Abstract
The genetic code in a eukaryotic system has been expanded by the engineering of Escherichia coli tyrosyl-tRNA synthetase (TyrRS) with the Y37V and Q195C mutations (37V195C), which specifically recognize 3-iodo-l-tyrosine rather than l-tyrosine. In the present study, we determined the 3-iodo-l-tyrosine- and l-tyrosine-bound structures of the 37V195C mutant of the E. coli TyrRS catalytic domain at 2.0-Å resolution. The γ-methyl group of Val-37 and the sulfur atom of Cys-195 make van der Waals contacts with the iodine atom of 3-iodo-l-tyrosine. The Val-37 and Cys-195 side chains are rigidly fixed by the neighboring residues forming the hydrophobic core of the TyrRS. The major roles of the two mutations are different for the 3-iodo-l-tyrosine-selective recognition in the first step of the aminoacylation reaction (the amino acid activation step): the Y37V mutation eliminates the fatal steric repulsion with the iodine atom, and the Q195C mutation reduces the l-tyrosine misrecognition. The structure of the 37V195C mutant TyrRS complexed with an l-tyrosyladenylate analogue was also solved, indicating that the 3-iodo-l-tyrosine and l-tyrosine side chains are similarly discriminated in the second step (the aminoacyl transfer step). These results demonstrate that the amino acid-binding pocket on the 37V195C mutant is optimized for specific 3-iodo-l-tyrosine recognition.
Keywords: crystal structure, tyrosyl-tRNA synthetase, 3-iodo-l-tyrosine
Proteins containing nonnatural amino acids, or “alloproteins” (1), are expected to expand the structural, functional, and chemical diversity of proteins. For the efficient production of alloproteins with site-specifically incorporated nonnatural amino acids, the natural genetic code system has been expanded with engineered pairs of aminoacyl-tRNA synthetases and tRNAs (2–12). After all, the esterification of an engineered tRNA with a nonnatural amino acid requires alterations in the amino acid recognition of the aminoacyl-tRNA synthetase.
In the past decade, archaeal/eukaryal and bacterial tyrosyl-tRNA synthetases (TyrRSs) have been engineered for the incorporation of nonnatural amino acids into proteins by using bacterial and eukaryal translation systems, respectively (2–5, 8–11). Schultz and coworkers (2, 5, 8–12) have succeeded in expanding the genetic codes of Escherichia coli and yeast by using a variety of nonnatural amino acid-specific mutants of the TyrRSs from Methanococcus jannaschii and E. coli, respectively. As we reported (3), E. coli TyrRS with the Y37V and Q195C mutations (37V195C mutant) efficiently recognizes 3-iodo-l-tyrosine, which the wild-type TyrRS cannot recognize, and this nonnatural component is incorporated into proteins in response to the amber codon, both in vitro (a plant translation system) (3) and in vivo (mammalian cells) (4). The alloproteins produced by these systems will be used as molecular switches for signaling pathways, as photocrosslinkers, fluorescently labeled probes, or heavy-atom derivatives for phasing in x-ray structure determinations.
To date, a number of 3D structures of native TyrRS molecules have been determined (13–19), revealing the substrate recognition mechanisms of the natural enzymes. In the present study, we solved the structures of the 37V195C mutant E. coli TyrRS catalytic domain (19) complexed with 3-iodo-l-tyrosine and with the misrecognized ligand, l-tyrosine, at resolutions of 2.0 Å. These high-resolution structures revealed the structural basis of the 3-iodo-l-tyrosine recognition. The iodine atom of 3-iodo-l-tyrosine is strictly recognized with the side chains of Val-37 and Cys-195 by van der Waals interactions, and the roles of these two residues differ during the selective recognition in the first step (the amino acid activation step). We also determined the structure of the 37V195C TyrRS complexed with the tyrosyladenylate analogue, 5′-O-[N-(l-tyrosyl)sulfamoyl]adenosine (Tyr-AMS), which revealed that the second step (the aminoacyl transfer step) discriminates between the substrates in the same way as the first step. Thus, we demonstrated that the 37V195C mutant has an optimized 3-iodo-l-tyrosine-binding pocket for specific recognition.
Materials and Methods
Preparation and Crystallization of the Truncated E. coli TyrRS Enzymes. The mutant E. coli TyrRS catalytic domains (residues 1–322 of the full-length 424-residue protein) were prepared as described in ref. 19. They were dialyzed against 20 mM Tris–Cl buffer (pH 7.5) containing 50 mM NaCl, 10 mM 2-mercaptoethanol, and saturated l-tyrosine, saturated 3-iodo-l-tyrosine (Sigma), or 2 mM Tyr-AMS (TyrSA, RNA-TEC, Leuven, Belgium) and were concentrated to 8–10 mg/ml with a Vivaspin concentrator (Vivascience, Hannover, Germany).
