Google Scholar]
- Chandler J.M., Lagasse E. Cancerous stem cells: deviant stem cells with cancer-causing misbehavior. Stem Cell Res. Ther. 2010;1:13. doi: 10.1186/scrt13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- Cohen S.J., Punt C.J., Iannotti N., Saidman B.H., Sabbath K.D., Gabrail N.Y., Picus J., Morse M., Mitchell E., Miller M.C., Doyle G.V., Tissing H., Terstappen L.W., Meropol N.J. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- da Silva-Diz V., Simon-Extremera P., Bernat-Peguera A., De Sostoa J., Urpi M., Penin R.M., Sidelnikova D.P., Bermejo O., Vinals J.M., Rodolosse A., Gonzalez-Suarez E., Moruno A.G., Pujana M.A., Esteller M., Villanueva A., Vinals F., Munoz P. Cancer stem-like cells act via distinct signaling pathways in promoting late stages of malignant progression. Cancer Res. 2016;76:1245–1259. doi: 10.1158/0008-5472.CAN-15-1631. [DOI] [PubMed] [Google Scholar]
- Dalerba P., Dylla S.J., Park I.K., Liu R., Wang X., Cho R.W., Hoey T., Gurney A., Huang E.H., Simeone D.M., Shelton A.A., Parmiani G., Castelli C., Clarke M.F. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Gastl G., Spizzo G., Obrist P., Dunser M., Mikuz G. Ep-CAM overexpression in breast cancer as a predictor of survival. Lancet. 2000;356:1981–1982. doi: 10.1016/S0140-6736(00)03312-2. [DOI] [PubMed] [Google Scholar]
- Goossens-Beumer I.J., Zeestraten E.C., Benard A., Christen T., Reimers M.S., Keijzer R., Sier C.F., Liefers G.J., Morreau H., Putter H., Vahrmeijer A.L., Van De Velde C.J., Kuppen P.J. Clinical prognostic value of combined analysis of Aldh1, Survivin, and EpCAM expression in colorectal cancer. Br. J. Cancer. 2014;110:2935–2944. doi: 10.1038/bjc.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosens M.J., Van Kempen L.C., Van De Velde C.J., Van Krieken J.H., Nagtegaal I.D. Loss of membranous ep-CAM in budding colorectal carcinoma cells. Mod. Pathol. 2007;20:221–232. doi: 10.1038/modpathol.3800733. [DOI] [PubMed] [Google Scholar]
- Gupta P.B., Chaffer C.L., Weinberg R.A. Cancer stem cells: mirage or reality? Nat. Med. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.M., Athanasiou T. Summary receiver operating characteristic curve analysis techniques in the evaluation of diagnostic tests. Ann. Thorac. Surg. 2005;79:16–20. doi: 10.1016/j.athoracsur.2004.09.040. [DOI] [PubMed] [Google Scholar]
- Karanikiotis C., Skiadas I., Karina M., Georgakopoulou S., Georgakopoulos E., Fountzilas G. A novel chromatographic method for Ep-CAM mRNA detection in peripheral blood and bone marrow of patients with metastatic colorectal cancer. Anticancer Res. 2005;25:319–323. [PubMed] [Google Scholar]
- Kim J.H., Bae J.M., Kim K.J., Rhee Y.Y., Kim Y., Cho N.Y., Lee H.S., Chang M.S., Kang G.H. Differential features of microsatellite-unstable colorectal carcinomas depending on EPCAM expression status. Korean J. Pathol. 2014;48:276–282. doi: 10.4132/KoreanJPathol.2014.48.4.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Bae J.M., Song Y.S., Cho N.Y., Lee H.S., Kang G.H. Clinicopathologic, molecular, and prognostic implications of the loss of EPCAM expression in colorectal carcinoma. Oncotarget. 2016;7:13372–13387. doi: 10.18632/oncotarget.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn S., Koch M., Nubel T., Ladwein M., Antolovic D., Klingbeil P., Hildebrand D., Moldenhauer G., Langbein L., Franke W.W., Weitz J., Zoller M. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol. Cancer Res. 2007;5:553–567. doi: 10.1158/1541-7786.MCR-06-0384. [DOI] [PubMed] [Google Scholar]
