Abstract
Riboswitches are highly structured RNA elements that control the expression of many bacterial genes by binding directly to small metabolite molecules with high specificity and affinity. In Bacillus subtilis, two classes of riboswitches have been described that discriminate between guanine and adenine despite an extremely high degree of homology both in their primary and secondary structure. We have identified intermolecular base triples between both purine ligands and their respective riboswitch RNAs by NMR spectroscopy. Here, specificity is mediated by the formation of a Watson–Crick base pair between the guanine ligand and a C residue or the adenine ligand and a U residue of the cognate riboswitch RNA, respectively. In addition, a second base-pairing interaction common to both riboswitch purine complexes involves a uridine residue of the RNA and the N3/N9 edge of the purine ligands. This base pairing is mediated by a previously undescribed hydrogen-bonding scheme that contributes to the affinity of the RNA–ligand interaction. The observed intermolecular hydrogen bonds between the purine ligands and the RNA rationalize the previously observed change in specificity upon a C to U mutation in the core of the purine riboswitch RNAs and the differences in the binding affinities for a number of purine analogs.
Keywords: base pairing, NMR, regulation of gene expression
Riboswitches have been identified as a new class of genetic control elements that modulate gene expression in bacteria, plants, and fungi (1, 2). Binding of small metabolite molecules to these highly structured RNA domains, mostly found in the 5′ untranslated regions (UTRs) of mRNAs, induces an allosteric rearrangement that results in the modulation of gene expression (reviewed in refs. 3–5; Fig. 1A). Riboswitches are composed of a ligand-binding domain and an expression platform that modulates either ribosome binding or transcription antitermination. So far, riboswitches have been reported for a number of different metabolites such as thiamine pyrophosphate, S-adenosylmethionine, FMN, lysine, coenzyme B12, glucosamine-6-phosphate, glycine (reviewed in refs. 3–5), and for the purine bases guanine (6) and adenine (7). All riboswitches bind their respective targets with high affinity and are able to discriminate even against very closely related compounds. For example, the guanine-specific riboswitch binds guanine with a Kd of ≈5 nM (6) but has no affinity for adenine. Adenine binding to the adenine-sensing riboswitch RNA is weaker with a Kd of ≈300 nM (7). Remarkably, adenine does not appear to be the optimal ligand for the RNA, because 2,6-diaminopurine binds much tighter (Kd ≈ 10 nM). Despite their different specificities, adenine- and guanine-responsive riboswitches share a highly conserved primary and secondary structure (7). The only significant difference is a single nucleotide in the core of the riboswitch RNA (Fig. 1B) conserved as a cytosine in all guanine-specific riboswitches and as a uridine in the adenine-specific riboswitches (7). Upon mutation of the cytosine to uridine, guanine-specific ribos-witches no longer bind to guanine but to adenine instead. The reverse is true for the adenine-specific riboswitches. This finding has led to the proposal that Watson–Crick-type hydrogen bonding between the ligand and the RNA at this position might be responsible for the specificity of these riboswitches (7).
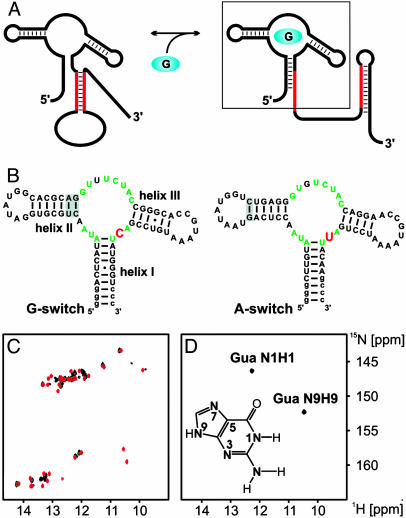
Fig. 1.
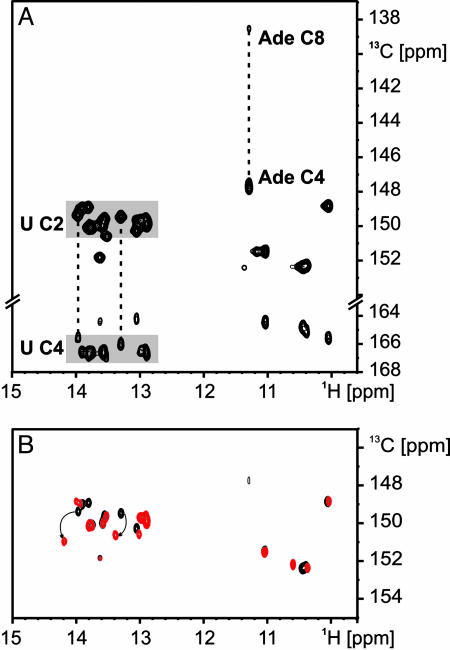
Guanine binding to the aptamer domain of the guanine-responsive riboswitch derived from the 5′ UTR of xpt-pbuX-mRNA in B. subtilis. (A) Conformational change induced by guanine binding to the riboswitch according to ref. 6. The ligand-binding domain is boxed. (B) Comparison of the secondary structures of the G-switch (Left) and A-switch (Right) RNA ligand-binding domains. Core residues conserved in all purine-binding riboswitch sequences are indicated in green (6, 7). The nucleotide that is conserved as a cytosine in all of the guanine-binding riboswitches and as a uridine in all of the adenine-binding riboswitches is shown in red. Extra residues not found in the original sequence and introduced to facilitate in vitro transcription are denoted in lowercase letters. Mutations in helical regions of the RNAs introduced to stabilize the RNA secondary structure are shaded gray. (C) Overlay of the imino regions of 1H, 15N-HSQC spectra of the uniformly 15N-labeled G-switch RNA in its free form (black) and bound to unlabeled guanine (red) at 283 K. (D) Imino region of an 1H, 15N-HSQC spectrum of 15N-labeled guanine bound to only cytidine-13C, 15N-labeled G-switch RNA. In Inset, the numbering scheme for purines is given with guanine as the example.