Crystallization of the TyrRS proteins was performed by the hanging-drop vapor diffusion technique. Crystallization of the mutant protein complexes with l-tyrosine and 3-iodo-l-tyrosine was performed at 4°C by using 3 μl of the protein solution and 3 μl of the reservoir solution, containing 100 mM Hepes–Na buffer (pH 7.5) with 2% (vol/vol) polyethylene glycol (PEG) 400 and 2 M ammonium sulfate. Crystallization of the mutant protein complexes with Tyr-AMS was performed at 20°C by using 3 μl of the protein solution and 3 μl of the reservoir solution, containing 85 mM Hepes–Na buffer (pH 7.5), 1.7% (vol/vol) PEG 400, and 1.7 M ammonium sulfate. Under both conditions, rounded cubic crystals appeared within a week and reached maximal dimensions (0.5 × 0.5 × 0.5 mm3)in ≈2 weeks.
X-Ray Data Collection and Structure Determination. The crystals were soaked in a cryoprotectant solution containing 100 mM Hepes–Na buffer (pH 7.5), 20 mM magnesium chloride, 15% (wt/vol) polyethylene glycol 4000, and 20% (vol/vol) ethylene glycol and were flash-frozen at 100 K. All of the data sets were collected at the beamline BL41XU at SPring-8 (Harima, Japan). The crystals of the mutant TyrRS·3-iodo-l-tyrosine and mutant TyrRS·l-tyrosine complexes belong to the space group P3121 with a = b = 82.9 Å and c = 93.4 Å and a = b = 82.9 Å and c = 93.0 Å, respectively. The crystals of the mutant TyrRS·Tyr-AMS complexes belong to the space group P3221 with a = b = 83.6 Å and c = 183 Å. The data were processed with denzo and scalepack (20). The statistics for all of the data sets are listed in Table 1.
Table 1. X-ray data collection and refinement statistics.
| 37V195C·3-IY | 37V195C·tyrosine | 37V195C·Tyr-AMS | |
|---|---|---|---|
| Data collection | |||
| Wavelength, Å | 0.9704 | 0.9704 | 1.0000 |
| Resolution, Å | 50–2.0 | 50–2.0 | 50–2.7 |
| Measured reflections | 214,474 | 209,249 | 139,612 |
| Unique reflections | 25,329 | 25,225 | 20,149 |
| Completeness,* % | 98.4 (98.0) | 98.5 (97.5) | 95.3 (93.4) |
| I/σ (I)* | 34.1 (5.4) | 30.2 (4.2) | 15.7 (2.9) |
| Rsym,† % | 3.6 (15.4) | 5.9 (24.9) | 8.0 (32.4) |
| Refinement statistics | |||
| Resolution, Å | 50–2.0 | 50–2.0 | 50–2.7 |
| Protein atoms | 2,536 | 2,528 | 5,022 |
| Substrates per subunit | 1 | 1 | 1 |
| Water oxygens | 302 | 286 | 115 |
| Rcryst,‡ % | 16.8 | 18.7 | 21.7 |
| Rfree,§ % | 21.9 | 22.4 | 28.2 |
| rms deviations | |||
| Bond length, Å | 0.020 | 0.008 | 0.007 |
| Bond angle, ° | 1.80 | 1.40 | 1.30 |
3-IY, 3-iodo-l-tyrosine.
Numbers in parentheses are for the last shell
Rsym = Σhkl|Iavg – Ii|/ΣhklIi
Rcryst = Σhkl‖Fo| – |Fc‖/Σhkl|Fo|
Rfree = Rcryst using 10% of Fo sequestered before refinement
We determined the TyrRS structures by molecular replacement with the program molrep (21, 22), using the wild-type TyrRS monomer structure (19) for the TyrRS·3-iodo-l-tyrosine/l-tyrosine complexes and the biologically active dimer form for the mutant TyrRS·Tyr-AMS complex as the starting models. The 3-iodo-l-tyrosine and Tyr-AMS models were generated by the Dundee prodrg2 server (23), and the mutant TyrRS model was automatically generated from that of the wild type by cns (24). The final models of the TyrRS·substrate complexes were built manually by using the program o (25).
