Abstract
SoxR is a global regulator contributing to multidrug resistance in Enterobacteriaceae. However, the contribution of SoxR to antibiotic resistance and fitness in Acinetobacter baumannii has not yet been studied. Comparisons of molecular characteristics were performed between 32 multidrug-resistant A. baumannii isolates and 11 susceptible isolates. A soxR overexpression mutant was constructed, and its resistance phenotype was analyzed. The impact of SoxR on efflux pump gene expression was measured at the transcription level. The effect of SoxR on the growth and fitness of A. baumannii was analyzed using a growth rate assay and an in vitro competition assay. The frequency of the Gly39Ser mutation in soxR was higher in multidrug-resistant A. baumannii, whereas the soxS gene was absent in all strains analyzed. SoxR overexpression led to increased susceptibility to chloramphenicol (4-fold), tetracycline (2-fold), tigecycline (2-fold), ciprofloxacin (2-fold), amikacin (2-fold), and trimethoprim (2-fold), but it did not influence imipenem susceptibility. Decreased expression of abeS (3.8-fold), abeM (1.3-fold), adeJ (2.4-fold), and adeG (2.5-fold) were correlated with soxR overexpression (P < .05). However, the expression levels of adeB and craA showed no obvious difference in the soxR-overexpression mutant. Competitive growth test results showed that soxR overexpression led to a lower growth rate, which was associated with a significant fitness cost in vitro. These results reveal that the global regulator SoxR is a negative regulator of efflux pump gene expression, and contributes to antibiotic resistance and fitness in A. baumannii.
Keywords: Acinetobacter baumannii, antibiotic resistance, efflux pump, global regulator, SoxR
1. Introduction
Acinetobacter baumannii is an important opportunistic pathogen that commonly causes nosocomial infections, such as ventilator-associated pneumonia and skin, soft tissue, wound, and bloodstream infections.[1] Multidrug-resistant A. baumannii strains have been increasingly reported worldwide, which raises serious concerns about the limited antimicrobial treatment options available.[2] SoxR is a global repressor protein involved in bacterial antibiotic resistance, and it contributes to multidrug-resistant phenotypes in Enterobacteriaceae.[3,4] SoxR protein induces the overexpression of efflux systems by activating the expression of the soxS gene in Enterobacteriaceae, or direct regulation in Pseudomonas aeruginosa.[5] However, the contribution of SoxR to antibiotic resistance and fitness in A. baumannii has not yet been studied. Here we focused on the role of the global regulator SoxR in A. baumannii. Comparisons of molecular characteristics were performed between carbapenem-resistant A. baumannii (CRAB) and carbapenem-susceptible A. baumannii (CSAB) isolates. A soxR-overexpression mutant was constructed, and the resistance phenotype and expression of multiple efflux pump genes were measured. We also analyzed the contribution of soxR overexpression to the growth and fitness of A. baumannii.
2. Materials and methods
2.1. Bacterial strain isolation and susceptibility testing
Forty-three A. baumannii clinical isolates were collected from the Chinese Antimicrobial Resistance Surveillance of Nosocomial Infections in 2011, including 11 carbapenem-susceptible isolates and 32 carbapenem-resistant isolates. A. baumannii was primarily identified using the VITEK2 system (bioMérieux, Marcy l’ toile, France), and confirmed by the presence of a blaOXA51-like gene. Susceptibility testing of antimicrobials was performed by the agar dilution method as described previously.[6] The study was approved by the Institutional Review Board of Peking University People's Hospital.
2.2. Polymerase chain reaction (PCR) and nucleotide sequencing
PCR amplification was performed using a 7300 thermocycler (Applied Biosystems, Foster City, CA). Primers were designed to amplify the full-length soxR and soxS sequences. Sequencing of the products was performed by an ABI 3730 DNA analyzer (Applied Biosystems) and analyzed using CLC sequence viewer software (CLC bio, Aarhus, Denmark).