Groundbreaking recent work has demonstrated the capability of NMR spectroscopy to solve the solution structures of RNAs approaching the size of many of the riboswitches (8, 9). However, NMR structure determination of RNAs in the size range of 70–100 nt often requires multiple selectively or segmentally labeled (10) samples and works best for molecules that can be subdivided into smaller domains that can be studied individually before the whole molecule is tackled in a “divide and conquer” approach (8, 9). However, selective isotope labeling in biomolecular complexes often allows the detailed characterization of intermolecular interactions without the need for a full structure determination even in very large complexes (11–13). In addition, even in larger RNAs it is possible to characterize the secondary structure or structural rearrangements (14–16) by directly analyzing the hydrogen-bonding patterns. Hydrogen bonds involving a NH-donor group and a nitrogen acceptor group can be directly observed in 2hJNN-HNN-COSY experiments (17, 18) that correlates nitrogen atoms by through-hydrogen bond 2hJNN-couplings.
We therefore used NMR spectroscopy in conjunction with appropriate isotope labeling of both the ligand and the RNA to characterize the intermolecular interactions between the purine ligands and the ligand-binding domains of the purine-responsive riboswitch RNAs. Specifically, we investigated the binding of guanine to the ligand-binding domain of the guanine-responsive riboswitch residing in the 5′ UTR of the xpt-pbuX-mRNA (G-switch RNA) and the binding of adenine to the ligand-binding domain of the adenine-responsive riboswitch residing in the 5′ UTR of the ydhL-mRNA (A-switch RNA), both from Bacillus subtilis. By directly observing hydrogen bonds between the ligand and the RNA, we show that in both cases a previously undescribed intermolecular base triple is formed. In either complex, an intermolecular Watson–Crick-type base pair between the purine ligand and the riboswitch RNA is responsible for the selectivity of the ligand recognition, whereas a conserved unusual base-pairing interaction involving the N3/N9 edge of the purine ligand and a uridine of the riboswitch RNA contributes to the affinity of the interaction.
Materials and Methods
Preparation of Labeled Guanine and Adenine. 15N- and 13C, 15N-labeled guanine was prepared by heating commercially available 15N-GTP or 13C, 15N-GTP (Silantes, Munich) in 1 M HCl for 120 min at 373 K. The reaction mixture was cooled and neutralized with a 1 M NaHCO3 solution. The precipitate was washed twice with water and ethanol and dried. We prepared 13C, 15N-labeled adenine by heating commercially available 13C, 15N-ATP (Silantes) in 1 M HCl for 120 min at 373 K. The reaction mixture was cooled, neutralized with a 1 M NaHCO3 solution, and fractionated by HPLC on a preparative C18 column (Vydac, Hesperia, CA, catalog no. 218TP510) equilibrated with water by employing an acetonitrile gradient. Unlabeled guanine, adenine, and 2,6-diaminopurine were purchased from Sigma.
Preparation of RNAs and RNA–Ligand Complexes. 15N- and 15N, 13C-labeled nucleotides were purchased from Silantes, and unlabeled NTPs were purchased from Sigma. All RNAs were prepared by in vitro transcription with T7 RNA polymerase from linearized plasmid DNA templates. The RNAs were purified as described in ref. 19 and finally exchanged into NMR buffer (25 mM KPO4, pH 6.2/50 mM KCl) by using Centricon-10 microconcentrators (Amicon).
For the formation of the guanine/G-switch RNA complexes, a25 μM stock solution of guanine in NMR buffer was mixed with a20 μM G-switch RNA solution in NMR buffer, and the mixture was concentrated to NMR concentrations by using Centricon-10 microconcentrators (Amicon). The adenine/A-switch RNA complex was prepared by adding small aliquots of a 5 mM stock solution of 13C, 15N-labeled adenine in NMR buffer to a 1 mM A-switch RNA solution. The final concentration of the guanine- and adenine–RNA complexes was ≈800–900 μM. The 2,6-diaminopurine–RNA complex sample had a concentration of 120 μM.
