Abstract
Beekeeping has been a highly valued industry in Taiwan. As a result, many subspecies of Apis mellifera have been introduced to Taiwan since 1911, leading to the hybridization of different subspecies. In order to know the matrilineal origins of Taiwan A. mellifera, a total of 280 samples collected from 33 apiaries throughout the island were examined. Using PCR-RFLP of four mitochondrial gene fragments, i.e., the non-coding region between tRNAleu and cytochrome c oxidase subunit II (intergenic tRNAleu-COII), cytochrome b (Cyt b), large subunit rRNA (Ls rRNA) and cytochrome c oxidase subunit I (COI), we only found two haplotypes exist in 280 samples. Haplotypes ababa and bbbaa account for 87% of these Western bees belonged to the Eastern European (C) lineage and 13% belonged to the Middle East (Z) lineage, respectively, with the latter being totally absent in northern Taiwan. African (A) and Mellifera (M) lineages, officially imported once in 1990s and 1930s respectively, were not detected. The identification of subspecies of A. mellifera and survey of their distribution on the island are expected to facilitate efficient breeding programs and establish a more booming beekeeping industry.
Keywords: Apis mellifera, Mitochondrial DNA, RFLP
1. Introduction
Taiwan is located among the western pacific rim. To the north lie Japan; to the south are the Philippine Islands; to the west is Mainland China. The beekeeping industry in Taiwan owes its popularity and economic importance to the tropical/subtropical climate which provides an abundant supply of rich and diverse nectar plants (An et al., 2004). In addition to help pollinating crops, the 120,000 managed colonies of Apis mellifera on this island produced in 2015 ca. 12,000 tons of varietal honey as well as 430 tons of royal jelly (ranking the second in the world) (Bogdanov, 2015).
Official records have shown that A. mellifera subspecies, including armeniaca, capensis, carnica, caucasica, cypria, ligustica and mellifera were imported to improve the yield of various bee products during the period of 1911–1997 (Supplementary Information Table 1) (An et al., 2004, Sung et al., 2006). Since then, none has been imported officially. In Taiwan, these Western bees of some apiaries are known to give high yield of royal jelly while bees from other yards are better honey producer. It has been generally assumed that A. mellifera in Taiwan is composed of hybrids of different subspecies. Nevertheless, no study has been carried out up to the present to elucidate the makeup of honey bees managed by the beekeepers. We believe that identification of honey bee subspecies via molecular characterization is essential for more advanced management in apiculture in Taiwan.
Twenty-eight subspecies of A. mellifera have been identified morphologically. They are classified, according to their geographic origins, into four branches/lineages, i.e., Africa (A), Western Europe (M), South-Eastern Europe (C) and Middle East (O) (Ruttner, 1988, Engel, 1999, Gupta, 2014). However, phylogeographical studies have shown that variations of morphological characters of these subspecies resulted from adaption to local environments often complicate their proper identification. Therefore, molecular methods using DNA markers, such as mitochondrial DNA (mtDNA), microsatellites and single nucleotide polymorphisms (SNPs), have been developed. Among them, analysis of restriction site polymorphisms of mtDNA fragments, characterized by their maternal inheritance, has been widely used to identify subspecies of honey bees. The length polymorphism of tRNAleu-COII region of mtDNA, due to the absence/presence of P element and different copy numbers of Q element, which correlates with their evolutionary lineages, has been frequently used to classify A. mellifera lineages (Garnery et al., 1992, Garnery et al., 1993).
Among the five mitochondrial lineages identified in A. mellifera (Meixner et al., 2013), A, M and C roughly match with the morphological branches A, M and C (Cornuet and Garnery, 1991, Garnery et al., 1992). The original 4th mitochondrial lineage O from the Near East (Franck et al., 2000, Palmer et al., 2000) was split into mitochondrial C and Z lineages (Meixner et al., 2013), with a sub-lineage of A embedded in the latter (Alburaki et al., 2011, Alburaki et al., 2013). The 5th mitochondrial lineage Y has been identified in Ethiopia (Franck et al., 2001).