2.3. Gene expression analysis using real-time reverse transcription PCR (RT-PCR)
Primers targeting adeB, adeJ, adeG, craA, abeM, and the housekeeping gene rpoB were used as described previously.[7–9] Primers were designed in this study for soxR (5′-ATGGATATTGGTGAAGTCG-3′ and 5′-TTAAAGTTTTGTTGGCTGAT-3′) and abeS (5′-TTTGGTCAGGCGCAGGTATT-3′ and 5′-ACCAATGCAGGCAGCTAAGT-3′). Bacteria were grown aerobically in Luria–Bertani (LB) broth until mid-log phase. DNase-treated RNA templates were prepared using the RNeasy Kit (Qiagen, Hilden, Germany). cDNA was generated from total RNA using random primer hexamers. RT-PCRs were performed using a 7300 thermocycler (Applied Biosystems) with a SYBR green PCR master mix (TaKaRa, Tokyo, Japan). The PCR program consisted of 5 seconds at 95°C, followed by 40 cycles of 15 seconds at 95°C and 31 seconds at 58°C. Each sample was run in triplicate.
2.4. Construction of the soxR-overexpression strain
The full-length soxR gene was amplified by PCR from 1 CRAB isolate. The PCR product was ligated into pWH1266, which is an Escherichia coli–Acinetobacter shuttle plasmid,[10] using the In-Fusion HD Cloning kit (Clontech, Saint-Germain-en-Laye, France) and transformed into E. coli JM109 (TaKaRa). Plasmids were isolated from transformants and used to transform electrocompetent A. baumannii, which were selected on Mueller–Hinton agar containing 100 μg/mL ticarcillin.
2.5. Growth rate assay
Bacteria were inoculated into 5 mL of Mueller–Hinton broth and incubated overnight at 37°C. Overnight cultures of A. baumannii strains were diluted 1:100 in Mueller–Hinton broth, and growth curves were performed in triplicate by incubating the cultures for 24 hours at 37°C with shaking at 200 rpm. Bacterial growth was monitored by measuring the optical density of the culture at 620 nm.
2.6. In vitro competition assay
In vitro competition assays were performed as described previously.[11] Briefly, the A. baumannii wild-type strain and the soxR-overexpression strain were cultured separately overnight in LB broth at 37°C. The bacteria were diluted 1:100, and equivalent numbers of the wild-type strain and the soxR-overexpression strain were pooled and cultured together at 37°C. At 0, 4, and 24 hours, aliquots of the mixed bacteria were diluted with sterile phosphate-buffered saline and plated onto 2 LB agar plates, one of which contained 100 μg/mL ticarcillin. Colony-forming units (CFU) were counted after 24 hours of incubation at 37°C. The competitive index (CI) was determined as follows: CI = (soxR-overexpression CFU/wild-type CFU)/(soxR-overexpression CFU/inoculated wild-type CFU).
2.7. Statistical analysis
SPSS 17.0 for Windows (SPSS Inc, Chicago, IL) was used for all statistical analyses. Comparative analyses were executed by χ2 or Fisher exact tests for categorical variables, and by the Student's t test for continuous variables. All tests were 2-tailed, and P values <.05 were considered statistically significant.
3. Results
3.1. Correlation between soxR mutations and antibiotic resistance in A. baumannii
To assess the correlation between antibiotic resistance and the global regulator SoxR, soxR mutations were investigated. In most CRAB isolates (29/32, 90.63%), a mutation in soxR (G115A) resulted in an amino acid substitution (Gly39Ser) in SoxR (Table 1). In 11 CSAB isolates, the Gly39Ser mutation was only identified in 1 strain, and the frequency (9.1%) was lower than that of the CRAB strains (P < .01). However, soxS was absent in all strains analyzed (Table 1).
Table 1.
Correlation between soxR mutation and antibiotic resistance in A. baumannii.