NMR Spectroscopy. NMR experiments were performed on Bruker DRX 600 MHz, AV 700 MHz, and AV 800 MHz spectrometers equipped with cryogenic probes and z-axis gradients. All spectra were processed and analyzed by using the Bruker nmrsuite (xwinnmr 3.5) and xeasy (20). NMR spectra were recorded in 90% H2O/10% D2O at a temperature of 283 K by using the watergate (21) water suppression scheme including water flip-back pulses (22). 1H, 15N-heteronuclear single quantum correlation (HSQC)-, 2JHN-1H, 15N-HSQC-, 1H, 13C-HSQC-, H(N)C-, and 15N-edited 2D- or 3D-NOESY experiments were carried out by using standard pulse sequences (23). The 2hJNN-HNN-COSY experiments were performed by using the pulse sequence described in ref. 17 with the following modifications. The 15N carrier frequency was kept at 153 ppm for the 1H, 15N-insensitive nuclei enhanced by polarization transfer steps and at 194 ppm during the NN-COSY step. The nitrogen 180°-square pulses during the NN-COSY step were replaced by 2-ms adiabatic smoothed CHIRP pulses (24). The 1H carrier frequency was kept at the H2O resonance. A mixing time of 20 ms was used for the NN-COSY transfer. A total of 512 scans per t1 time increment were collected.
Results
Binding of Guanine to the G-Switch RNA. A 73-nt RNA corresponding to the ligand-binding domain of the guanine-responsive riboswitch in the 5′ UTR of the xpt-pbuX-mRNA (G-switch RNA) from B. subtilis was prepared by in vitro transcription in either uniformly 15N-labeled form or in a cytosine-13C, 15N-labeled form. The RNA contains an extension of helix I to allow efficient transcription and a mutation in helix II where an unstable A:C base pair is replaced by a more stable Watson–Crick base pair (Fig. 1B) as found in many of the homologous sequences (ref. 6 and also Fig. 7, which is published as supporting information on the PNAS web site). This RNA displays a well resolved set of NMR signals in the imino region of a 1H, 15N-HSQC spectrum already in its free form (Fig. 1C, black). The addition of a stoichiometric amount of unlabeled guanine led to the appearance of new signals in the 1H, 15N-HSQC spectrum with proton chemical shifts in the region of 10–12 ppm and to specific changes in the chemical shifts of other signals (Fig. 1C, red). This result indicated the formation of a stable guanine–RNA complex associated with a structural reorganization of the RNA involving the formation of numerous noncanonical base pairs. The 1H, 15N-HSQC spectra of the wild-type RNA (WTG-switch) in complex with guanine indicated an identical overall fold for the two complexes. Most importantly, the mutations in helix II in the G-switch RNA did not perturb the chemical environment of the bound guanine because the chemical shifts of the ligand were the same in both complexes (Fig. 8, which is published as supporting information on the PNAS web site).
Guanine free in solution exists as a mixture of different tautomeric forms (25). Its nitrogen-bound protons are in fast exchange with the solvent, and, therefore, their NMR signals were not observable (data not shown). In contrast, the bound guanine showed two signals in the imino region of a 1H, 15N-HSQC spectrum that could be observed selectively in a complex of 15N-labeled guanine and only cytidine-13C, 15N-labeled RNA (Fig. 1D). By using HSQC- and HCN-type experiments and chemical shift arguments (Fig. 9, which is published as supporting information on the PNAS web site) and in agreement with the observed intermolecular nuclear Overhauser effects (NOEs) (see below), these two signals were assigned to the H1N1 imino group and the H9N9 group of guanine.
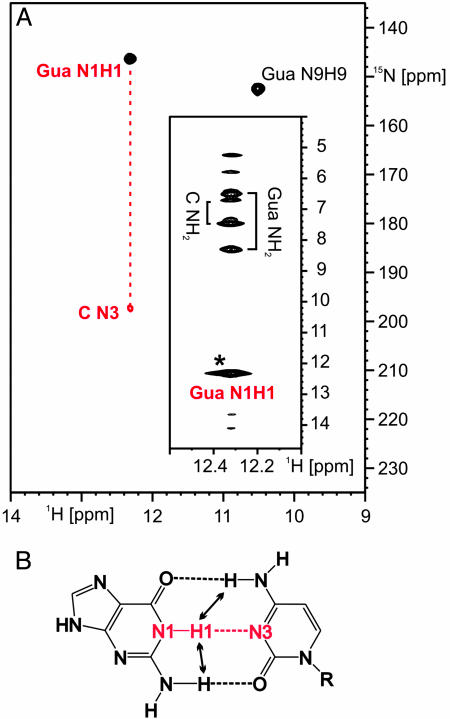
A Watson–Crick G:C Base Pair Between the Guanine Ligand and the G-Switch RNA. The presence of two imino group signals for the bound guanine already indicated that these sites must be protected from exchange with the solvent by hydrogen bonding in the complex. The signal of the N1H1 imino group in the Watson–Crick face of the bound guanine displayed proton and nitrogen chemical shifts (12.3 and 146.3 ppm, respectively) that were well within the typical chemical shift ranges for G:C Watson–Crick base pairs found in RNA (23). A 2D-HNN-COSY experiment recorded on a complex of 15N-labeled guanine and only cytidine-13C, 15N-labeled RNA yielded a single correlation between the H1N1 imino group of the bound G and a cytidine N3 nitrogen (Fig. 2A) as identified by its chemical shift of 197.3 ppm (26). Because of the labeling scheme used, this result represents an intermolecular hydrogen bond, and the correlation between the guanine H1N1 imino group and a N3 nitrogen of a C demonstrates a Watson–Crick-type hydrogen bonding scheme. In addition, the amino group of the bound guanine is involved in a hydrogen bond because its two protons have distinct chemical shifts (8.3 and 6.5 ppm; Fig. 2 A Inset). As expected for a Watson–Crick G:C base pair, the H1N1 imino group of the bound guanine shows strong NOE cross peaks to a cytidine amino group in a 2D-15N-edited NOESY spectrum (Fig. 2 A Inset).