The existence of honey bees has been threatened by multiple factors, such as pesticide usage on crops, fragmentation and loss of habitat as well as pathogens and parasites (Antunez et al., 2009, Rosenkranz et al., 2010, Sanchez-Bayo et al., 2016). Colony Collapse Disorder (CCD), a combination of deadly effects, has widely occurred across Europe and America (Stokstad, 2007, van der Zee et al., 2012, Lee et al., 2015). Besides, the natural biodiversity of honey bees may be declined by the selective management of those subspecies that bring forth profits to the beekeepers. In view of the fact that CCD has not been observed in Taiwan and high yield of honey/royal jelly has been achieved, we intended to establish the matrilineal origins of these precious honey bees by analyzing four fragments of mtDNA, i.e., the non-coding region between tRNAleu and cytochrome c oxidase subunit II (intergenic tRNAleu-COII), cytochrome b (Cyt b), large subunit rRNA (Ls rRNA) and cytochrome c oxidase subunit I (COI) by PCR-RFLP and sequencing the tRNAleu-COII fragments of samples collected throughout the island.
2. Materials and methods
2.1. Sampling
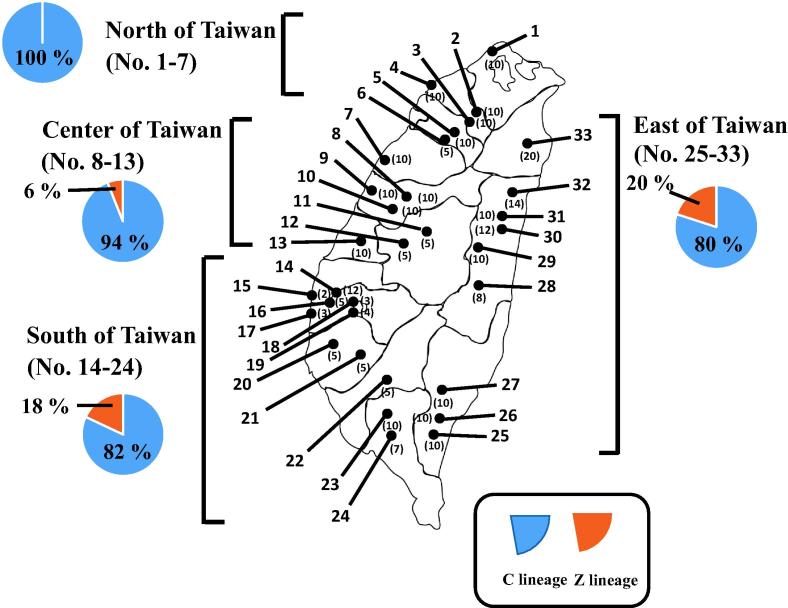
A. mellifera samples representing 280 colonies were collected from northern (No. 1–7 apiary), central (No. 8–13 apiary), southern (No. 14–24 apiary) and eastern (No. 25–33 apiary) regions of Taiwan during September 2015–February 2016 (Fig. 1 and Supplementary Information Table 2). Five workers from each colony were preserved in 99% ethanol and stored at room temperature. One individual worker per colony was subjected for total DNA extraction.
Figure 1.
The locations of thirty-three apiaries throughout Taiwan with the haplotype frequency. Total 280 A. mellifera colonies were sampled from northern (No. 1–7), central (No. 8–19), southern (No. 20–24) and eastern (No. 25–33) regions of Taiwan. Numbers in brackets represent number of colonies sampled at each apiary. The pie charts in four regions represent the haplotype frequency.
2.2. Total DNA extraction
Total DNA extraction was performed using Easy DNA High-Speed Extraction Tissue kit (Saturn Biotech, Australia) and following the manufacturer’s protocol. One foreleg from one individual bee was removed and dried for 5 min at 50 °C. Then it was homogenized in 100 μL mixture solution of Solution 1A, Solution 1B and Solution 2, and incubated for 30 min at 95 °C. After that, samples were spun down to remove cell debris and the supernatants with DNA were subjected to mtDNA analysis.
