Abstract
ArgBP2, and its brain-specific splice variant, nArgBP2, are interactors and substrates of Abl/Arg tyrosine kinases and of the ubiquitin ligase Cbl. They are members of a family of adaptor proteins that colocalize with actin on stress fibers and at cell-adhesion sites, including neuronal synapses. We show here that their NH2-terminal region, which contains a sorbin homology domain domain, interacts with spectrin, and we identify binding proteins for their COOH-terminal SH3 domains. All these binding partners participate in the regulation of the actin cytoskeleton. These include dynamin, synaptojanin, and WAVE isoforms, as well as WAVE regulatory proteins. At least two of the ArgBP2/nArgBP2 binding partners, synaptojanin 2B and WAVE2, undergo ubiquitination and Abl-dependent tyrosine phosphorylation. ArgBP2/nArgBP2 knockdown in astrocytes produces a redistribution of focal adhesion proteins and an increase in peripheral actin ruffles, whereas nArgBP2 overexpression produces a collapse of the actin cytoskeleton. Thus, ArgBP2/nArgBP2 is a scaffold protein that control the balance between adhesion and motility by coordinating the function of multiple signaling pathways converging on the actin cytoskeleton.
Keywords: cell adhesion, dynamin, ruffles, synapse, synaptojanin
ArgBP2 was identified as a ubiquitously expressed substrate and interactor of the Arg and Abl protein-tyrosine kinases and found to be concentrated on actin stress fibers (1). Subsequently, nArgBP2, a nervous-system-specific splice variant of ArgBP2, was described as a binding partner of the postsynaptic protein SAPAP and reported to be localized postsynaptically in cerebellar Purkinje cells (2). ArgBP2/nArgBP2 belong to a family of adaptor proteins that includes vinexin and CAP/ponsin and that are thought to participate in the regulation of cell adhesion, actin cytoskeleton organization, and signaling downstream of growth factor receptors (3). Members of this protein family are characterized by a sorbin homology (SoHo) domain in the NH2-terminal region and three SH3 domains (referred to henceforth as SH3-1, SH3-2, and SH3-3) in the COOH-terminal region. In addition, nArgBP2, but not ArgBP2 or other family members, comprises a central Zn+2 finger motif (3). The SoHo domain (≈50 aa) is so called because it is contained in a fragment of ArgBP2 (amino acids 163–338 of the rat protein) that was first identified as a soluble peptide (sorbin) in the lumen of the pig intestine, possibly as a result of cell lysis and proteolysis of ArgBP2 (4). The SoHo domain, which in the protein CAP binds flotillin (5), remains poorly characterized. Binding partners of the SH3 domain region of ArgBP2 include, beside Abl and Arg, the ubiquitin ligase Cbl, the protein kinase Pyk2, and actin adaptors found at focal adhesions and adhering junctions, such as vinculin and afadin (1, 2, 6, 7). Thus, ArgBP2/nArgBP2 and other family members may function to bring in close proximity to each other proteins that must act together in the regulation of cell adhesion and signaling. For example ArgBP2 negatively regulates Abl kinase by mediating the formation of a complex with Abl and Cbl, thus promoting ubiquitination and degradation of Abl (6).
ArgBP2/nArgBP2 may be a key mediator of Arg/Abl kinase effects on actin function in brain (8, 9), thus contributing to Arg/Abl actions on CNS development (10) and synapse plasticity (11, 12). Recently, ArgBP2 and nArgBP2 were identified as major brain binding partners for synaptojanin 1 and 2 (13) (Y. Nemoto and P.D.C., unpublished observations), polyphosphoinositide phosphatases whose main substrate is phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] (14, 15). Thus, ArgBP2/nArgBP2 may provide a link between Arg/Abl function and PI(4,5)P2 metabolism in the regulation of actin dynamics. In this study, we shed light on the properties and potential function of ArgBP2/nArgBP2.
Materials and Methods
DNA Constructs. Full-length myc-tagged rat nArgBP2 cDNA (amino acids 1–1196) and Flag-tagged rat synaptojanin 2b2 were generated by Y. Nemoto in our laboratory (16) (Y. Nemoto and P.D.C., unpublished observations). HA-tagged nArgBP2 was produced by subcloning the full-length sequence in pcDNAI-HA by PCR. An nArgBP2 construct lacking the SH3 domains (nArgBP2-ΔSH3; amino acids 1–865) was subcloned in both pGEX-6P-1 and pEGFP-C2 by PCR. GFP-tagged nArgBP2 was obtained by insertion in the nArgBP2-pcDNAI of the GFP coding sequence, in frame with the first methionine of nArgBP2. The sorbin region (amino acids 163–338) and the three SH3 domains of nArgBP2 were PCR-amplified and cloned into pEGFP-C2, pGEX-2TK, or pGEX-6P-1. All constructs made by PCR were sequence-verified. The following reagents were generous gifts: YFP-Actin inducible adenovirus, from P. Keller (Max Planck Institute, Dresden, Germany); Abl in a mammalian expression vector, from A. Koleske (Yale University); c-Cbl cDNA, from I. Dikic (Goethe University, Frankfurt); and Flag-tagged WAVE-2, from A. Zucconi (Tor Vergata University, Rome). Flag-tagged human synaptojanin 1 and other constructs were from our laboratory.