Fig. 2.
Watson–Crick-type hydrogen bonding between guanine and the G-switch RNA. (A) Imino region of a HNN-COSY experiment using 13C, 15N-C-only labeled RNA and 15N-labeled guanine. The correlation between the guanine H1N1 imino group and a cytosine N3 of the RNA is indicated by a red dashed line. (Inset) Strip for the H1N1 imino group of guanine from a 15N-edited-NOESY experiment. An asterisk denotes the diagonal signal. (B) Hydrogen-bonding scheme for a Watson–Crick G:C base pair with the observed hydrogen bond highlighted and arrows denoting the observed NOE contacts that are typical for a Watson–Crick G:C base pair.
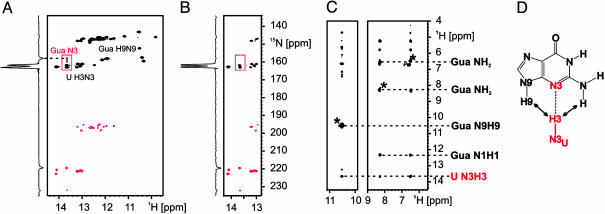
Hydrogen Bonding to the N3 of the Bound Guanine. To characterize additional intermolecular hydrogen bond interactions between the bound guanine and the G-switch RNA, 2D-HNN-COSY experiments were performed for complexes of uniformly 15N-labeled RNA with either 13C, 15N-labeled or unlabeled guanine. In the first case, both intra- and intermolecular hydrogen bonds could be observed, whereas in the latter case, only intramolecular hydrogen bonds of the RNA were detectable. The comparison of the two HNN-COSY spectra revealed a correlation between an uridine H3N3 imino group of the RNA with a nitrogen at 157.9 ppm for the complex containing the 13C, 15N-labeled guanine that was absent in the spectrum of the complex with the unlabeled ligand (Fig. 3 A and B) and therefore must be due to an intermolecular hydrogen bond. The 15N chemical shift of 157.9 ppm identifies this nitrogen as the N3 of the bound guanine (26), indicating the presence of an intermolecular hydrogen bond with a uridine H3N3 imino group as the donor group and the N3 nitrogen of guanine as the acceptor. Such a hydrogen bond brings the donor uridine H3N3 imino group into close proximity to both the guanine H9N9 imino and the guanine amino group. Accordingly, strong NOEs are observable between the uridine H3N3 imino group and the guanine H9N9 imino and amino group (Fig. 3C).
Fig. 3.
Hydrogen bonding between the N3/N9 edge of guanine and the G-switch RNA. (A) Imino region of a HNN-COSY experiment using uniformly 15N-labeled RNA and 15N, 13C-labeled guanine. The correlation between the guanine N3 nitrogen and a uridine H3N3 imino group of the RNA is highlighted with a red box. The position of the H9N9 imino group signal of the bound guanine is also indicated. The signal for the guanine H1N1 imino group is overlapped with signals of the RNA. (B) Region of a HNN-COSY experiment by using uniformly 15N-labeled RNA and unlabeled guanine. The correlation between the guanine N3 nitrogen and a uridine H3N3 imino group of the RNA observed in A is absent. One-dimensional slices at a 1H chemical shift of 13.6 ppm are given for the two HNN-COSY experiments in A and B to illustrate the signal-to-noise ratio in the two experiments. (C) Strip for the H9N9 imino group and the amino group of the bound guanine from a 15N-edited-NOESY experiment with the assignment of intermolecular NOEs between the ligand and the U H3N3 imino group of the RNA. The diagonal peaks are indicated with an asterisk. (D) Hydrogen-bonding scheme for the G:U base-pairing interaction with the observed hydrogen bond highlighted in red and arrows denoting the observed intermolecular NOE contacts.
For the H9N9 imino group of the bound guanine, no correlation was observed in the HNN-COSY spectrum. Because the protection from exchange with the solvent and the chemical shifts strongly suggests that this group is also hydrogen bonded, this finding indicated that in this case the hydrogen bond acceptor was an oxygen rather than a nitrogen. Remarkably, the formation of the hydrogen bond between the uridine H3N3 imino group of the RNA and the guanine N3 nitrogen positions one of the uridine carbonyl groups near the guanine H9N9 imino group (see below). An identical hydrogen-bonding pattern is observed in HNN-COSY experiments for the WT G-switch RNA bound to guanine (Fig. 10, which is published as supporting information on the PNAS web site).