2.3. PCR amplification of mtDNA fragment
The fragments of the intergenic tRNAleu-COII region, Cyt b, Ls rRNA and COI of mitochondria were selected for PCR amplification with primer pairs of tRNAleu-COII region (Garnery et al., 1993), Cyt b (Crozier et al., 1991), Ls rRNA (with slight modification from Hall and Smith (1991)) and COI (Hall and Smith, 1991) (Table 1). PCR were performed using the thermal profile: 98 °C for 30 s, 35 cycles of 98 °C for 10 s, 40/45/50 °C for 15 s, 72 °C for 50 s and finished with 72 °C for 5 min. Each PCR reaction contains 5× PCR buffer, 0.1 μM of each primer pair, 10 mM dNTP, 2 μL of DNA and 0.25 μL of Phusion™ DNA Polymerase (NEB, USA) to make a final volume of 25 μL.
Table 1.
Primers used in this study.
| Mitochondrial genes | Primers | Tm (°C) | References |
|---|---|---|---|
| tRNAleu-COII | Forward (Position 3363): 5′ GGCAGAATAAGTGCATTG 3′ | 50 | Garnery et al. (1993) |
| Reverse (Position 3934): 5′ CAATATCATTGATGACC 3′ | |||
| Cyt b | Forward (Position 11400): 5′ TATGTACTACCATGAGGACAAATATC 3′ | 45 | Crozier et al. (1991) |
| Reverse (Position 11884): 5′ ATTACACCTCCTAATTTATTAGGAAT 3′ | |||
| Ls rRNA | Forward (Position 13708): 5′ CTATAGGGTCTTATCGTCCCATA 3′ | 40 | Hall and Smith (1991) |
| Reverse (Position 14443): 5′ GTACCTTTTGTATCAGGGTTGA 3′ | |||
| COI |
Forward (Position 1908): 5′ TTAAGATCCCCAGGATCATG 3′ | 45 |
Hall and Smith (1991) |
| Reverse (Position 2951): 5′ TGCAAATACTGCACCTATTG 3′ | |||
(The position numbers of all primers are based on the mitochondrial sequence of A. mellifera ligustica L06178).
2.4. Digestion with restriction enzymes
Each PCR amplification product was digested with the appropriate restriction enzyme (Table 2) in a final volume of 20 μL, containing 10× reaction buffer, 17 μL of DNA and 1 μL of restriction enzyme (NEB, USA), with overnight incubation at 37 °C.
Table 2.
Summary of restriction enzyme digested patterns of four mitochondrial lineages.
| Mitochondrial genes | Restriction enzymes | Lineages of A. mellifera |
References | |||
|---|---|---|---|---|---|---|
| M | A | C | O | |||
| tRNAleu-COII | DraI | + | + | + | + | Garnery et al. (1993) |
| Cyt b | BglII | + | − | + | + | Crozier et al. (1991) |
| HinfI | + | + | − | + | Suppasat et al. (2007) | |
| Ls rRNA | EcoRI | − | − | + | − | Zaitoun et al. (2008) |
| Suppasat et al. (2007) | ||||||
| COI | HinCII | + | − | − | − | Hall and Smith (1991) |
2.5. Electrophoresis
The fragment sizes of PCR amplification and restriction enzyme digestion products were analyzed by the QIAxcel Advanced system (Qiagen, USA). DNA fragments were separated in QIAxcel gel cartridge using QIAxcel Method AM420 and the collected signal data were converted to gel image by the QIAxcel ScreenGel Software. The size marker and alignment marker used in this system are 100 bp/2.5 kp and 15 bp/5 kb, respectively.
2.6. Cloning and sequencing
The amplified gene fragments of tRNAleu-COII were cloned into pGEM-T vector (Promega, USA) and sequenced from both directions. The resulting sequences were compared to published sequences available in Genbank. DNA sequence alignment was performed, using the Clustal W algorithm of Vector NTI® software V10 (Invitrogen, USA). In the phylogenetic analysis, the nucleotide sequences were aligned to construct a phylogenetic tree by using the neighbor-joining method in the MEGA6 program with a Tamura 3-Parameter model, pairwise deletion of gaps, and 1000 bootstrap replicates (Tamura et al., 2013).