3.2. Contribution of soxR overexpression to A. baumannii antibiotic resistance
To decipher the role of SoxR in A. baumannii, the soxR-overexpression mutant AB26/soxR was constructed. Susceptibility testing was performed with AB26/soxR, and the parental strain AB26 was used as a control (Table 2). The data showed that soxR overexpression resulted in increased susceptibility to chloramphenicol (4-fold), tetracycline (2-fold), tigecycline (2-fold), ciprofloxacin (2-fold), amikacin (2-fold), and trimethoprim (2-fold), but it did not influence imipenem susceptibility.
Table 2.
Antibiotic susceptibility change of soxR-overexpression strain.

3.3. Influence of soxR overexpression on efflux pump gene expression
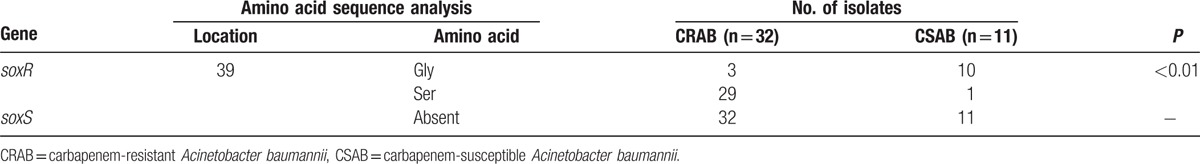
Multiple efflux pump genes, which correlate with antibiotic resistance in A. baumannii, including resistance-nodulation-division (RND) family genes (adeB, adeJ, and adeG), the small multidrug resistance (SMR) family gene abeS, the multidrug and toxic compound extrusion (MATE) gene abeM, and the major facilitator superfamily gene craA were investigated. As shown in Figure 1, the expression of abeS, abeM, adeJ, and adeG was decreased in AB26/soxR with statistical significance (P < .05). The expression level of abeS was decreased 3.8-fold in soxR-overexpression isolates, which showed the most obvious downregulation. However, no significant difference was detected in craA or adeB expression among soxR-overexpression isolates (data not shown).
Figure 1.
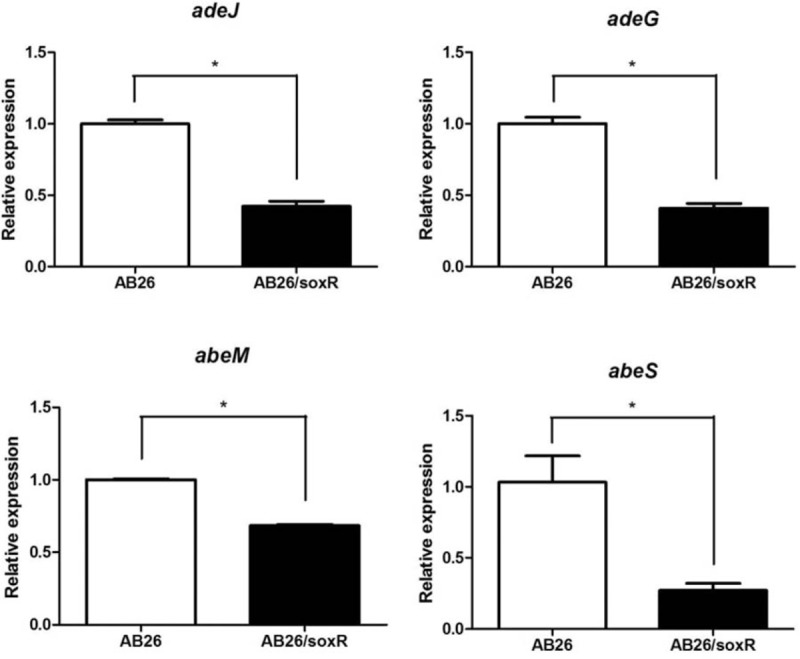
Relative expression of efflux pump genes in A. baumannii. ∗, P < .05. AB26, the wild-type strain; AB26/soxR, the soxR-overexpression strain.