The refinement of these structures was carried out by using cns. The refinement included rigid body refinement and several rounds of simulated annealing refinement, using a starting temperature of 2,500 K, as well as individual B-factor refinement. From each data set, 10% of the starting data were randomly chosen and set aside for crossvalidation. After the majority of these models were built, the water molecules were picked automatically, as implemented in cns, and several rounds of refinement were executed. Then, the water molecules were selected manually. The refinements for the mutant TyrRS·3-iodo-l-tyrosine and the mutant TyrRS·l-tyrosine were converged to final R factors of 16.8% and 18.7%, with Rfree values of 21.9% and 22.4%, respectively (50- to 2.0-Å-resolution range). The refinements for the mutant TyrRS·Tyr-AMS were converged to final Rcryst and Rfree of 24.6% and 27.7%, respectively (50- to 2.7-Å-resolution range).
All of the structural images were generated by the program cuemol (http://cuemol.sourceforge.jp/en) and were rendered in the program pov-ray (www.povray.org). Superpositions of the structures were produced by the program lsqkab (22).
Preparation of TyrRS Mutants and Aminoacylation Assays. The expression plasmids bearing the full-length TyrRS mutants were generated from the plasmid for the wild-type TyrRS (3) by using the PCR technique. The sequences of the genes were confirmed by DNA sequencing. The mutant proteins were purified as described in ref. 3. E. coli tRNATyr transcripts were also prepared as described in ref. 3. Aminoacylation reactions were carried out at 37°C in 100 mM Tris–Cl buffer (pH 7.5) containing 15 mM magnesium chloride, 40 mM potassium chloride, 0.05 mg·ml-1 BSA, 1 mM DTT, 4 mM ATP, 100 μM 3-iodo-l-tyrosine/l-tyrosine, 10 μM tRNATyr, and 100–500 nM enzyme. Analyses of aminoacylated tRNAs were carried out by acidic PAGE as described in ref. 3.
Results and Discussion
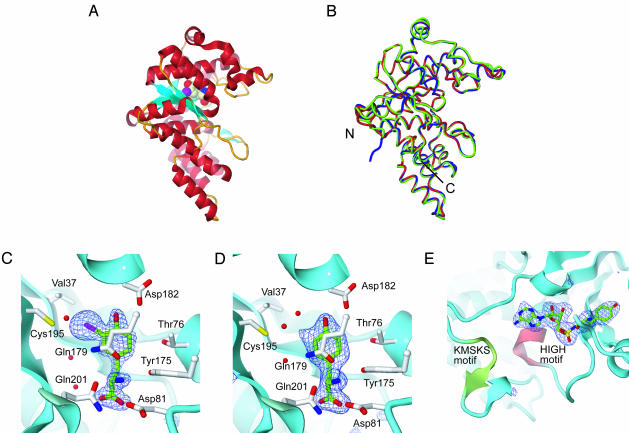
Overall Structures. The crystal structures of the E. coli 37V195C mutant TyrRS catalytic domain (residues 1–322) complexed with 3-iodo-l-tyrosine and l-tyrosine were solved and refined to final R factors of 16.8% and 18.7% (Rfree = 21.9% and 22.4%), respectively, at resolutions of 2.0 Å (Fig. 1). Each asymmetric unit contains one TyrRS subunit, including the Rossmann-fold catalytic domain, and one amino acid molecule. In each structure, the two subunits are related by a crystallographic twofold symmetry axis and form a homodimer like those of the B. stearothermophilus TyrRS structure (14). As expected, these structures and the wild-type E. coli TyrRS·l-tyrosine structure (19) also superpose well on each other, with rms deviations for the main-chain atoms of <1 Å (Fig. 1B).
Fig. 1.
Structures of the 37V195C mutant E. coli TyrRS complexed with 3-iodo-l-tyrosine/l-tyrosine. (A) The ribbon model of the monomeric structure of the mutant TyrRS·3-iodo-l-tyrosine complex. The 3-iodo-l-tyrosine is shown by a Corey–Pauling–Koltun model. The α-helices and β-strands are shown in red and cyan, respectively. (B) The superposed structures of wild-type TyrRS·l-tyrosine (green), 37V195C TyrRS·3-iodo-l-tyrosine (red), and 37V195C TyrRS·l-tyrosine (blue). The N and C termini are marked. (C–E) FO - FC simulated-annealing omit electron density maps (3.5 σ) around the amino acid-binding sites of the mutant TyrRS complexed with 3-iodo-l-tyrosine (C), l-tyrosine (D), and Tyr-AMS (E). The carbon atoms of the substrate, nitrogen, oxygen, sulfur, and iodine atoms are shown by green, blue, red, yellow, and purple sticks, respectively. Water oxygens are shown as red spheres. The main-chain structures are shown by cyan ribbons. The HIGH and KMSKS motifs are shown by red and green ribbons, respectively.