3. Results and discussion
3.1. PCR amplification of mtDNA fragments of A. mellifera
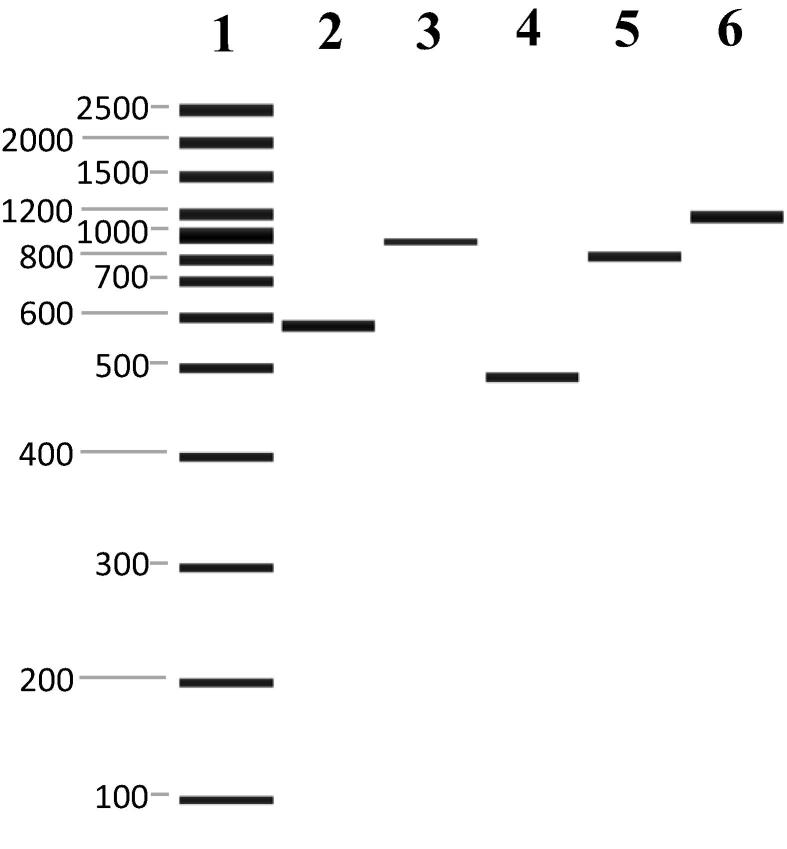
All four mtDNA fragments i.e., the intergenic tRNAleu-COII region, Cyt b, Ls rRNA, and COI were successfully amplified for all samples (Fig. 2). Two amplicon sizes, i.e., 580 and 930 bp for the tRNAleu-COII region were observed. The amplicon of ca. 580 bp appeared in 245 samples while 930 bp amplicon was observed in the remaining 35 samples. This result is similar to that of a study of Thailand Western bees which produced two amplicon sizes 573 and 837 bp of tRNAleu-COII fragments (Suppasat et al., 2007). The details of sequencing analysis of this region are provided in the following section. The amplicon sizes of Cyt b, Ls rRNA and COI are ca. 500, 800, and 1100 bp respectively, matching with those from previous studies (Crozier et al., 1991, Hall and Smith, 1991).
Figure 2.
PCR amplification of four mtDNA fragments of A. mellifera. Four gene fragments, i.e., tRNAleu-COII, Cyt b, Ls rRNA and COI fragments were amplified from mtDNA. Lane 1: DNA size marker (100 bp/2.5 kp); Lane 2 & 3: tRNAleu-COII fragments; Lane 4: Cyt b; Lane 5: Ls rRNA; Lane 6: COI.