Antibodies. Rabbit polyclonal anti-nArgBP2 were raised in our laboratory against a GST fusion protein of rat nArgBP2-ΔSH3 (amino acids 1–865) and against a synthetic peptide corresponding to amino acids 34–59 of the protein. Sources of monoclonal antibodies were as follows: anti-β-catenin (Transduction Laboratories, Lexington, KY), anti-vinculin (Sigma), anti-paxillin (Transduction Laboratories), anti-α2 spectrin (Chemicon), anti-PSD95 (Upstate Biotechnology, Lake Placid, NY), anti-dynamin 1 (Hudy1, Upstate Biotechnology), anti-phosphotyrosine (Cell Signaling Technology, Beverly, MA), anti-HA (rat) (3F-10) (Roche), and anti-Flag (M2 and M5) (Sigma). Polyclonal antibodies were obtained as follows: goat anti-dynamin 2 (C-18, Santa Cruz Biotechnology), rabbit anti-synaptojanin 1 (OCK) (our laboratory), rabbit anti-synaptojanin 2 (Santa Cruz Biotechnology), rabbit anti-Abl (Santa Cruz Biotechnology), rabbit anti-Cbl (Santa Cruz Biotechnology), goat anti-WAVE2 (Santa Cruz Biotechnology), and rabbit pan anti-WAVE (kind gift of J. D. Scott, Oregon Health and Science University, Portland).
Cell Culture and Immunofluorescence. CHO-K1, HEK 293, and COS-7 cells were purchased from American Type Culture Collection. Primary cultures of astrocytes were prepared as described in ref. 17. Primary cultures from hippocampal neurons were prepared and processed for immunostaining as reported in ref. 17. For immunofluorescence of nArgBP2, cells were fixed at –20°C for 5 min in 100% methanol.
RNA-Mediated Interference. RNA interference (RNAi)-mediated nArgBP2 knock-down was performed by transfecting chemically synthesized small interfering RNA (siRNA) duplexes (IDT) (250 nM) with Oligofectamine (Invitrogen). The targeted sequence of mouse and rat nArgBP2 was 5′-GGACUGGUACAAGACAAUGTT-3′. After transfection, cells were cultured for 72 h.
Videomicroscopy and Image Analysis. Control and RNAi-treated astrocytes were serum-starved for 24 h and infected with YFP-actin adenovirus for 12–16 h before analysis. Epi-fluorescence imaging of actin dynamics was performed on an Olympus BX-51 microscope equipped with a ×20 water lens (0.95 numerical aperture). Epi-illumination was accomplished with a Polychrome IV monochrometer (TILL Photonics, Planegg, Germany) set to 505 nm that was electronically shuttered between acquisitions (typically 200 msec). Images were acquired with an Imago QE-cooled charge-coupled device camera by using a CFP/YFP filter cube (Chroma Technology, Rockingham, VT) and analyzed with tillvision (TILL Photonics). A “show-tracks” macro was used for Fig. 4C to visualize differences in moving objects. This tool allows determining differences between user-defined frames (10 in our case) over the entire movie (120 frames, 5-sec intervals) and conflates these differences into a single projection, which was then quantified. The final single image was inverted and scaled to display differences as a range of grays, with darker pixels representing higher motility.
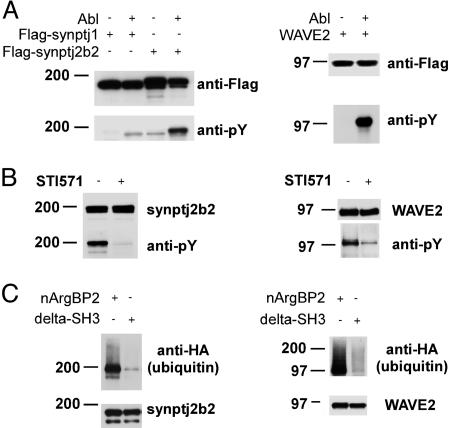
Fig. 4.
Tyrosine phosphorylation and ubiquitination of synaptojanin 2 and WAVE2. (A) Synaptojanin 2b and WAVE2 are phosphorylated by Abl. HEK 293T cells were transfected with plasmids encoding Flag-tagged synaptojanin 1, synaptojanin 2b, WAVE2, and Abl as indicated. Anti-Flag immunoprecipitates were probed with anti-phosphotyrosine antibody. (B) Tyrosine phosphorylation of synaptojanin 2b and WAVE2 is inhibited by the Abl/Arg kinase inhibitor STI571. HEK293T cells were transfected with plasmids encoding Flag-tagged WAVE2 or synaptojanin 2b and Abl in the presence or in the absence of 10 μM Abl kinase inhibitor STI571. Anti-Flag immunoprecipitates were probed with anti-phosphotyrosine antibody. (C) Ubiquitination of synaptojanin 2b and WAVE2. HEK 293T cells were transfected with Flag-tagged synaptojanin 2b or WAVE-2 plus Abl, Cbl, HA-ubiquitin, and either full-length nArgBP2 or nArgBP2 lacking the SH3 domain region (nArgBP2-ΔSH3) as indicated. Anti-Flag immunoprecipitates were probed by Western blotting with anti-HA antibodies (ubiquitin).
Miscellaneous Procedures. Tissue homogenization and GST pull-down experiments were carried out as described in ref. 18. Cell transfections were performed with Lipofectamine 2000 (Invitrogen). Immunoprecipitations were performed from cell lysates generated in RIPA buffer (20 mM Tris, pH 7.5/100 mM NaCl/50 mM NaF/1% Nonidet P-40/0.1% DOC/0.1% SDS) in the presence of a protease inhibitor mixture (Roche), 1 mM orthovanadate, and (for studies of ubiquitination) 1 μM YU101 (a proteasome inhibitor; Calbiochem), using Sigma anti-Flag M2 agarose beads. SDS/PAGE and Western blotting were performed according to standard protocols. MALDI-MS was carried out by the Yale Keck Protein Chemistry Facility (http://keck.med.yale.edu/prochem). In vitro translated proteins were prepared according to the manufacturer's protocols (TnT Quick Coupled Transcription/Translation System, Invitrogen).