The crystal structure of the 37V195C mutant enzyme complexed with Tyr-AMS was also solved by the same method and was refined to a final R factor of 24.6% (Rfree = 27.7%) at a resolution of 2.7 Å. These crystals belong to the space group P3221, in contrast to the protein·amino acid complex crystals, which belong to P3121. Each asymmetric unit is the biologically active homodimeric form. The subunits of their structures also superpose well on those of the TyrRS·l-tyrosine/3-iodo-l-tyrosine structures. In both structures, the substrate is bound to the Rossmann-fold domain of each subunit (Fig. 1 A) and is well ordered, as shown in Fig. 1 C–E.
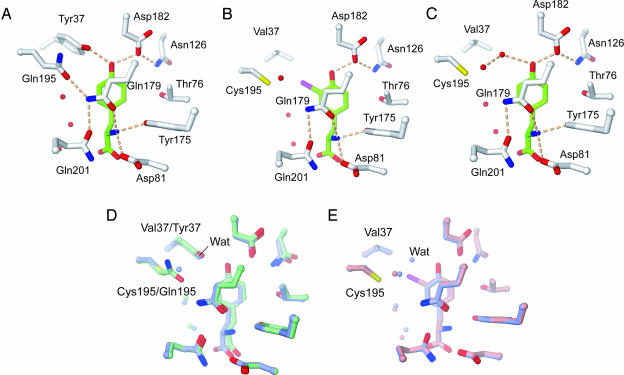
Hydrogen Bonds of the 37V195C Mutant TyrRS with 3-Iodo-l-tyrosine and l-tyrosine. In the 37V195C mutant, the substitution of Val and Cys for the wild-type Tyr-37 and Gln-195 residues, respectively, creates the space for the bulky iodine atom of 3-iodo-l-tyrosine (described in detail below) but removes the hydrogen bonds between Tyr-37 and the substrate l-tyrosine and between Gln-195 and Gln-179 in the wild-type complex (Fig. 2 A–C). The other hydrogen-bonding schemes of the substrate-binding pocket of the E. coli TyrRS 37V195C mutant with 3-iodo-l-tyrosine (Fig. 2B) and l-tyrosine (Fig. 2C) are the same as those of the wild-type E. coli TyrRS (Fig. 2 A) (19) and the B. stearothermophilus TyrRS (13) with l-tyrosine. In the E. coli TyrRS 37V195C mutant, Asp-81, Tyr-175, and Gln-179 hydrogen-bond with the α-amino group of the substrate amino acid (Fig. 2 B and C), as in the wild-type TyrRS (Fig. 2 A) (19). However, the position of one of the carboxyl group oxygens of l-tyrosine is slightly shifted in the 37V195C mutant, compared with the wild-type TyrRS (Fig. 2D). This shift is due to the 37V195C mutation and may have caused the almost 250-fold decrease in the kcat value for the l-tyrosine activation reaction (3). However, the carboxyl groups of 3-iodo-l-tyrosine and l-tyrosine in the 37V195C mutant TyrRS structures superpose better (Fig. 2E). The kcat value for 3-iodo-l-tyrosine is ≈10-fold higher than that for l-tyrosine (3).
Fig. 2.
Comparison of the substrate-binding site structures of E. coli TyrRSs. (A–C) The amino acid-binding site of the wild-type TyrRS·l-tyrosine (A) (19), 37V195C TyrRS·3-iodo-l-tyrosine (B), and 37V195C TyrRS·l-tyrosine (C) complexes. The colors of the atoms are the same as in Fig. 1 C and D. The hydrogen bonds are indicated by pink broken lines. (D) Superposed 37V195C (light blue) and wild-type (green) TyrRS·l-tyrosine structures. The water molecules are shown by balls. The water molecule that hydrogen-bonds to the l-tyrosine is labeled “Wat.” (E) Superposed 37V195C mutant TyrRS structures complexed with 3-iodo-l-tyrosine (pink) and l-tyrosine (light blue).