3.2. RFLP of four mtDNA regions in 280 samples of A. mellifera
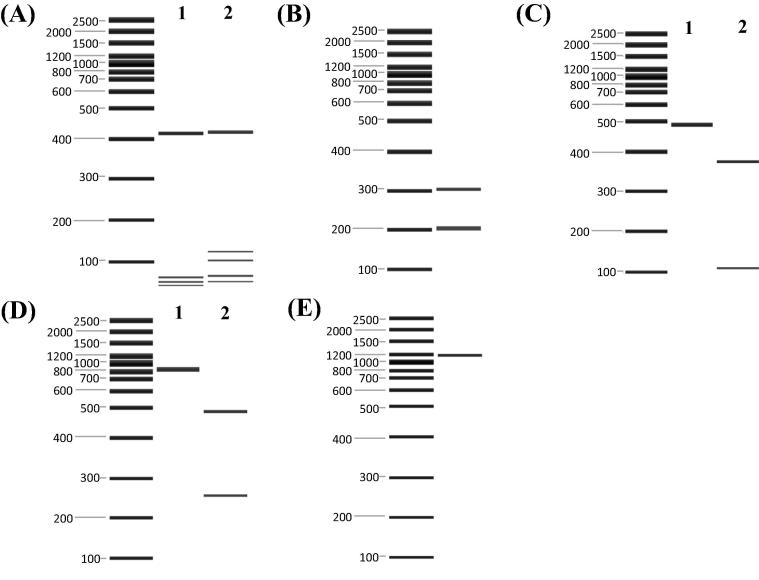
Table 2 summarizes the patterns of restriction enzyme digests of four mtDNA regions used in the characterization of mitochondrial lineages of A. mellifera. The presence of a BglII restriction site in Cyt b fragment in all 280 samples excludes A lineage (pattern ‘b’, Fig.3B); and the absence of HinCII site in COI fragment of all samples indicates that none of them belongs to M lineage (pattern ‘a’, Fig.3E) (Crozier et al., 1991, Hall and Smith, 1991). This leaves only the possibility that these 280 samples belonged to either lineage C or Z; and the presence/absence of two restriction sites, HinfI site in Cyt b fragment and EcoRI site in Ls rRNA fragment, is used to differentiate these two lineages (Suppasat et al., 2007, Zaitoun et al., 2008). Thirty-five samples, with HinfI site in Cyt b fragment (pattern ‘b’) and a lack of EcoRI site in Ls rRNA fragment (pattern ‘a’), were designated as mitochondrial lineage Z (Lane 2 and Lane 1 in Fig.3C and D respectively); and the remaining 245 samples, having EcoRI site in Ls rRNA fragment (pattern ‘b’) but not HinfI site in Cyt b fragment (pattern ‘a’), belonged to C lineage (Lane 1 and Lane 2 in Fig.3C and D respectively). Additionally, the digestion patterns of 580 and 930 bp fragments of tRNAleu-COII with DraI (pattern ‘a’ and ‘b’, Fig.3A) match with the result of Thailand Western bees ThaiA and ThaiB, which have been demonstrated as East Mediterranean (i.e., lineage C) and Middle East (i.e., lineage Z) lineage respectively (Suppasat et al., 2007).
Figure 3.
Restriction enzyme digested patterns of four mtDNA fragments of A. mellifera. (A) tRNAleu-COII fragments digested by DraI; (B) Cyt b digested by BglII; (C) Cyt b digested by HinfI; (D) Ls rRNA digested by EcoRI; (E) COI digested by HinCII. DNA size marker (100 bp/2.5 kp) was used in this work.
The patterns generated from all five restriction digests were combined into composite haplotypes. The composite haplotypes indicate the restriction patterns of five PCR product/restriction enzyme combinations: [tRNAleu-COII/DraI] [Cyt b/BglII] [Cyt b/HinfI] [Ls RNA/EcoRI] [COI/HinCII]. Only two composite haplotypes, i.e., ababa and bbbaa, were found in the 280 colonies of Taiwan A. mellifera, which further refer to lineage C and Z, respectively.