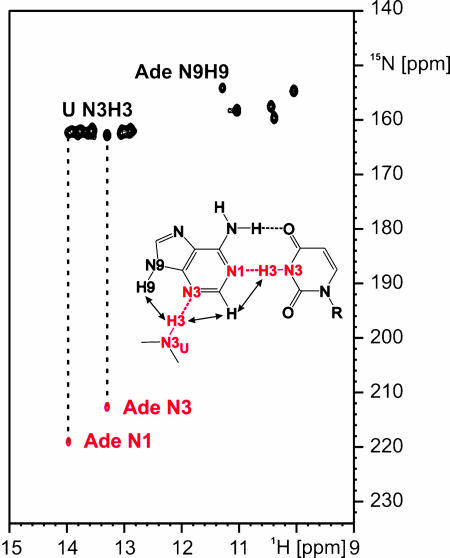
Intermolecular Hydrogen Bonds Between Adenine and the A-Switch RNA. A 71-nt RNA corresponding to the ligand-binding domain of the adenine-responsive riboswitch in the 5′ UTR of the ydhL-mRNA from B. subtilis (A-switch RNA) was prepared by in vitro transcription in only uridine-13C, 15N-labeled form. The addition of 13C, 15N-labeled adenine to this RNA resulted in changes in the imino region of the 1H, 15N-HSQC spectrum of the RNA similar to those described for the G-switch RNA upon addition of guanine. This result indicated the formation of a stable adenine–RNA complex in slow exchange on the NMR time scale (Fig. 11, which is published as supporting information on the PNAS web site) in agreement with the published Kd for the adenine–A-switch complex (7). One of the imino resonances, only observable in the complex, was identified as the adenine H9N9 imino group based on the chemical shifts of the directly attached carbon atoms (Figs. 4A and 5A). The HNN-COSY experiment performed with the complex of 13C, 15N-labeled adenine and only uridine-13C, 15N-labeled A-switch RNA revealed two correlations involving two uridine H3N3 imino groups and the N1 and N3 nitrogens of the bound adenine, respectively (Fig. 4). Therefore, two intermolecular hydrogen bonds were formed. One corresponds to a Watson–Crick-like hydrogen bond with the H3N3 imino group of uridine as the donor and the adenine N1 nitrogen as the acceptor group. In the second one, another uridine H3N3 imino group serves as the donor group whereas the adenine N3 nitrogen is the hydrogen bond acceptor group. Both uridine imino groups displayed strong NOEs to the H2 proton of the bound adenine. In addition, the uridine imino group hydrogen bonded to the adenine N3 showed a strong NOE to the adenine H9N9 imino group (Fig. 12, which is published as supporting information on the PNAS web site).
Fig. 4.
Hydrogen bonding between adenine and the A-switch RNA. Imino region of a HNN-COSY experiment using only uridine-13C, 15N-labeled RNA and 15N, 13C-labeled adenine. The correlations between two uridine H3N3 imino groups of the A-switch RNA and the N1 and N3 nitrogens of the bound adenine, respectively, are indicated by dashed lines. (Inset) Hydrogen-bonding scheme for the intermolecular base-pairing interactions between adenine and the A-switch RNA with the observed hydrogen bonds highlighted and arrows denoting the observed NOE contacts.
Fig. 5.
Carbonyl carbon chemical shifts as indicators of hydrogen bonds. (A) A 2D-H(N)C spectrum of the only uridine 13C, 15N-labeled A-switch RNA in complex with 13C, 15N-labeled adenine. The chemical shift ranges observed for the C2 and C4 carbonyl groups of uridines in Watson–Crick A:U base pairs are shaded in gray. The two uridines involved in intermolecular hydrogen bonds with the ligand (dashed lines) have C2 and C4 chemical shifts close to those for uridines involved in canonical Watson–Crick A:U base pairs. In addition, the correlations between the H9N9 imino group and the C4 and C8 carbons of the bound adenine are indicated. (B) Superimposition of the C2 carbonyl region of 2D-H(N)C spectra of the only uridine 13C, 15N-labeled A-switch RNA in complex with either adenine (black) or 2,6-diaminopurine (red). The chemical shift changes observed for the C2 carbons of the two uridines involved in intermolecular hydrogen bonding with the ligand are indicated by arrows.
Uridine C2 and C4 Carbonyl Groups as Hydrogen Bond Acceptors. Despite being protected from exchange with the solvent and therefore most likely hydrogen bonded, the adenine H9N9 imino group showed no correlation in the HNN-COSY experiment, strongly suggesting an oxygen as the likely hydrogen bond acceptor for this group. Interestingly, hydrogen bonding between the adenine N3 nitrogen and the uridine H3N3 imino group brought one of the uridine carbonyl groups into close spatial proximity to the adenine H9N9 imino group. The direct detection of hydrogen bonds with imino groups as the donor and a carbonyl oxygen as acceptor is virtually impossible in larger RNAs because of the very small size of the corresponding 3hJNCO- and 4hJNN-coupling constants (27, 28). However, the uridine C2 and C4 carbonyl carbon chemical shifts themselves are sensitive reporters of hydrogen bonding (29). The C2 and C4 carbonyl carbons of both uridines that form intermolecular hydrogen bonds with the adenine have chemical shifts similar to those for uridines involved in regular intramolecular Watson–Crick A:U base pairs (Fig. 5A). This finding indicates that in both uridines the C4 position is involved in a hydrogen bond, whereas the C2 position is not. For the uridine that forms a hydrogen bond with the N1 nitrogen of adenine, this result is in agreement with the observed intermolecular Watson–Crick A:U base pair. For the other uridine, this result implies its C4 carbonyl group as the likely hydrogen bond acceptor for the adenine H9N9 imino group (Fig. 6A). In 2,6-diaminopurine, an amino group replaced the proton attached to the 2 position of the adenine. Therefore, 2,6-diaminopurine potentially could form two additional hydrogen bonds with the C2 carbonyl groups of the two uridines (Fig. 6B). Accordingly, replacement of the adenine ligand with 2,6-diaminopurine caused chemical shift changes toward higher ppm (Fig. 5B) for the C2 carbonyl groups of the two uridines hydrogen bonded to the ligand, whereas the C2 carbonyl chemical shifts of the other uridines in the A-switch RNA remained virtually unperturbed (Fig. 5B).