Asp-182 hydrogen-bonds with the phenolic hydroxyl group of 3-iodo-l-tyrosine and l-tyrosine (Fig. 2). The hydroxyl group recognition is more dependent on this hydrogen bond in the 37V195C mutant than in the wild type, because another hydrogen bond with Tyr-37 (Fig. 2 A) has been removed. Actually, in the 37V195C TyrRS mutant, additional mutations that affect the hydrogen bonding, such as the replacement of Asp-182 by Asn or Leu and/or the replacement of Asn-126 by Asp, drastically decrease the activity for 3-iodo-l-tyrosine (Fig. 5, which is published as supporting information on the PNAS web site). The presence of the iodine atom in 3-iodo-l-tyrosine may have affected the strength of the hydrogen bond between its phenolic hydroxyl group and Asp-182. The distances between the oxygen atoms of the hydroxyl group and the Asp-182 side chain are nearly the same in the 3-iodo-l-tyrosine- and l-tyrosine-bound 37V195C mutant TyrRS structures (Fig. 2E). Nevertheless, the hydrogen bond for 3-iodo-l-tyrosine seems to be stronger than that for l-tyrosine, because the iodine atom of 3-iodo-l-tyrosine attracts electrons in the phenyl ring and the acidity of the phenolic hydroxyl group is higher than that of l-tyrosine. However, the mutations of Asp-182 and Asn-126 showed that the hydrogen bond of Asp-182 is dispensable for binding the hydroxyl group of l-tyrosine (Fig. 5). A water molecule exists in the 37V195C·l-tyrosine structure in place of the iodine atom in the 37V195C TyrRS mutant·3-iodo-l-tyrosine structure (Fig. 2C). This water molecule superposes on the Tyr-37 hydroxyl group of the wild-type enzyme (Fig. 2D). Interestingly, such a water molecule also is seen in the structure of the Y34F mutant of B. stearothermophilus TyrRS, which has only slightly lower activity than the wild type (26).
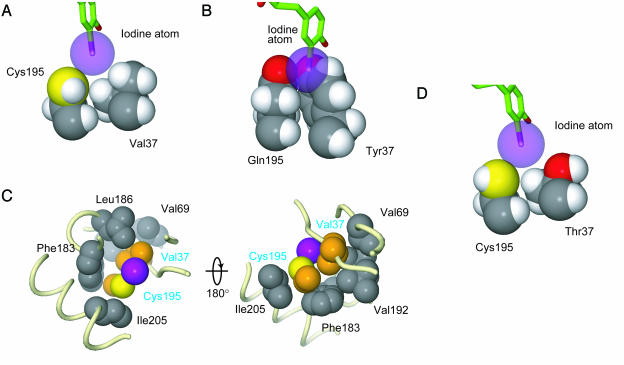
Recognition of the 3-Iodo Group by the 37V195C Mutant TyrRS. The wild-type TyrRS never recognizes 3-iodo-l-tyrosine as the substrate (3). However, the Tyr-37→Val and Gln-195→Cys mutations expand the binding pocket, thus creating the space that accommodates the large iodine atom of 3-iodo-l-tyrosine (Fig. 2B). The iodine atom of 3-iodo-l-tyrosine fits snugly with residues 37 and 195 of the 37V195C mutant TyrRS (Fig. 3A). The side chain of Val-37 is directed toward the iodo group, and the γ-methyl group of Val-37 makes van der Waals contacts with the iodine atom (Fig. 3A). Although the hydrogen atoms are not actually visible in the electron density map, the distance between the iodine atom and these hydrogen atoms just allows the rotation of these Cγ hydrogen atoms, without causing steric hindrance with the iodine atom. However, Cys-195 interacts with the iodine atom of 3-iodo-l-tyrosine by its sulfur atom, not the hydrogen atom of the thiol group. The sulfur atom also just contacts the iodine atom of 3-iodo-l-tyrosine by the van der Waals radius (Fig. 3A). The hydrogen atom of the thiol group does not clash with the iodine atom in any possible configuration (Fig. 3 A and D).
Fig. 3.
van der Waals contacts among the side chains of the TyrRSs and the iodine atom. (A) van der Waals contacts among the iodine atom, Val-37, and Cys-195 in the 37V195C TyrRS·3-iodo-l-tyrosine complex. (B) The model of the wild-type TyrRS complexed with 3-iodo-l-tyrosine. The iodine atom clashes with Tyr-37 and Gln-195. (C) Top and bottom views of the residues surrounding and supporting Val-37 and Cys-195 in the 37V195C TyrRS·3-iodo-l-tyrosine structure. The carbon atoms of Val-37 and Cys-195 are colored orange. The main chain is shown by tubes. (D) The model of the 37T195C mutant TyrRS·3-iodo-l-tyrosine complex. All of the side chains are shown by Corey–Pauling–Koltun models. The van der Waals radii of the hydrogen, carbon, oxygen, sulfur, and iodine atoms are 1.2, 1.7, 1.52, 1.8, and 1.98 Å, respectively. The hydrogen and carbon atoms are colored white and gray, respectively, and the colors of the other atoms are the same as in Fig. 1 C and D. The hydrogen atoms were generated by the program cns (24).