3.3. Sequence analysis of tRNAleu-COII region
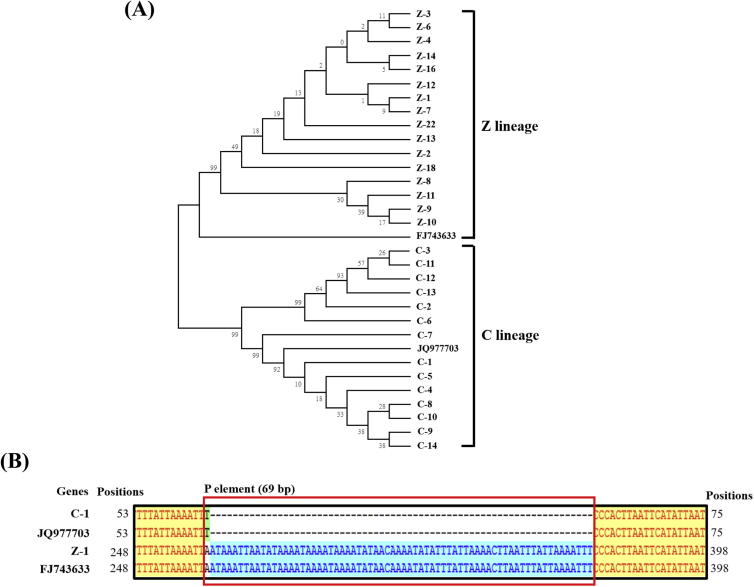
Since PCR amplification of tRNAleu-COII fragment from mtDNA produced one amplicon (580 bp) in all 245 samples of lineage C and a amplicon size of 930 bp in all 35 samples of Z lineage, we decided to sequence the tRNAleu-COII region in some selected samples from both lineages, i.e., 14 samples of C lineages from 14 cities (one sample per city) and 16 samples of Z lineages from Center, South and East of Taiwan. A bootstrap consensus tree was constructed using aforementioned 30 samples with two reference genes (JQ977703 and FJ743633), and can be clustered into two groups, C and Z lineages (Fig.4A). The blast result shows that the 580 bp amplicon was identical to the corresponding sequence of lineage C (JQ977703) (Fig.4B) and the sequence identity among 14 samples of C lineage is 98%. The sequence of 930 bp amplicon was identical to lineage O (FJ743633) which subsequently merged into lineage Z (Franck et al., 2000, Franck et al., 2001, Alburaki et al., 2011) and the sequence identity among 16 samples of Z lineages is 98%. The presence in this amplicon of a 69 bp P element and the RFLP data presented above fully confirm that these 35 samples belong to lineage Z (Fig.4B).
Figure 4.
Phylogenetic analysis of tRNAleu-COII gene fragments. (A) The tRNAleu-COII gene fragments of 32 samples, including 14 samples of C lineages, 16 samples of Z lineages, C lineage reference gene (JQ977703) and Z lineage reference gene (FJ743633) were aligned to construct a phylogenetic tree using the neighbor-joining method in the MEGA6 program with a Tamura 3-Parameter model, pairwise deletion of gaps, and 1000 bootstrap replicates. (B) Alignment analysis of 580 and 930 bp fragments of tRNAleu-COII. Highly conserved residues are marked in yellow and the P element is framed in red color.
3.4. Distribution and abundance of A. mellifera lineages C and Z
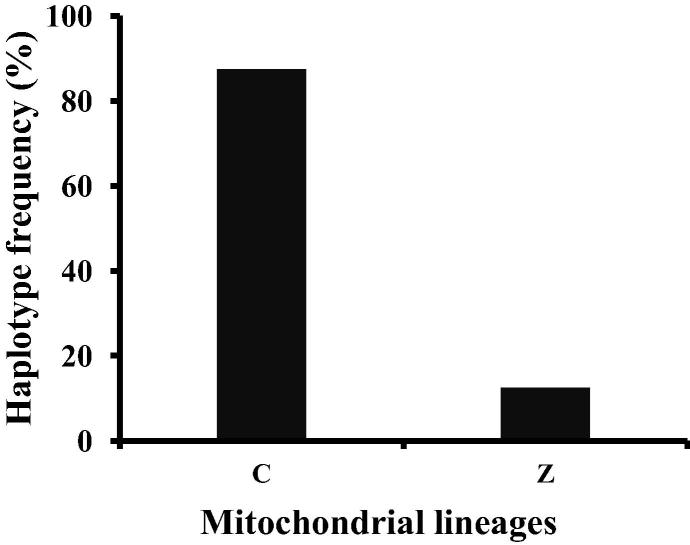
According to Ruttner’s scenario, the haplotype of the 17 imported A. mellifera subspecies in Taiwan consisted of 70% of C lineage, 18% of O lineage, 6% of M lineage and 6% of A lineage (Supplementary Information Fig. 1A) (Ruttner, 1988). Converting into the mitochondrial classification system and taking into consideration of the reclassification of subspecies O lineage into C and Z lineages, the haplotype frequencies of Taiwan Western bee would be 76% of C lineage, 6% of Z lineage, 6% of M lineage and 6% of A lineage (Supplementary Information Fig. 1B) (Meixner et al., 2013). In 280 samples examined in this study, 245 and 35 samples were identified, respectively, as mitochondrial C and Z lineages (Fig. 5). The bees of C lineage distribute throughout the entire island, while the bees of Z lineage, absent in the northern region, are mainly present in the southern and eastern regions (Fig. 1).