Results
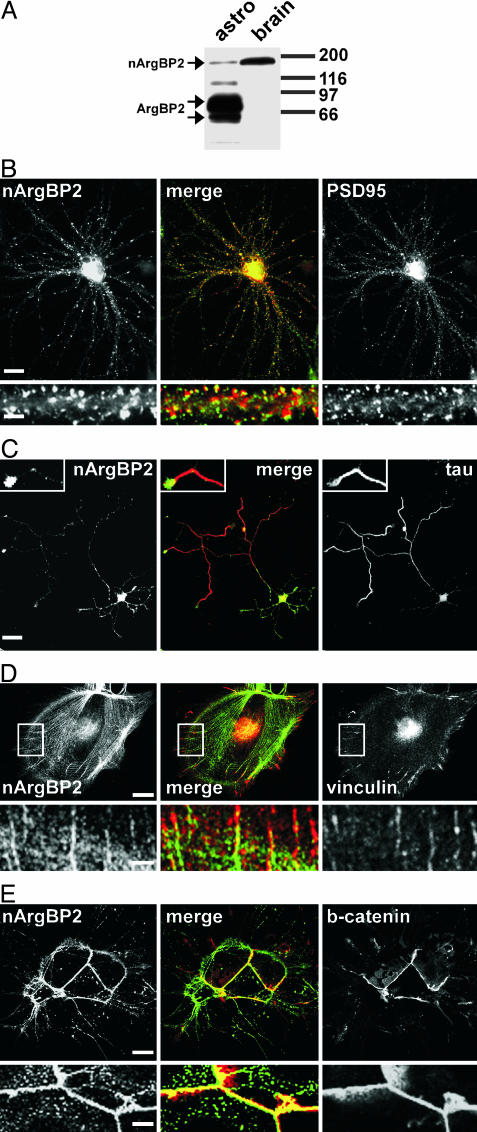
Localization of ArgBP2/nArgBP2 in Cells of the Nervous System. Antibodies directed against a protein region common to both ArgBP2 and nArgBP2 (amino acids 1–865 of rat nArgBP2) were used to stain by immunofluorescence neurons and astrocytes in primary culture. Prior Western blot analysis revealed that, as expected, nArgBP2 was by far the predominant species in brain, whereas ArgBP2 (which is represented by two bands migrating at in the 60- to 70-kDa range) predominated in nonneuronal cells, including cultured astrocytes (Fig. 1A and data not shown). In mature neuronal cultures (21 days in vitro), immunoreactivity had the typical punctate staining characteristic of synapses, consistent with the previously reported presence of nArgBP2 in dendritic spines (2) (Fig. 1B). However, in contrast to PSD95, a marker of dendritic tips, nArgBP2 had a more diffuse distribution throughout the spine, similar to that of actin (19) (Fig. 1B). In developing isolated neurons (5 days in vitro), immunoreactivity was present at similar levels in the distal portions of both axons and dendrites (Fig. 1C). This result suggests that the high levels of nArgBP2 present in spines are the result of progressive concentration rather than of selective targeting and makes plausible the presence of a pool of nArgBP2 at the presynapse even in mature neurons.
Fig. 1.
Expression of ArgBP2/nArgBP2 in brain and in primary cultures of brain cells. (A) ArgBP2/nArgBP2 immunoreactivity in total homogenates of astrocytes in primary culture and of brain tissue as revealed by Western blotting. nArgBP2 is the prevalent isoform in total brain, whereas the two ArgBP2 bands predominate in cultured astrocytes. (B and C) Localization of endogenous nArgBP2 in cultured hippocampal neurons. In mature (21 days in vitro) neurons (B), nArgBP2 is concentrated at synapses, primarily in dendritic spines, as revealed by double immunofluorescence for nArgBP2 and PSD95. A dendritic shaft is shown at higher magnification under each field. (Scale bars: 20 μm for the main fields; 5 μm for the small fields.) In developing isolated neurons (3–5 days in vitro)(C), nArgBP2 is localized throughout the dendritic arbor but is also present in axons (counterstained with the axonal marker tau) where it is concentrated in axon terminals. (Scale bars: 30 μm for the main field; 10 μm for the Insets.) (D and E) Localization of endogenous ArgBP2 in astrocytes. ArgBP2 is localized on stress fibers and at focal adhesions as demonstrated by counterstain with antibodies directed against vinculin, a focal adhesion marker (D). The portion enclosed by a white rectangle is shown at high magnification in Lower. (Scale bars: Upper, 20 μm; Lower, 4 μm.) In confluent cultures (E), ArgBP2 is concentrated at sites of cell–cell contact, where it colocalizes with β-catenin. (Scale bars: Upper, 10 μm; Lower, 2 μm.)
In astrocytes, ArgBP2 immunoreactivity was localized on stress fibers, as demonstrated by colocalization with phalloidin (a marker of F-actin; data not shown), in agreement with previous studies on other nonneuronal cell types (1). In addition, it was concentrated together with vinculin at sites of cell adhesion (Fig. 1D). When glial cells were tightly apposed to each other, ArgBP2 immunoreactivity colocalized with β-catenin at sites of cell–cell contact, revealing a concentration at adherens junctions (Fig. 1E). These findings are consistent with the known interaction of vinexin–CAP/ponsin–ArgBP2 family proteins with components of the actin cytoskeleton and cell-adhesion structures (3).