3.4. Effect of SoxR on the growth and fitness of A. baumannii
To determine whether soxR overexpression affects the bacterial growth rate, growth curves were performed for the soxR-overexpression strain AB26/soxR and the wild-type strain AB26. However, the growth rate of AB26/soxR did not differ significantly from that of AB26. In vitro competition experiments were also performed to determine the relative growth rates of each of the strains (Fig. 2). The growth rate of AB26/soxR was lower than that of AB26 when cultured together. In addition, soxR overexpression was associated with a significant fitness cost in vitro. Measurements were performed at 4 and 24 hours, and a significant decrease in the growth of the AB26/soxR strain was observed. After a 4-hour co-culture, the CI was 0.723 (P < .05), and after a 24-hour co-culture, the CI was even lower (0.299) and highly significant (P < .01).
Figure 2.
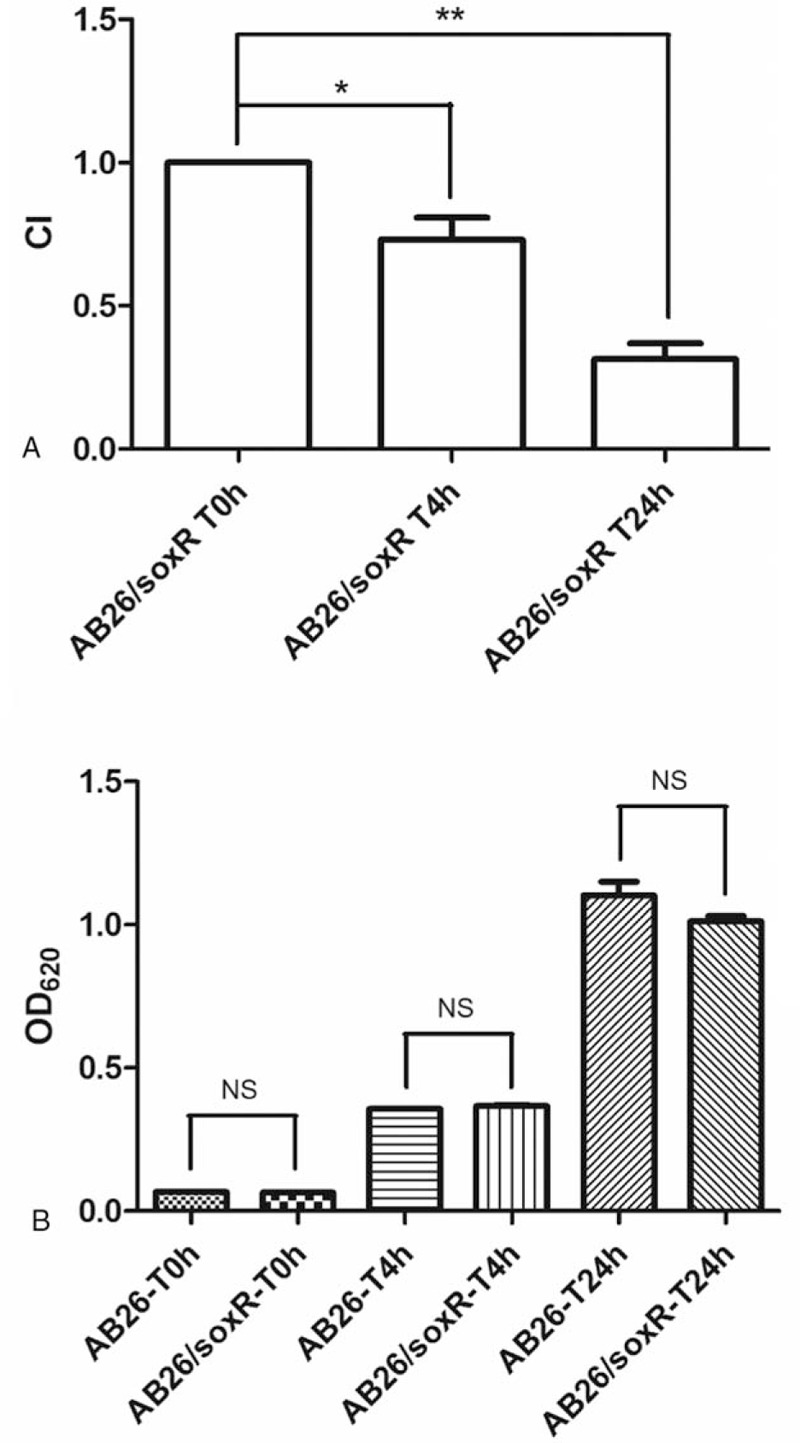
Relative in vitro competition indexes of the soxR-overexpression strain. (A) In vitro competition indexes at 4 and 24 hours. ∗, P < .05. ∗∗, P < .01. (B) Growth rate of the soxR-overexpression strain and the wild-type strain. AB26, the wild-type strain; AB26/soxR, the soxR-overexpression strain. NS = no significant difference.