Fig. 6.
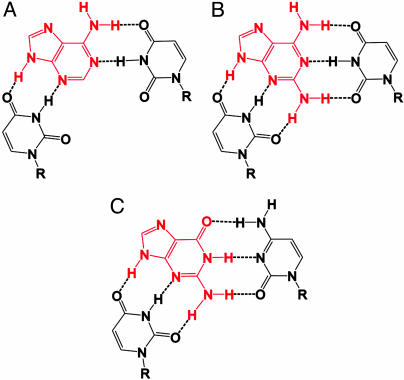
Ligand recognition by an intermolecular base triple in the purine-responsive riboswitches. (A) Intermolecular base-pairing interactions between adenine (red) and two uridines in the adenine-responsive riboswitch. (B) Two additional hydrogen bonds in the complex between 2,6-diaminopurine (red) and the adenine-responsive riboswitch. (C) Intermolecular base-pairing interactions between guanine (red) and nucleotides of the guanine-responsive riboswitch.
Remarkably, a similar hydrogen bonding scheme could be envisioned for the amino group in the 2 position of the guanine and the C2 carbonyl groups of the cytidine and the uridine that forms intermolecular hydrogen bonds with the N3 nitrogen of the ligand in the G-switch RNA (Fig. 6C). Accordingly, the C2 carbonyl of this uridine was shifted relative to the nonhydrogen-bonded C2 carbonyl groups of uridines in regular Watson–Crick base pairs toward higher ppm (data not shown).
Hydrogen Bonding to the N7 Nitrogen of the Ligand. No hydrogen bonds between the RNA and the N7 nitrogen of the bound purine ligands were detected in the HNN-COSY experiments. Furthermore, no strong NOE crosspeaks between the H8 position of the purine ligand and functional groups of the RNA were observed (data not shown). We compared 1H, 15N-HSQC spectra of the G-switch RNA/guanine complex with spectra of G-switch RNA bound to 7-deazaguanine. Despite the replacement of the N7 nitrogen by a CH group, 7-deazaguanine still bound to G-switch RNA, albeit with lower affinity (6), and in agreement with the published binding constants formed a stable 1:1 complex in slow exchange on the NMR time scale. The replacement of the potential hydrogen bonding acceptor N7 with a CH group should disrupt possible hydrogen bonds to this position. NH and NH2 groups of the RNA hydrogen bonded to the N7 nitrogen should become less protected against exchange with the solvent, and, consequently, their NMR signals should broaden significantly and shift to lower ppm values or even become undetectable. The comparison of the imino and amino regions of 1H, 15N-HSQC spectra of G-switch RNA bound to either 7-deazaguanine or guanine (Fig. 13, which is published as supporting information on the PNAS web site) revealed the same number of signals in both complexes with no obvious differences in line width. A few signals of imino groups in Watson–Crick base pairs experienced changes in their chemical shifts. However, these signals were still within the chemical shift range of imino groups in Watson–Crick base pairs. Thus, the replacement of the nitrogen N7 with a CH group in 7-deazaguanine apparently does not disrupt hydrogen bonds to RNA imino and amino groups.
Discussion
The ligand-binding domains of the guanine- and adenine-responsive riboswitch RNAs bind to their respective ligands guanine and adenine in a very similar manner. In both cases, two base-pairing interactions were observed between the bound purine and the riboswitch RNA. One was an intermolecular Watson–Crick base pair: a G:C base pair in the case of guanine bound to the guanine-responsive riboswitch and a A:U base pair in the case of adenine bound to the adenine-responsive riboswitch. The other was a previously undescribed base-pairing interaction between the N3/N9 edge of the purine ligand and a uridine of the RNA displaying two intermolecular hydrogen bonds between the H9N9 imino group and the N3 nitrogen of the purine and the C4 carbonyl group and the H3N3 imino group of a uridine residue in the RNA, respectively. The formation of an intermolecular Watson–Crick base pair is the basis for the discrimination between different purine ligands by the different riboswitches. In contrast, in the base-pairing interaction of the RNA with the N3/N9 edge of the ligand, a structural feature common to both adenine and guanine was recognized by the RNA. The combination of these two recognition elements therefore rationalizes the previous finding (7) that a single C to U mutation can switch the specificity of the guanine-sensitive riboswitch to adenine and vice versa without being detrimental to the affinity of the interaction. The observed base-pairing scheme between the purines and the RNA also explains why, not adenine itself, but the related 2,6-diaminopurine is the tighter binding ligand for the adenine-responsive riboswitch RNA because the 2-amino group of 2,6-diaminopurine can form two additional hydrogen bonds with two C2 carbonyl groups of the RNA. A related argument can be made in the case of 2-aminopurine, which binds to the A-switch RNA with an affinity similar to that of adenine (7). The loss of a Watson–Crick-type hydrogen bond to the uridine due to the absence of the 6-amino group in the ligand is compensated for by the formation of additional hydrogen bonds involving the 2-amino group. Similarly, the reduced binding affinity of hypoxanthine compared with guanine to the guanine-responsive riboswitch can be attributed to the absence of the 2-amino group in hypoxanthine.