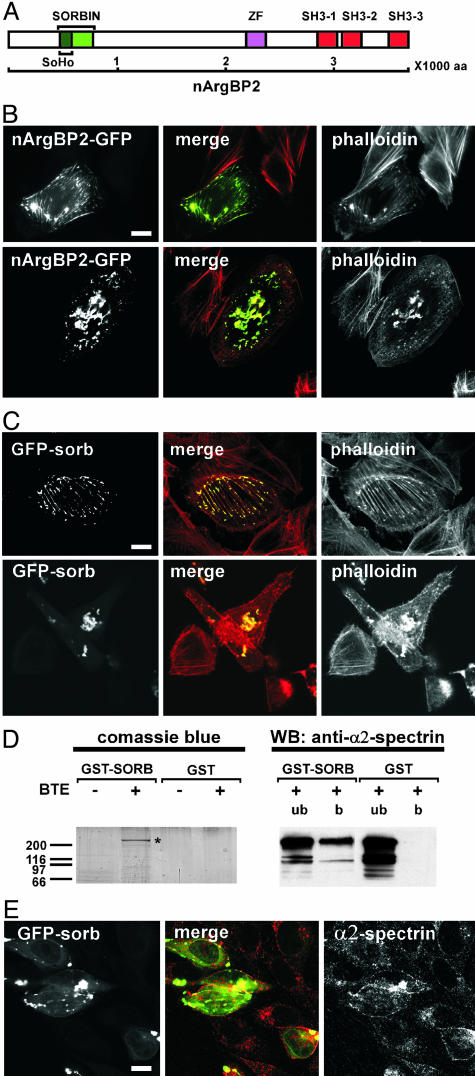
Overexpression of nArgBP2 Disrupts the Actin Cytoskeleton. Targeting of nArgBP2 to actin-rich structures and adhesion sites was confirmed by the localization of GFP-tagged nArgBP2 expressed at low/moderate levels in either astrocytes (data not shown) or fibroblastic cell lines (Fig. 2B). At higher level of expression, GFP-nArgBP2 induced a coalescence of F-actin into large aggregates and colocalized with actin into these aggregates (Fig. 2C). Expression of the nArgBP2 sorbin fragment (amino acids 163–338) (Fig. 2A), which also colocalized with F-actin when expressed at low levels, was sufficient to produce this effect (Fig. 2C).
Fig. 2.
Effect of nArgBP2 overexpression on the actin and spectrin cytoskeleton. (A) Domain cartoon of nArgBP2. SoHo, sorbin homology domain; ZF, zinc-finger domain; SH3, Src homology domain 3. (B and C) Expression of GFP-nArgBP2 (B) or GFP-sorbin (C) disrupts the actin cytoskeleton in CHO cells. In all transfected cells, both constructs colocalize with a pool F actin (phalloidin stain). At a low level of expression they colocalize on stress fibers and focal adhesions (Upper). In cells expressing higher levels of these proteins, F-actin coalesces into aggregates (Lower). (Scale bar: 10 μm.) (D) The sorbin fragment interacts with α2-spectrin. (Left) Material affinity-purified from a rat brain Triton X-100 extract (BTE) by GST-sorbin or GST was resolved by SDS/PAGE and stained with Coomassie blue. The band indicated by an asterisk was identified by MALDI as α2-spectrin. (Right) Anti-α2-spectrin Western blotting of unbound and bound material from the affinity purification (b, bound material; ub, unbound material). (E) Endogenous α2-spectrin relocalizes in sorbin aggregates produced by expression of GFP-sorbin. (Scale bar: 10 μm.)
The Sorbin Region of ArgBP2/nArgBP2 Interacts with α2-spectrin. To gain some insight into the mechanism(s) responsible for the striking effect of the sorbin region on the actin cytoskeleton, we searched for its potential interactors. Pull-down experiments from a Triton X-100 rat brain extract were performed by using GST-sorbin as bait. The major protein retained by the GST fusion protein was identified by MALDI-MS as α2-spectrin (α-fodrin) (Fig. 2D). This result was validated by Western blotting of the affinity-purified band with anti-α2-spectrin antibodies. Furthermore, in cells expressing a GFP fusion of the sorbin region, endogenous α2-spectrin partially redistributed from its predominant diffuse localization at the cell periphery to sorbin-containing aggregates (Fig. 2E). We note that another substrate and interactor of the Abl kinase family, the protein ArgBP1 (also called Abi or E3B1), also interacts with spectrin (20). Considering the critical role of spectrin as a component of the membrane-associated cytoskeleton, alteration of α2-spectrin function, for example by competition with other spectrin binding partners, may be the primary event leading to F-actin aggregation in cells overexpressing nArgBP2 or expressing its sorbin domain.
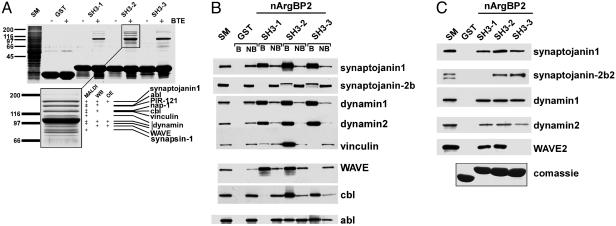
The SH3 Domains of ArgBP2/nArgBP2 Bind Regulatory Components of the Actin Cytoskeleton. Additional links between ArgBP2/nArgBP2 and the machinery controlling the actin cytoskeleton emerged from the analysis of the interactors of its SH3 domains. GST fusion proteins of each of the three SH3 domains (SH3-1, SH3-2, and SH3-3) were used in pull-down assays from Triton X-100 extracts of rat brains, and bound material was analyzed by SDS/PAGE and Western blotting, and, in the case of the material affinity purified by SH3-2, also by MALDI. The three SH3 domains had a partially overlapping set of binding partners, with SH3-2 displaying the most numerous interactors (Fig. 3A). These included synaptojanin 1 and 2 (13, 16) (Y. Nemoto and P.D.C., unpublished observations) and other previously reported interactors, such as vinculin (2), Abl (1), and Cbl (6), but also new binding partners. The major new interactor was identified as dynamin (both dynamin 1 and dynamin 2), a GTPase implicated in endocytosis and also in the regulation of actin dynamics (21–23) (Fig. 3 A and B). Other interactors were WAVE 1 and 2, and components of a WAVE regulatory complex (PIR121 and Nap1) (24, 25) (Fig. 3 A and B and data not shown). Synapsin, a presynaptically enriched actin-binding protein (26), was also identified in the bound material. The interactions of synaptojanin 1 and 2, dynamin 1 and 2, and WAVE2 were confirmed by pull-downs of in vitro-translated proteins on GST-SH3 fusion proteins. Synaptojanin 1 and the dynamins bound all three SH3 domains, whereas synaptojanin 2 bound only SH3-2 and SH3-3, and WAVE2 bound only SH3-1 and SH3-2 (Fig. 3C). Collectively, the interactions of the SH3 region of nArgBP2 provide evidence for a role of nArgBP2 in the regulation of actin function.