Figure 5.
Haplotype frequency of 280 A. mellifera samples. The two composite haplotypes, i.e., ababa and bbbaa, refer to the mitochondrial C and Z lineage, respectively.
4. Conclusions
This is the first investigation of the origin of Western bees in Taiwan. The population of Taiwan Western bee mainly consists of Eastern Europe lineage which widely distributes throughout Taiwan. Additionally Taiwan Western bee population also contains a small proportion of Middle East lineage. According to haplotype frequency analysis, the Middle East lineage has increased from the initial 6% to 12.5%, indicating that this lineage population has expanded in Taiwan. The reason for the absence of Western Europe and Africa lineages is uncertain, though one might presume that it was related to the importation of low number of colonies or other beekeeping issues. Further studies will be performed to address the issues of expansion and distribution of Middle East lineage in Taiwan.
Acknowledgements
This work was supported by grant 104-2321-B-005-018 from the Ministry of Science and Technology, Taiwan, ROC. We thank Prof. Wu-Chun Tu for the help on collection of samples from east region of Taiwan and beekeepers of 33 apiaries for providing bee samples. We also thank Dr. Chih-Ning Sun for helpful discussions and critical reading of this manuscript.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.sjbs.2016.12.008.
Appendix A. Supplementary data
Haplotype frequency of the introduced A. mellifera in Taiwan. Haplotype frequency was calculated according to morphometric lineages (A) and mitochondrial lineages (B).
Table 1. Importation history of Western bees in Taiwan (Adapted from Sung et al., 2006). Table 2. The details of 33 apiaries and sampling numbers.
References
- Alburaki M., Bertrand B., Legout H., Moulin S., Alburaki A., Sheppard W.S., Garnery L. A fifth major genetic group among honeybees revealed in Syria. BMC Genet. 2013;14:117–127. doi: 10.1186/1471-2156-14-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alburaki M., Moulin S., Legout H., Alburaki A., Garnery L. Mitochondrial structure of Eastern honeybee populations from Syria, Lebanon and Iraq. Apidologie. 2011;42:628–641. [Google Scholar]
- An J.K., Ho K.K., Chen Y.W. Taipei National Institute for Compilation and Translation; 2004. Apiculture; p. 524. [Google Scholar]
- Antunez K., Martin-Hernandez R., Prieto L., Meana A., Zunino P., Higes M. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia) Environ. Microbiol. 2009;11:2284–2290. doi: 10.1111/j.1462-2920.2009.01953.x. [DOI] [PubMed] [Google Scholar]
- Bogdanov, S., 2015. The Royal Jelly Book. Bee Product Science, pp. 1–14.
- Cornuet J.M., Garnery L. Mitochondrial DNA variability in honeybees and its phylogeographic implications. Apidologie. 1991;22:627–642. [Google Scholar]
- Crozier Y.C., Koulianos S., Crozier R.H. An improved test for Africanized honeybee mitochondrial DNA. Experientia. 1991;47:968–969. doi: 10.1007/BF01929894. [DOI] [PubMed] [Google Scholar]
- Engel M.S. The taxonomy of recent and fossil honey bees (hymenoptera: apidae; apis) J. Hymenoptera Res. 1999;8:165–196. [Google Scholar]
- Franck P., Garnery L., Loiseau A., Oldroyd B.P., Hepburn H.R., Solignac M., Cornuet J.M. Genetic diversity of the honeybee in Africa: microsatellite and mitochondrial data. Heredity. 2001;86:420–430. doi: 10.1046/j.1365-2540.2001.00842.x. [DOI] [PubMed] [Google Scholar]
- Franck P., Garnery L., Solignac M., Cornuet J.-M. Molecular confirmation of a fourth lineage in honey bees from the Near East. Apidologie. 2000;31:167–180. [Google Scholar]
- Garnery L., Cornuet J.M., Solignac M. Evolutionary history of the honey bee Apis mellifera inferred from mitochondrial DNA analysis. Mol. Ecol. 1992;1:145–154. doi: 10.1111/j.1365-294x.1992.tb00170.x. [DOI] [PubMed] [Google Scholar]
- Garnery L., Solignac M., Celebrano G., Cornuet J.M. A simple test using restricted PCR-amplified mitochondrial DNA to study the genetic structure of Apis mellifera L. Experientia. 1993;49:1016–1021. [Google Scholar]
- Gupta, R.K., 2014. Taxonomy and distribution of different honeybee species. In: Beekeeping for Poverty Alleviation and Livelihood Security, vol. 1, pp. 63–103.