Interestingly, in our experiments we could not detect interactions between the Hoogsteen edge of the purine ligand and the RNA. The Hoogsteen edges of guanine and adenine are different from each other. Two hydrogen-bond acceptor groups (N7 and O6) are found there in guanine, whereas adenine possesses a hydrogen-bond acceptor group (N7) and a hydrogen-bond donor, the 6-amino group, at this position. Yet, only one nucleotide systematically differs in the core sequences of guanine- and adenine-specific riboswitches (6, 7): the one that is involved in the formation of the intermolecular Watson–Crick base pair with the ligand. If, however, the two purine ligands interacted through their Hoogsteen edges with their respective RNAs in a similar fashion, this interaction would require additional systematic differences in the core sequences of the guanine- and adenine-specific riboswitches. Such differences have not been observed so far (6, 7). Alternatively, RNA–ligand interactions involving the Hoogsteen edge of the bound purine could be limited to the N7 position of the purine ring. The HNN-COSY experiments and the experiments with the 7-deazaguanine-bound G-switch RNA strongly suggest that no NH or NH2 group is involved in hydrogen bonding to the N7 position of the ligand. Therefore, the N7 nitrogen is either not recognized by the RNA or, alternatively, forms a hydrogen bond with a 2′-OH group of the RNA. The higher affinity of the RNA observed for guanine as compared with 7-deazaguanine (6) favors the latter possibility.
The base-pairing scheme between the purine ligand and the riboswitch RNAs described here has not been observed previously in any other RNA or RNA–ligand complex. The artificial RNA aptamers for ATP that have a lower affinity for the ligand do not interact with the adenine nucleotide through a classical Watson–Crick base pair (30, 31). In the case of the xanthine/guanine RNA-aptamer binding studies with analogues indicate that neither the functional group in the 2 position nor the N3 nitrogen of the purine ring is relevant for binding (32). In the guanosine-binding aptamer RNA, the N7 nitrogen of the Hoogsteen edge and the Watson–Crick face of the base appear to be major determinants of affinity (33). Therefore, the unique recognition mode for guanine and adenine of the guanine- and adenine-sensitive riboswitch RNAs described here apparently reflects the unique requirements for selectivity and affinity in the cellular environment essential for performing the natural regulatory functions of the riboswitches. In this respect, it is particularly useful to use the H9N9 imino group of the purines for binding because this group is not available for hydrogen bonding in all of the nucleoside and nucleotide derivatives that are abundant in the cell.
Note Added in Proof. The findings reported here are in agreement with recent x-ray data of the hypoxanthine–G switch complex described by Batey et al. (34) and of the guanine–G switch and adenine–A switch complexes described by Serganov et al. (35).
Supplementary Material
Acknowledgments
We thank Sabine Häfner and Matthias Görlach (both of the Institute of Molecular Biotechnology, Jena, Germany) for a generous gift of T7-RNA-polymerase and Elke Stirnal for excellent technical assistance. We thank Hashim Al-Hashimi and Boris Fürtig for inspiring discussions. This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 579 for RNA–ligand interactions.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HSQC, heteronuclear sequential quantum correlation; NOE, nuclear Overhauser effects.