Moreover, the conformations of residues 37 and 195 are fixed rigidly by van der Waals interactions with the surrounding residues (Fig. 3C). Val-37 and Cys-195 reinforce each other (Fig. 3A). In addition, Val-37 is tightly supported by Val-69 on the neighboring β-strand and by Leu-186 and Val-192. Cys-195 also is braced through interactions with Phe-183 and Ile-205, although the interactions seem to be less tight than those of Val-37 with Val-69, Leu-186, and Val-192. Actually, this supporting manner is conserved between the wild-type TyrRS and the 37V195C mutant TyrRS, and the Cβ atoms of residues 37 and 195 in both structures, as well as the main-chain atoms, can superpose on each other (Fig. 2D).
Thus, these van der Waals interactions and conformational limitations of residues 37 and 195 form the rigid pocket for the iodo group, and Asp-182 and the other residues anchor 3-iodo-l-tyrosine in the proper location by hydrogen bonds for efficient recognition in the 37V195C mutant TyrRS. Therefore, this binding pocket is highly specific to 3-iodo-l-tyrosine, and other 3-substituted tyrosine analogues, such as 3-amino-l-tyrosine, 3-nitro-l-tyrosine, 3-dl-methoxytyrosine, and l-3-(2-naphthyl)-alanine, cannot be recognized by the 37V195C mutant TyrRS (Fig. 6, which is published as supporting information on the PNAS web site). Similarly, O-methyl-l-tyrosine was not recognized by the mutant TyrRS.
Distinct Roles of Val-37 and Cys-195 in 3-Iodo-l-tyrosine Recognition. The roles of the Val-37 and Cys-195 mutations differ in the selective 3-iodo-l-tyrosine recognition by the E. coli TyrRS, whereas both Val-37 and Cys-195 contact 3-iodo-l-tyrosine by van der Waals interactions.
The 3-iodo-l-tyrosine-bound model of the wild-type TyrRS (Fig. 3B) shows that Tyr-37 produces a severe van der Waals clash between the iodo group and the phenyl ring. The distance between the nearest carbon atom of the phenyl ring and the iodine atom is 1.87 Å, which is shorter than the van der Waals radius of the iodine atom (1.98 Å). By contrast, only the oxygen atom of the Gln-195 carbonyl group overlaps with the iodine atom by 0.89 Å in the van der Waals radius. Furthermore, the oxygen atom of Gln-195 is not likely to overlap with the iodine atom in the Y37V mutant, because the Gln side chain can sufficiently rotate if the Tyr-37 side chain is not there (Fig. 3B). Therefore, the contribution of the Y37V mutation is much larger than that of the Q195C mutation in terms of eliminating the fatal clash. Actually, the Y37V single mutation facilitates the efficient activation of 3-iodo-l-tyrosine, which is never detected with the wild-type TyrRS (3). The efficiency of the 3-iodo-l-tyrosine activation was as high as 53% of that of the l-tyrosine activation (3).
Furthermore, the amino acid that replaces Tyr at position 37 should be Val for the 3-iodo-l-tyrosine recognition, considering the activities of other mutants for the two substrates (3). In contrast to the 37V195C mutant, 37L195C has only a little 3-iodo-l-tyrosine activation activity, and 37I195C has almost none (3). These replacements would cause a steric clash between the iodine atom of 3-iodo-l-tyrosine and the side chain of residue 37. The 37A195C mutant also has 3-fold lower activity than the 37V195C mutant (3), because the van der Waals contact between Ala-37 and the iodine atom would be weaker than that of Val-37 for this distant interaction. Interestingly, even a slight change of the residue size, as in the V37T replacement, also impaired the activity for 3-iodo-l-tyrosine relative to tyrosine by ≈8-fold (data not shown), probably for the same reason (Fig. 3D).
However, the Q195C mutation eliminates the hydrogen bond between Gln-195 and Gln-179 (as shown in Fig. 2), which may destabilize the substrate-binding pocket. Actually, the Q195C single mutant TyrRS activates l-tyrosine more weakly than the wild type and very slightly activates 3-iodo-l-tyrosine (3). When the Q195C mutation was added to the Y37V mutant TyrRS, the activity for l-tyrosine was impaired ≈200-fold, whereas the activity for 3-iodo-l-tyrosine was impaired only 10-fold (3). Thus, in the 37V195C double mutant, the destabilization effect due to the loss of the Gln-195–Gln-179 hydrogen bond seems to be stronger with l-tyrosine than with 3-iodo-l-tyrosine. As described above, the iodine group of 3-iodo-l-tyrosine is fixed by the van der Waals interactions in the pocket of the 37V195C mutant TyrRS, but this pocket is too large to anchor the l-tyrosine side chain. Therefore, 3-iodo-l-tyrosine is likely to be more tightly fixed in the active site than l-tyrosine, which might be the reason for the higher activity for the former.