Fig. 3.
The SH3 domains of nArgBP2 bind to actin regulatory proteins. (A) GST or GST-nArgBP2 SH3 domains were used as baits in affinity-purification experiments from a rat brain Triton X-100 extract (BTE, brain Triton X-100 extract). Material retained by the GST fusion proteins, as well as baits only, were resolved by SDS/PAGE and stained with Coomassie blue (Upper). (Lower) Higher-magnification view of the protein bands retained by SH3-2, and methods used for their identification: MALDI-MS, Western blotting (WB), and overlay essay (OE). (B) Western blot analysis of proteins retained by GST-nArgBP2 SH3 domains. SM, starting material; B, bound material; NB, unbound material. (C) GST or GST-nArgBP2 SH3 domains were used as baits in affinity purification experiments from in vitro-translated binding partners. GST was used as negative control. GST fusion proteins used in the pull down essay were resolved by SDS/PAGE and stained with Coomassie blue (Lower).
Synotaptojanin 2 and WAVE2 Are Tyrosine-Phosphorylated by Abl and Ubiquitinated. It was shown that ArgBP2 mediates, via its SH3 domains, the formation of a complex containing Abl and Cbl (6), which promotes the Abl-dependent phosphorylation of Cbl and the ubiquitination by Cbl of both Abl and ArgBP2 (6). We investigated whether other ArgBP2 binding partners beside Cbl undergo Abl-dependent phosphorylation and ubiquitination. Flagged-tagged versions of synaptojanin 1, synaptojanin 2b, dynamin 1 and 2, and WAVE2 were expressed in HEK 293 cells with and without Abl cotransfection. nArgBP2 binding partners were then immunoprecipitated from cell lysates and their state of phosphorylation was revealed by Western blotting with anti-phosphotyrosine antibody. Synaptojanin 2b and WAVE2 were strongly tyrosine-phosphorylated when coexpressed with Abl (Fig. 4A). The tyrosine phosphorylation of synaptojanin 1 was much less efficient (Fig. 4A), and no phosphotyrosine signal was detected on dynamin (data not shown). To confirm this result, cells cotransfected with Abl and either synaptojanin 2b or WAVE2 were incubated in the presence or in absence of the Abl/Arg inhibitor STI571 (10 μM). The tyrosine phosphorylation of both synaptojanin 2b and WAVE2 was strikingly reduced by this drug (Fig. 4B).
To determine whether nArgBP2 also facilitates the ubiquitination of either synaptojanin 2b or WAVE2, Flag-tagged versions of the two proteins were expressed in HEK 293 cells together with Abl, Cbl, HA-ubiquitin, and full-length nArgBP2 or a nArgBP2 construct devoid of the SH3 domains (nArgBP2-ΔSH3). Synaptojanin 2 and WAVE2 were then immunoprecipitated from cell lysates, and their ubiquitination state was revealed by Western blotting with anti-HA antibodies. Synaptojanin 2b and WAVE2 were ubiquitinated both in the presence and the absence of overexpressed nArgBP2 (Fig. 4C and data not shown). Strikingly, however, this reaction was very strongly inhibited in the presence of nArgBP2-ΔSH3. Possibly, endogenous ArgBP2 family members are sufficient to facilitate the occurrence of the reaction, and nArgBP2-ΔSH3 has a dominant negative effect by competing interactions of the NH2-terminal region of these endogenous proteins (Fig. 4C).
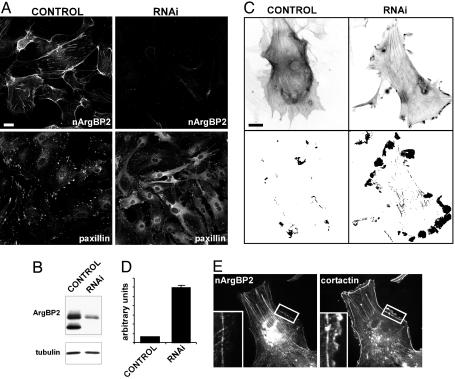
Knock-Down of ArgBP2/nArgBP2 Enhances Peripheral Ruffling. The interactions of ArgBP2/nArgBP2, together with the well established link between Abl/Arg and actin, suggest a physiological role of ArgBP2/nArgBP2 in the regulation of the actin cytoskeleton. An RNAi-based approach was used on astrocytes in primary culture to assess the consequence of ArgBP2/nArgBP2 knock-down on cell structure and motility. First, a siRNA pair that produced an efficient depletion of the protein was selected. Three days after transfection with this siRNA, a strong reduction of ArgBP2/nArgBP2 immunoreactivity was observed by both immunofluorescence and Western blotting, whereas no changes were observed in mock transfected cells (Fig. 5 A Upper and B). In depleted cells, an overall change of cell morphology was observed, with cells appearing more elongated and with numerous protrusions (Fig. 5 A Lower and data not shown). Immunofluorescence for two focal adhesion markers, paxillin (27) and focal adhesion kinase (FAK) (28), demonstrated an increase of their cytosolic pools (Fig. 5 A Lower and data not shown), suggesting their partial dissociation from focal adhesion complexes.