- Lau J., Ioannidis J.P., Schmid C.H. Quantitative synthesis in systematic reviews. Ann. Intern. Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- Liao M.Y., Kuo M.Y., Lu T.Y., Wang Y.P., Wu H.C. Generation of an anti-EpCAM antibody and epigenetic regulation of EpCAM in colorectal cancer. Int. J. Oncol. 2015;46:1788–1800. doi: 10.3892/ijo.2015.2876. [DOI] [PubMed] [Google Scholar]
- Linnenbach A.J., Wojcierowski J., Wu S.A., Pyrc J.J., Ross A.H., Dietzschold B., Speicher D., Koprowski H. Sequence investigation of the major gastrointestinal tumor-associated antigen gene family, GA733. Proc. Natl. Acad. Sci. U. S. A. 1989;86:27–31. doi: 10.1073/pnas.86.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinov S.V., Bakker H.A., Gourevitch M.M., Velders M.P., Warnaar S.O. Evidence for a role of the epithelial glycoprotein 40 (Ep-CAM) in epithelial cell-cell adhesion. Cell Adhes. Commun. 1994;2:417–428. doi: 10.3109/15419069409004452. [DOI] [PubMed] [Google Scholar]
- Lugli A., Iezzi G., Hostettler I., Muraro M.G., Mele V., Tornillo L., Carafa V., Spagnoli G., Terracciano L., Zlobec I. Prognostic impact of the expression of putative cancer stem cell markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer. Br. J. Cancer. 2010;103:382–390. doi: 10.1038/sj.bjc.6605762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetzel D., Denzel S., Mack B., Canis M., Went P., Benk M., Kieu C., Papior P., Baeuerle P.A., Munz M., Gires O. Nuclear signalling by tumour-associated antigen EpCAM. Nat. Cell Biol. 2009;11:162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- Majumdar A., Curley S.A., Wu X., Brown P., Hwang J.P., Shetty K., Yao Z.X., He A.R., Li S., Katz L., Farci P., Mishra L. Hepatic stem cells and transforming growth factor beta in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2012;9:530–538. doi: 10.1038/nrgastro.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D.G., Group, P Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldenhauer G., Salnikov A.V., Luttgau S., Herr I., Anderl J., Faulstich H. Therapeutic potential of amanitin-conjugated anti-epithelial cell adhesion molecule monoclonal antibody against pancreatic carcinoma. J. Natl. Cancer Inst. 2012;104:622–634. doi: 10.1093/jnci/djs140. [DOI] [PubMed] [Google Scholar]
- Morel A.P., Lievre M., Thomas C., Hinkal G., Ansieau S., Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosolits S., Markovic K., Frodin J.E., Virving L., Magnusson C.G., Steinitz M., Fagerberg J., Mellstedt H. Vaccination with Ep-CAM protein or anti-idiotypic antibody induces Th1-biased response against MHC class I- and II-restricted Ep-CAM epitopes in colorectal carcinoma patients. Clin. Cancer Res. 2004;10:5391–5402. doi: 10.1158/1078-0432.CCR-04-0425. [DOI] [PubMed] [Google Scholar]
- Munz M., Kieu C., Mack B., Schmitt B., Zeidler R., Gires O. The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene. 2004;23:5748–5758. doi: 10.1038/sj.onc.1207610. [DOI] [PubMed] [Google Scholar]
- Nagrath S., Sequist L.V., Maheswaran S., Bell D.W., Irimia D., Ulkus L., Smith M.R., Kwak E.L., Digumarthy S., Muzikansky A., Ryan P., Balis U.J., Tompkins R.G., Haber D.A., Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.V., Vanner R., Dirks P., Eaves C.J. Cancer stem cells: an evolving concept. Nat. Rev. Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- Nicolazzo C., Massimi I., Lotti L.V., Vespa S., Raimondi C., Pulcinelli F.M., Gradilone A., Gazzaniga P. Impact of chronic exposure to bevacizumab on EpCAM-based detection of circulating tumor cells. Chin. J. Cancer Res. 2015;27:491–496. doi: 10.3978/j.issn.1000-9604.2015.04.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paret C., Hildebrand D., Weitz J., Kopp-Schneider A., Kuhn A., Beer A., Hautmann R., Zoller M. C4.4A as a candidate marker in the diagnosis of colorectal cancer. Br. J. Cancer. 2007;97:1146–1156. doi: 10.1038/sj.bjc.6604012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patman G. Colorectal cancer: targeting the root of colorectal cancer—eliminating cancer stem cells. Nat. Rev. Gastroenterol. Hepatol. 2016;13:2. doi: 10.1038/nrgastro.2015.209. [DOI] [PubMed] [Google Scholar]