Intriguingly, the replacements of the Cys-195 residue in the 37V195C mutant TyrRS increased the unfavorable l-tyrosine binding. The 37V195A mutant misactivates the l-tyrosine somewhat more than the 37V195C mutant (3), probably because the space formed by the smaller Ala-195 residue would be filled with a water molecule and the resultant hydrogen-bond network would compensate for the lost Gln-195–Gln-179 hydrogen bond, stabilizing the l-tyrosine binding. The 37V195N mutant also misactivates the l-tyrosine ≈2 times more than 37V195C for the same reason: Asn-195 can hydrogen-bond with Gln-179, as Gln-195 does in the wild-type enzyme. Moreover, even the 37V195S mutant TyrRS misactivates l-tyrosine at almost the same rate as 3-iodo-l-tyrosine (3). A Ser side chain can make a hydrogen bond, whereas a Cys residue cannot. Thus, the Ser-195 hydroxyl group would hydrogen-bond to the nearby water molecule, which would stabilize the tyrosine binding by a hydrogen-bond network. The Ala, Ser, and Asn replacements of Cys-195 also slightly impaired the 3-iodo-l-tyrosine activation activity (3), probably because the carbon atoms replacing the sulfur atom of Cys-195 remain slightly farther from the iodine atom, thus weakening the van der Waals interactions, even if residue 195 accommodates the substrate (Fig. 3C). Taken together, these results demonstrate that Val-37 and Cys-195 of the 37V195C mutant are optimal for specific 3-iodo-l-tyrosine recognition with their distinct roles.
Conserved Tyr-AMS Binding Manner. According to the previous study by Kiga et al. (3), the l-tyrosylation activity of the 37V195C mutant TyrRS is not very high, such that 3-iodo-l-tyrosine (300 μM) was selectively (>95%) incorporated into the protein in the presence of l-tyrosine (80 μM). We confirmed the aminoacylation activity of the 37V195C mutant by a time-course acidic PAGE analysis. This analysis revealed that the 37V195C mutant aminoacylates its cognate tRNA with l-tyrosine 10-fold more slowly than with 3-iodo-l-tyrosine, in the presence of each amino acid at concentrations of 100 μM (data not shown).
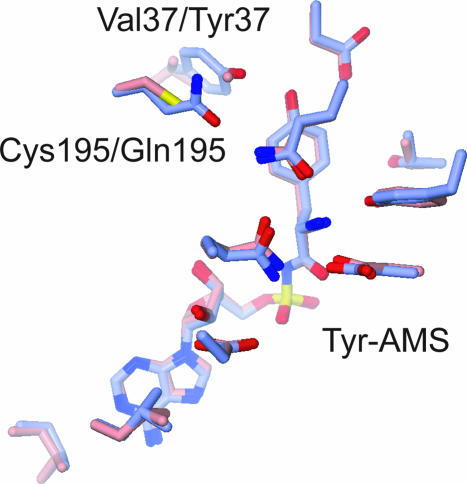
The comparison of the 37V195C TyrRS mutant·Tyr-AMS structure with the wild-type TyrRS·Tyr-AMS structure (19) revealed that the Tyr-AMS-binding sites of these structures superpose well and that the manners of the amino acid-moiety recognition are strongly conserved in both structures (Fig. 4). Therefore, the 3-iodo-l-tyrosine and l-tyrosine side chains are similarly discriminated in the aminoacyl transfer step, as in the amino acid activation step, and this discrimination mechanism is the basis for the selective recognition of 3-iodo-l-tyrosine by the 37V195C TyrRS mutant.
Fig. 4.
Superposed 37V195C (pink) and wild-type (light blue) (19) TyrRS·Tyr-AMS structures. The nitrogen, oxygen, and sulfur atoms are colored blue, red, and yellow, respectively.
Supplementary Material
Acknowledgments
We acknowledge the contributions of Drs. N. Ito (RIKEN), M. Kawamoto, and H. Sakai (Japan Synchrotron Radiation Research Institute, Hyogo, Japan) for the synchrotron data collection at BL41XU in SPring-8. We thank Dr. D. Kiga (University of Tokyo, Tokyo) for helpful advice. We also thank C. Saito and Prof. S. Takahashi (Japan Women's University, Tokyo) for help in the preparation of the TyrRS proteins. This work was supported in part by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (to S.Y.), and the RIKEN Structural Genomics/Proteomics Initiative, the National Project on Protein Structural and Functional Analyses of MEXT (S.Y.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TyrRS, tyrosyl-tRNA synthetase; 37V195C, Escherichia coli TyrRS with the Y37V and Q195C mutations; Tyr-AMS, 5′-O-[N-(l-tyrosyl)sulfamoyl]adenosine.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1WQ3, 1WQ4, and 1VBN).