Fig. 5.
RNAi mediated knock-down of nArgBP2. (A and B) siRNA knock-down of endogenous nArgBP2 in mouse astrocytes. Immunofluorescence (A) reveals a striking reduction of nArgBP2 immunoreactivity (Upper) and partial redistribution of paxillin immunoreactivity (Lower). (Scale bar: 20 μm.) The strong reduction in ArgBP2 immunoreactivity is confirmed by Western blot analysis (B). (C and D) Prominent presence of actin ruffles in nArgBP2 knocked-down astrocytes. Mouse astrocytes transfected with either vehicle alone (control) or siRNAs (RNAi) were infected with a YFP-actin-encoding adenovirus. Inverted images of YFP fluorescence are shown in C Upper. Graphic display of motility in the same two fields during a 10-min observation are shown in C Lower, where pixel differences are presented as grayscale images (see Materials and Methods). Dark areas represent sites of motility. (Scale bars: 20 μm.) Quantification of motility in ruffles containing areas (see Materials and Methods) from eight different movies for each condition is shown in D.(E) ArgBP2 immunoreactivity is absent form the leading edge of a ruffle (Inset), which is positive for cortactin.
Live imaging of serum-starved astrocytes expressing YFP-actin revealed little motility of actin-positive structures after transfection with control siRNA. In these cells, almost no peripheral ruffles were visible (Fig. 5C). In contrast, after ArgBP2/nArgBP2 knockdown, peripheral actin ruffles were very prominent and highly dynamic in astrocytes, despite the absence of serum, as revealed both by observation of time-lapse movies and by quantitative analysis of the motility (Fig. 5 C and D). Interestingly, in serum-stimulated astrocytes, where ruffles are abundant, ArgBP2/nArgBP2 was never concentrated in ruffles (revealed by cortactin staining) (Fig. 5E), thus strengthening a putative role of nArgBP2 in adhesion rather than in motility. Collectively, these findings suggest that a balance between adhesion and motility may be shifted toward motility in the absence of ArgBP2/nArgBP2.
Discussion
Our results provide evidence for a role of ArgBP2/nArgBP2 as a scaffold protein that integrates the function of multiple regulatory pathways converging primarily on the regulation of the actin cytoskeleton.
The sorbin region of ArgBP2/nArgBP2 binds α2-spectrin (α-fodrin) and may therefore play an important role in targeting ArgBP2/nArgBP2 to the cell surface. SH3 domain-mediated interactions may function, instead, to coordinate the actions of a variety of proteins implicated in actin function, signaling, and membrane trafficking. In addition to components of cell-adhesion sites, such as vinculin and afadin (2), the SH3 region of ArgBP2/nArgBP2 binds proteins thought to play a key role in the generation of cell ruffles and lamellipodia, although ArgBP2 itself is not present in these structures. The most prominent such protein is dynamin, possibly because of its abundance in the brain extract used for affinity purification. Dynamin, a GTPase well known for its role in the fission reaction of endocytosis (29, 30), has also been implicated in the regulation of actin dynamics (22, 23, 31, 32), in particular at peripheral ruffles (33). A second protein is WAVE, a member of the WASP-WAVE family of proteins that promote Arp2/3-mediated actin polymerization (34). As shown recently, the recruitment of WAVE to the plasma membrane is mediated, at least in part, by GTP-bound Rac1 via an actin regulatory complex that comprises PIR121, Nap1, Abi-1, and HSPC300 (24, 25). Thus, the recovery of PIR121 and NAP1 in GST-SH3 pull-downs may be indirect and mediated by WAVE. The synaptojanins, two other interactors of the SH3 domains of ArgBP2/nArgBP2, may also regulate ruffle formation via the dephosphorylation of PI(4,5)P2 and phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] (35, 36). These phosphoinositides function as coreceptors for many actin regulatory proteins as well as important cofactors in the activation of WASP-WAVE family proteins (34, 37). Furthermore, synaptojanin 2 binds Rac-GTP and colocalizes with Rac-GTP at the plasma membrane of ruffles (16, 35), thus indicating another point of convergence of regulatory pathways controlled by ArgBP2/nArgBP2.
Among the previously known interactors of the SH3 domains of nArgBP2, the tyrosine kinase Abl and the ubiquitin ligase Cbl function as regulators of cell adhesion and actin-based cell ruffling via their enzymatic activities (9, 38–40). Interestingly, both WAVE and synaptojanin 2 undergo ubiquitination and Abl-dependent tyrosine phosphorylation. Furthermore, Abl was reported to be negatively regulated by PI(4,5)P2, the substrate of synaptojanin (41).
Collectively, these interactions may explain the phenotype produced by the knock-down of ArgBP2/nArgBP2: increased peripheral ruffles and the partial destabilization of cell adhesion, as reflected by the increased cytosolic pools of cell-adhesion components. Lack of ArgBP2/nArgBP2 may impair a negative control of proteins involved in ruffle formation. ArgBP2/nArgBP2 may achieve this negative control either by sequestering such proteins or by regulating them via the enzymatic actions of Abl and Cbl. Thus, a key and general function of ArgBP2/nArgBP2 may be to help controlling the balance between cell adhesion and motility by integrating the function of multiple regulatory pathways. Via local actions, ArgBP2/nArgBP2 may also control vesicular trafficking reactions, primarily endocytosis, as suggested by its binding to dynamin, which mediates fission, and to synaptojanin, which participates in clathrin adaptors uncoating (14). This possibility fits with the strong evidence for a critical role of actin in endocytosis (42, 43).