- Raimondi C., Nicolazzo C., Gradilone A. Circulating tumor cells isolation: the "post-EpCAM era". Chin. J. Cancer Res. 2015;27:461–470. doi: 10.3978/j.issn.1000-9604.2015.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitsma J.B., Glas A.S., Rutjes A.W., Scholten R.J., Bossuyt P.M., Zwinderman A.H. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J. Clin. Epidemiol. 2005;58:982–990. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Sampetrean O., Saya H. Characteristics of glioma stem cells. Brain Tumor Pathol. 2013;30:209–214. doi: 10.1007/s10014-013-0141-5. [DOI] [PubMed] [Google Scholar]
- Schmidt M., Scheulen M.E., Dittrich C., Obrist P., Marschner N., Dirix L., Schmidt M., Ruttinger D., Schuler M., Reinhardt C., Awada A. An open-label, randomized phase II study of adecatumumab, a fully human anti-EpCAM antibody, as monotherapy in patients with metastatic breast cancer. Ann. Oncol. 2010;21:275–282. doi: 10.1093/annonc/mdp314. [DOI] [PubMed] [Google Scholar]
- Subramanian N., Kanwar J.R., Kanwar R.K., Sreemanthula J., Biswas J., Khetan V., Krishnakumar S. EpCAM aptamer-siRNA chimera targets and regress epithelial cancer. PLoS One. 2015;10:e0132407. doi: 10.1371/journal.pone.0132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi S., Okumura T., Wang T.C. Gastric cancer stem cells. J. Clin. Oncol. 2008;26:2876–2882. doi: 10.1200/JCO.2007.15.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Van Cutsem E., Nordlinger B., Cervantes A., GROUP, E. G. W Advanced colorectal cancer: ESMO clinical practice guidelines for treatment. Ann. Oncol. 2010;21(Suppl. 5):v93–v97. doi: 10.1093/annonc/mdq222. [DOI] [PubMed] [Google Scholar]
- Varga M., Obrist P., Schneeberger S., Muhlmann G., Felgel-Farnholz C., Fong D., Zitt M., Brunhuber T., Schafer G., Gastl G., Spizzo G. Overexpression of epithelial cell adhesion molecule antigen in gallbladder carcinoma is an independent marker for poor survival. Clin. Cancer Res. 2004;10:3131–3136. doi: 10.1158/1078-0432.ccr-03-0528. [DOI] [PubMed] [Google Scholar]
- Went P., Vasei M., Bubendorf L., Terracciano L., Tornillo L., Riede U., Kononen J., Simon R., Sauter G., Baeuerle P.A. Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br. J. Cancer. 2006;94:128–135. doi: 10.1038/sj.bjc.6602924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.F., Ngai P., Ho D.W., Yu W.C., Ng M.N., Lau C.K., Li M.L., Tam K.H., Lam C.T., Poon R.T., Fan S.T. Identification of local and circulating cancer stem cells in human liver cancer. Hepatology. 2008;47:919–928. doi: 10.1002/hep.22082. [DOI] [PubMed] [Google Scholar]
- Zeuner A., Todaro M., Stassi G., De Maria R. Colorectal cancer stem cells: from the crypt to the clinic. Cell Stem Cell. 2014;15:692–705. doi: 10.1016/j.stem.2014.11.012. [DOI] [PubMed] [Google Scholar]
- Zhou F.Q., Qi Y.M., Xu H., Wang Q.Y., Gao X.S., Guo H.G. Expression of EpCAM and Wnt/beta-catenin in human colon cancer. Genet. Mol. Res. 2015;14:4485–4494. doi: 10.4238/2015.May.4.6. [DOI] [PubMed] [Google Scholar]
- Zintzaras E., Ioannidis J.P. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21:3672–3673. doi: 10.1093/bioinformatics/bti536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA checklist.
Baseline characteristics of Ep-CAM overexpression or loss with the clinicopathological features of patients with CRC.