References
- 1.Koide, H., Yokoyama, S., Kawai, G., Ha, J. M., Oka, T., Kawai, S., Miyake, T., Fuwa, T. & Miyazawa, T. (1988) Proc. Natl. Acad. Sci. USA 85, 6237-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang, L., Brock, A., Herberich, B. & Schultz, P. G. (2001) Science 292, 498-500. [DOI] [PubMed] [Google Scholar]
- 3.Kiga, D., Sakamoto, K., Kodama, K., Kigawa, T., Matsuda, T., Yabuki, T., Shirouzu, M., Harada, Y., Nakayama, H., Takio, K., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 9715-9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto, K., Hayashi, A., Sakamoto, A., Kiga, D., Nakayama, H., Soma, A., Kobayashi, T., Kitabatake, M., Takio, K., Saito, K., et al. (2002) Nucleic Acids Res. 30, 4692-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin, J. W., Cropp, T. A., Anderson, J. C., Mukherji, M., Zhang, Z. & Schultz, P. G. (2003) Science 301, 964-967. [DOI] [PubMed] [Google Scholar]
- 6.Zhang, Z., Alfonta, L., Tian, F., Bursulaya, B., Uryu, S., King, D. S. & Schultz, P. G. (2004) Proc. Natl. Acad. Sci. USA 101, 8882-8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson, J. C., Wu, N., Santoro, S. W., Lakshman, V., King, D. S. & Schultz, P. G. (2004) Proc. Natl. Acad. Sci. USA 101, 7566-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santoro, S. W., Wang, L., Herberich, B., King, D. S. & Schultz, P.G. (2002) Nat. Biotechnol. 20, 1044-1048. [DOI] [PubMed] [Google Scholar]
- 9.Chin, J. W., Santoro, S. W., Martin, A. B., King, D. S., Wang, L. & Schultz, P. G. (2002) J. Am. Chem. Soc. 124, 9026-9027. [DOI] [PubMed] [Google Scholar]
- 10.Chin, J. W., Martin, A. B., King, D. S., Wang, L. & Schultz, P. G. (2002) Proc. Natl. Acad. Sci. USA 99, 11020-11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, Z., Gildersleeve, J., Yang, Y. Y., Xu, R., Loo, J. A., Uryu, S., Wong, C. H. & Schultz, P. G. (2004) Science 303, 371-373. [DOI] [PubMed] [Google Scholar]
- 12.Wang, L., Brock, A. & Schultz, P. G. (2002) J. Am. Chem. Soc. 124, 1836-1837. [DOI] [PubMed] [Google Scholar]
- 13.Brick, P. & Blow, D. M. (1987) J. Mol. Biol. 194, 287-297. [DOI] [PubMed] [Google Scholar]
- 14.Brick, P., Bhat, T. N. & Blow, D. M. (1989) J. Mol. Biol. 208, 83-98. [DOI] [PubMed] [Google Scholar]
- 15.Qiu, X., Janson, C. A., Smith, W. W., Green, S. M., McDevitt, P., Johanson, K., Carter, P., Hibbs, M., Lewis., C., Chalker, A., et al. (2001) Protein Sci. 10, 2008-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang, X. L., Skene, R. J., McRee, D. E. & Schimmel, P. (2002) Proc. Natl. Acad. Sci. USA 99, 15369-15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yaremchuk, A., Kriklivyi, I., Tukalo, M. & Cusack, S. (2002) EMBO J. 21, 3829-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, T., Nureki, O., Ishitani, R., Yaremchuk, A., Tukalo, M., Cusack, S., Sakamoto, K. & Yokoyama, S. (2003) Nat. Struct. Biol. 10, 425-432. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, T., Takimura, T., Sekine, R., Vincent, K., Kamata, K., Sakamoto, K., Nishimura, S. & Yokoyama, S. (2005) J. Mol. Biol. 346, 105-117. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 21.Vagin, A. & Teplyakov, A. (1997) J. Appl. Crystallogr. 30, 1022-1025. [Google Scholar]
- 22.Collaborative Computational Project, No. 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 23.van Aalten, D. M., Bywater, R., Findlay, J. B., Hendlich, M., Hooft, R. W. & Vriend, G. (1996) J. Comput. Aided Mol. Des. 10, 255-262. [DOI] [PubMed] [Google Scholar]
- 24.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 25.Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 26.Fothergill, M. D. & Fersht, A. R. (1991) Biochemistry 30, 5157-5164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.