All these actions may also be important in the nervous system, where nArgBP2 is concentrated at synapses. Pilot experiments indicate that nArgBP2 knock-down in neurons has a strong inhibitory effect on synapse formation but enhances formation of filopodia, consistent with the hypothesis that its suppression enhances motility at the expense of adhesion. At synapses, some of the synaptic interactors of nArgBP2, such as the synaptojanin 1/2 and dynamin 1/2, are concentrated in the presynaptic side, whereas nArgBP2 is enriched postsynaptically. However, growing evidence indicates that many presynaptic proteins are also present postsynaptically and vice versa (44, 45). Accordingly, synaptojanin and dynamin isoforms are also present postsynaptically in dendrites (46–48). Conversely, as shown in this study by the analysis of isolated young neurons, nArgBP2 is also present in axons terminals, where it could bind a variety of presynaptic proteins, including synapsin, an interactor of its SH3 domains.
Acknowledgments
We thank Yasuo Nemoto for initial results leading to this project and for critical reagents and Anthony Koleske for discussion. This work was supported in part by National Institutes of Health Grants NS36251 (to P.D.C.) and CA46128 (to P.D.C. and D.T.) and a postdoctoral fellowship from Telethon (Italy) (to G.C.). S.C. was a Howard Hughes Medical Institute Postdoctoral Fellow supported by the Life Sciences Research Foundation.
Abbreviations: RNAi, RNA interference; siRNA, small interfering RNA.
References
- 1.Wang, B., Golemis, E. A. & Kruh, G. D. (1997) J. Biol. Chem. 272, 17542–17550. [DOI] [PubMed] [Google Scholar]
- 2.Kawabe, H., Hata, Y., Takeuchi, M., Ide, N., Mizoguchi, A. & Takai, Y. (1999) J. Biol. Chem. 274, 30914–30918. [DOI] [PubMed] [Google Scholar]
- 3.Kioka, N., Ueda, K. & Amachi, T. (2002) Cell Struct. Funct. 27, 1–7. [DOI] [PubMed] [Google Scholar]
- 4.Charpin, G., Chikh-Issa, A. R., Guignard, H., Jourdan, G., Dumas, C., Pansu, D. & Descroix-Vagne, M. (1992) Gastroenterology 103, 1568–1573. [DOI] [PubMed] [Google Scholar]
- 5.Baumann, C. A., Ribon, V., Kanzaki, M., Thurmond, D. C., Mora, S., Shigematsu, S., Bickel, P. E., Pessin, J. E. & Saltiel, A. R. (2000) Nature 407, 202–207. [DOI] [PubMed] [Google Scholar]
- 6.Soubeyran, P., Barac, A., Szymkiewicz, I. & Dikic, I. (2002) Biochem. J. 370, 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haglund, K., Ivankovic-Dikic, I., Shimokawa, N., Kruh, G. D. & Dikic, I. (2004) J. Cell Sci. 117, 2557–2568. [DOI] [PubMed] [Google Scholar]
- 8.Lanier, L. M. & Gertler, F. B. (2000) Curr. Opin. Neurobiol. 10, 80–87. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez, S. E., Krishnaswami, M., Miller, A. L. & Koleske, A. J. (2004) Trends Cell Biol. 14, 36–44. [DOI] [PubMed] [Google Scholar]
- 10.Koleske, A. J., Gifford, A. M., Scott, M. L., Nee, M., Bronson, R. T., Miczek, K. A. & Baltimore, D. (1998) Neuron 21, 1259–1272. [DOI] [PubMed] [Google Scholar]
- 11.Finn, A. J., Feng, G. & Pendergast, A. M. (2003) Nat. Neurosci. 6, 717–723. [DOI] [PubMed] [Google Scholar]
- 12.Moresco, E. M., Scheetz, A. J., Bornmann, W. G., Koleske, A. J. & Fitzsimonds, R. M. (2003) J. Neurophysiol. 89, 1678–1687. [DOI] [PubMed] [Google Scholar]
- 13.Zucconi, A., Dente, L., Santonico, E., Castagnoli, L. & Cesareni, G. (2001) J. Mol. Biol. 307, 1329–1339. [DOI] [PubMed] [Google Scholar]
- 14.Cremona, O., Di Paolo, G., Wenk, M. R., Luthi, A., Kim, W. T., Takei, K., Daniell, L., Nemoto, Y., Shears, S. B., Flavell, R. A., et al. (1999) Cell 99, 179–188. [DOI] [PubMed] [Google Scholar]
- 15.Chi, Y., Zhou, B., Wang, W. Q., Chung, S. K., Kwon, Y. U., Ahn, Y. H., Chang, Y. T., Tsujishita, Y., Hurley, J. H. & Zhang, Z. Y. (2004) J. Biol. Chem. 279, 44987–44995. [DOI] [PubMed] [Google Scholar]
- 16.Nemoto, Y., Wenk, M. R., Watanabe, M., Daniell, L., Murakami, T., Ringstad, N., Yamada, H., Takei, K. & De Camilli, P. (2001) J. Biol. Chem. 276, 41133–41142. [DOI] [PubMed] [Google Scholar]
- 17.Banker, G. A. (1980) Science 209, 809–810. [DOI] [PubMed] [Google Scholar]
- 18.Slepnev, V. I., Ochoa, G. C., Butler, M. H., Grabs, D. & Camilli, P. D. (1998) Science 281, 821–824. [DOI] [PubMed] [Google Scholar]