References
- 1.Barrick, J. E., Corbino, K. A., Winkler, W. C., Nahvi, A., Mandal, M., Collins, J., Lee, M., Roth, A., Sudarsan, N., Jona, I., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 6421-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sudarsan, N., Barrick, J. E. & Breaker, R. R. (2003) RNA 9, 644-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandal, M. & Breaker, R. R. (2004) Nat. Rev. Mol. Cell Biol. 5, 451-463. [DOI] [PubMed] [Google Scholar]
- 4.Nudler, E. & Mironov, A. S. (2004) Trends Biochem. Sci. 29, 11-17. [DOI] [PubMed] [Google Scholar]
- 5.Grundy, F. J. & Henkin, T. M. (2004) Curr. Opin. Microbiol. 7, 126-131. [DOI] [PubMed] [Google Scholar]
- 6.Mandal, M., Boese, B., Barrick, J. E., Winkler, W. C. & Breaker, R. R. (2003) Cell 113, 577-586. [DOI] [PubMed] [Google Scholar]
- 7.Mandal, M. & Breaker, R. R. (2003) Nat. Struct. Mol. Biol. 11, 29-35. [DOI] [PubMed] [Google Scholar]
- 8.Lukavsky, P. J., Kim, I., Otto, G. A. & Puglisi, J. D. (2003) Nat. Struct. Mol. Biol. 11, 1033-1038. [DOI] [PubMed] [Google Scholar]
- 9.D'Souza, V., Dey, A., Habib, D. & Summers, M. F. (2004) J. Mol. Biol. 337, 427-442. [DOI] [PubMed] [Google Scholar]
- 10.Kim, I., Lukavsky, P. J. & Puglisi, J. D. (2002) J. Am. Chem. Soc. 124, 9338-9339. [DOI] [PubMed] [Google Scholar]
- 11.Fiaux, J., Bertelsen, E. B., Horwich, A. L. & Wüthrich, K. (2002) Nature 418, 207-211. [DOI] [PubMed] [Google Scholar]
- 12.Rüdiger, S., Freund, S. M., Veprintsev, D. B. & Fersht, A. R. (2002) Proc. Natl. Acad. Sci. USA 99, 11085-11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McElroy, C., Manfredo, A., Wendt, A., Gollnick, P. & Foster, M. (2002) J. Mol. Biol. 323, 463-473. [DOI] [PubMed] [Google Scholar]
- 14.Yan, X., Kong, X., Xia, Y., Sze, K. H. & Zhu, G. (2000) J. Magn. Reson. 147, 357-360. [DOI] [PubMed] [Google Scholar]
- 15.Dingley, A. J., Steger, G., Esters, B., Riesner, D. & Grzesiek, S. (2003) J. Mol. Biol. 334, 751-767. [DOI] [PubMed] [Google Scholar]
- 16.Tisne, C., Roques, B. P. & Dardel, F. (2004) J. Biol. Chem. 279, 3588-3595. [DOI] [PubMed] [Google Scholar]
- 17.Dingley, A. J. & Grzesiek, S. (1998) J. Am. Chem. Soc. 120, 8293-8297. [Google Scholar]
- 18.Pervushin, K., Ono, A., Fernandez, C., Szyperski, T., Kainosho, M. & Wüthrich, K. (1998) Proc. Natl. Acad. Sci. USA 95, 14147-14151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoldt, M., Wöhnert, J., Görlach, M. & Brown, L. (1998) EMBO J. 17, 6377-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartels, C., Xia, T., Billeter, M., Güntert, P. & Wüthrich, K. (1995) J. Biomol. NMR 6, 1-10. [DOI] [PubMed] [Google Scholar]
- 21.Piotto, M., Saudek, V. & Sklenar, V. (1992) J. Biomol. NMR 2, 661-665. [DOI] [PubMed] [Google Scholar]
- 22.Grzesiek, S. & Bax, A. (1993) J. Am. Chem. Soc., 115, 12593-12594. [Google Scholar]
- 23.Wijmenga, S. S. & van Buuren, B. N. M. (1998) Prog. NMR Spectroscopy 32, 287-387. [Google Scholar]
- 24.Boehlen, J. M. & Bodenhausen, G. (1993) J. Magn. Reson. A 102, 293-301. [Google Scholar]
- 25.Jang, Y. H., Goddard, W. A., III, Noyes, K. T., Sowers, L. C., Hwang, S. & Chung, D. S. (2003) J. Phys. Chem. B 107, 344-357. [Google Scholar]
- 26.Markovski, V., Sullivan, G. R. & Roberts, J. D. (1977) J. Am. Chem. Soc. 99, 714-718. [DOI] [PubMed] [Google Scholar]
- 27.Dingley, A. J., Masse, J. E., Feigon, J. & Grzesiek, S. (2000) J. Biomol. NMR 16, 279-289. [DOI] [PubMed] [Google Scholar]
- 28.Liu, A., Majumdar, A., Hu, W., Kettani, A., Skripkin, E. & Patel, D. J. (2000) J. Am. Chem. Soc. 122, 3206-3210. [Google Scholar]
- 29.Fürtig, B., Richter, C., Wöhnert, J. & Schwalbe, H. (2003) ChemBioChem 4, 936-962. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, F., Kumar, R. A., Jones, R. A. & Patel, D. J. (1996) Nature 382, 183-186. [DOI] [PubMed] [Google Scholar]
- 31.Dieckmann, T., Suzuki, E., Nakamura, G. E. & Feigon, J. (1996) RNA 2, 628-640. [PMC free article] [PubMed] [Google Scholar]
- 32.Kiga, D., Futamura, Y., Sakamoto, K. & Yamamoto, S. (1998) Nucleic Acids Res. 26, 1755-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connell, G. J. & Yarus, M. (1994) Science 264, 1137-1141. [DOI] [PubMed] [Google Scholar]
- 34.Batey, R. T., Gilbert, S. D. & Montange, R. K. (2004) Nature 432, 411-415. [DOI] [PubMed] [Google Scholar]
- 35.Serganov, A., Yuan, Y. R., Pikovskaya, O., Polonskaia, A., Malinina, L., Phan, A. T., Hobartner, C., Micura, R., Breaker, R. R. & Patel, D. J. (2004) Chem. Biol. 11, 1729-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.