- Hall H.G., Smith D.R. Distinguishing African and European honeybee matrilines using amplified mitochondrial DNA. Proc. Natl. Acad. Sci. U.S.A. 1991;88:4548–4552. doi: 10.1073/pnas.88.10.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.V., Steinhauer N., Rennich K., Wilson M.E., Tarpy D.R., Caron D.M., Rose R., Delaplane K.S., Baylis K., Lengerich E.J., Pettis J., Skinner J.A., Wilkes J.T., Sagili R., vanEngelsdorp D. A national survey of managed honey bee 2013–2014 annual colony losses in the USA. Apidologie. 2015;46:292–305. [Google Scholar]
- Meixner M.D., Pinto M.A., Bouga M., Kryger P., Ivanova E., Fuchs S. Standard methods for characterising subspecies and ecotypes of Apis mellifera. J. Apic. Res. 2013;52:1–28. [Google Scholar]
- Palmer M., Smith D., Kaftanoglu O. Brief communication. Turkish honeybees: genetic variation and evidence for a fourth lineage of Apis mellifera mtDNA. J. Hered. 2000;91:42–46. doi: 10.1093/jhered/91.1.42. [DOI] [PubMed] [Google Scholar]
- Rosenkranz P., Aumeier P., Ziegelmann B. Biology and control of Varroa destructor. J. Invertebr. Pathol. 2010;103:96–119. doi: 10.1016/j.jip.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Ruttner F. Springer; Berlin: 1988. Biogeography and Taxonomy of Honeybees. [Google Scholar]
- Sanchez-Bayo F., Goulson D., Pennacchio F., Nazzi F., Goka K., Desneux N. Are bee diseases linked to pesticides? – a brief review. Environ. Int. 2016;89:7–11. doi: 10.1016/j.envint.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Stokstad E. The case of the empty hives. Science. 2007;316:970–972. doi: 10.1126/science.316.5827.970. [DOI] [PubMed] [Google Scholar]
- Sung I.-H., Chiang C.-H., Ho K.-K., Yamane S. A review on the history of beekeeping in taiwan and the development of its research works (hymenoptera, apidae, apinae) Formos. Entomol. 2006;8:129–147. [Google Scholar]
- Suppasat T., Smith D.R., Deowanish S., Wongsiri S. Matrilineal origins of Apis mellifera in Thailand. Apidologie. 2007;38:323–334. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee R., Pisa L., Andonov S., Brodschneider R., Charrière J.-D., Chlebo R., Coffey M.F., Crailsheim K., Dahle B., Gajda A., Gray A., Drazic M.M., Higes M., Kauko L., Kence A., Kence M., Kezic N., Kiprijanovska H., Kralj J., Kristiansen P., Martin Hernandez R., Mutinelli F., Nguyen B.K., Otten C., Özkırım A., Pernal S.F., Peterson M., Ramsay G., Santrac V., Soroker V., Topolska G., Uzunov A., Vejsnæs F., Wei S., Wilkins S. Managed honey bee colony losses in Canada, China, Europe, Israel and Turkey, for the winters of 2008–9 and 2009–10. J. Apic. Res. 2012;51:100–114. [Google Scholar]
- Zaitoun S., Hassawi D.S., Shahrour W. Origin of Jordanian honeybee Apis mellifera (Hymenoptera: Apidae) using amplified mitochondrial DNA. Eur. J. Entomol. 2008;105:41–44. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Haplotype frequency of the introduced A. mellifera in Taiwan. Haplotype frequency was calculated according to morphometric lineages (A) and mitochondrial lineages (B).
Table 1. Importation history of Western bees in Taiwan (Adapted from Sung et al., 2006). Table 2. The details of 33 apiaries and sampling numbers.