- 19.Fischer, M., Kaech, S., Knutti, D. & Matus, A. (1998) Neuron 20, 847–854. [DOI] [PubMed] [Google Scholar]
- 20.Ziemnicka-Kotula, D., Xu, J., Gu, H., Potempska, A., Kim, K. S., Jenkins, E. C., Trenkner, E. & Kotula, L. (1998) J. Biol. Chem. 273, 13681–13692. [DOI] [PubMed] [Google Scholar]
- 21.McNiven, M. A., Kim, L., Krueger, E. W., Orth, J. D., Cao, H. & Wong, T. W. (2000) J. Cell Biol. 151, 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schafer, D. A. (2004) Traffic 5, 463–469. [DOI] [PubMed] [Google Scholar]
- 23.Ochoa, G. C., Slepnev, V. I., Neff, L., Ringstad, N., Takei, K., Daniell, L., Kim, W., Cao, H., McNiven, M., Baron, R. & De Camilli, P. (2000) J. Cell Biol. 150, 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eden, S., Rohatgi, R., Podtelejnikov, A. V., Mann, M. & Kirschner, M. W. (2002) Nature 418, 790–793. [DOI] [PubMed] [Google Scholar]
- 25.Innocenti, M., Zucconi, A., Disanza, A., Frittoli, E., Areces, L. B., Steffen, A., Stradal, T. E., Fiore, P. P., Carlier, M. F. & Scita, G. (2004) Nat. Cell Biol. 6, 319–327. [DOI] [PubMed] [Google Scholar]
- 26.De Camilli, P., Benfenati, F., Valtorta, F. & Greengard, P. (1990) Annu. Rev. Cell Biol. 6, 433–460. [DOI] [PubMed] [Google Scholar]
- 27.Turner, C. E. (2000) Nat. Cell Biol. 2, E231–E236. [DOI] [PubMed] [Google Scholar]
- 28.Parsons, J. T. (2003) J. Cell Sci. 116, 1409–1416. [DOI] [PubMed] [Google Scholar]
- 29.Danino, D. & Hinshaw, J. E. (2001) Curr. Opin. Cell Biol. 13, 454–460. [DOI] [PubMed] [Google Scholar]
- 30.Takei, K., McPherson, P. S., Schmid, S. L. & De Camilli, P. (1995) Nature 374, 186–190. [DOI] [PubMed] [Google Scholar]
- 31.Orth, J. D., Krueger, E. W., Cao, H. & McNiven, M. A. (2002) Proc. Natl. Acad. Sci. USA 99, 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, E. & De Camilli, P. (2002) Proc. Natl. Acad. Sci. USA 99, 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buccione, R., Orth, J. D. & McNiven, M. A. (2004) Nat. Rev. Mol. Cell Biol. 5, 647–657. [DOI] [PubMed] [Google Scholar]
- 34.Miki, H. & Takenawa, T. (2003) J. Biochem. (Tokyo) 134, 309–313. [DOI] [PubMed] [Google Scholar]
- 35.Chuang, Y. Y., Tran, N. L., Rusk, N., Nakada, M., Berens, M. E. & Symons, M. (2004) Cancer Res. 64, 8271–8275. [DOI] [PubMed] [Google Scholar]
- 36.Oikawa, T., Yamaguchi, H., Itoh, T., Kato, M., Ijuin, T., Yamazaki, D., Suetsugu, S. & Takenawa, T. (2004) Nat. Cell Biol. 6, 420–426. [DOI] [PubMed] [Google Scholar]
- 37.Malecz, N., McCabe, P. C., Spaargaren, C., Qiu, R., Chuang, Y. & Symons, M. (2000) Curr. Biol. 10, 1383–1386. [DOI] [PubMed] [Google Scholar]
- 38.Plattner, R., Kadlec, L., DeMali, K. A., Kazlauskas, A. & Pendergast, A. M. (1999) Genes Dev. 13, 2400–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scaife, R. M., Courtneidge, S. A. & Langdon, W. Y. (2003) J. Cell Sci. 116, 463–473. [DOI] [PubMed] [Google Scholar]
- 40.Plattner, R., Irvin, B. J., Guo, S., Blackburn, K., Kazlauskas, A., Abraham, R. T., York, J. D. & Pendergast, A. M. (2003) Nat. Cell Biol. 5, 309–319. [DOI] [PubMed] [Google Scholar]
- 41.Engqvist-Goldstein, A. E. & Drubin, D. G. (2003) Annu. Rev. Cell Dev. Biol. 19, 287–332. [DOI] [PubMed] [Google Scholar]
- 42.Qualmann, B. & Kessels, M. M. (2002) Int. Rev. Cytol. 220, 93–144. [DOI] [PubMed] [Google Scholar]
- 43.Kessels, M. M., Engqvist-Goldstein, A. E., Drubin, D. G. & Qualmann, B. (2001) J. Cell Biol. 153, 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naisbitt, S., Kim, E., Tu, J. C., Xiao, B., Sala, C., Valtschanoff, J., Weinberg, R. J., Worley, P. F. & Sheng, M. (1999) Neuron 23, 569–582. [DOI] [PubMed] [Google Scholar]
- 45.McPherson, P. S., Garcia, E. P., Slepnev, V. I., David, C., Zhang, X., Grabs, D., Sossin, W. S., Bauerfeind, R., Nemoto, Y. & De Camilli, P. (1996) Nature 379, 353–357. [DOI] [PubMed] [Google Scholar]
- 46.Gray, N. W., Fourgeaud, L., Huang, B., Chen, J., Cao, H., Oswald, B. J., Hemar, A. & McNiven, M. A. (2003) Curr. Biol. 13, 510–515. [DOI] [PubMed] [Google Scholar]
- 47.Zhou, Q., Xiao, M. & Nicoll, R. A. (2001) Proc. Natl. Acad. Sci. USA 98, 1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]