4. DISCUSSION
SoxR is a global repressor protein involved in multidrug-resistance in Enterobacteriaceae.[3,4] SoxR protein is produced constitutively at a low level, and it activates the expression of the soxS gene in response to superoxide-generating agents.[12] The activity of SoxS is regulated only by its intracellular concentration, and it binds promoter regions of its target genes to recruit RNA polymerase.[12] High SoxS activity is attributable to a frameshift mutation that truncates soxR, and it is responsible for fluoroquinolone resistance in E. coli.[13] In Klebsiella pneumoniae, a mutation in soxR (C375G) results in an amino acid substitution (N125K) in SoxR protein. The soxR mutation induced the overexpression of efflux systems, which were phenotypically characterized by multidrug resistance.[5] Recent studies of the SoxR regulon in Pseudomonas indicate that SoxR plays an alternative role to that in the traditional SoxR–SoxS paradigm. In P. aeruginosa, SoxR can directly activates a 6-gene regulon, which includes a multidrug efflux pump system encoded by the mexGHI-ompD 4-gene operon, and a probable efflux pump gene PA3718.[14] In our study, we identified a correlation between decreased antibiotic susceptibility and the frequency of clinical strains carrying the Gly39Ser mutation in soxR. However, the soxS gene was absent in all A. baumannii strains in our study. We speculate that SoxR may directly regulate target genes, such as efflux pump genes, in A. baumannii. Because soxS upregulation is associated with antibiotic resistance in Enterobacteriaceae, and soxS was absent in all A. baumannii strains in our study, a soxR-overexpression mutant was constructed to analyze the contribution of SoxR to antibiotic resistance and fitness in A. baumannii.
Efflux-mediated resistance has been found in many bacterial genera.[15] Overexpression of the efflux system, responsible for reducing the accumulation of antibiotics, is an efficient mechanism for multidrug resistance. RND efflux systems (AdeABC, AdeIJK, and AdeFGH), which confer multidrug resistance when overexpressed, typically exhibit wide substrate ranges.[15] The AbeM and AbeS pumps that belong to the MATE- and SMR-type exporters, respectively, also accommodate many drug substrates.[16] The deletion of the SMR family gene abeS resulted in significant decreases in the minimum inhibitory concentrations of chloramphenicol, ciprofloxacin, and erythromycin in A. baumannii.[17] In our study, we investigated the effect of overexpressing soxR on the expression of several efflux pump genes, including the RND family genes adeB, adeJ, and adeG; the SMR family gene abeS; the MATE family gene abeM; and the major facilitator superfamily gene craA. The expression levels of the RND genes adeJ and adeG were 2.4- and 2.5-fold lower in isolates with overexpression of soxR. Meanwhile, real-time RT-PCR showed that abeS expression was 3.8-fold lower in the soxR-overexpression strain. The change in expression of multiple efflux pump genes suggests that SoxR may be involved in a complex regulatory network.
The inability to construct a soxR deletion mutant is a limitation of our study. Construction of the soxR deletion mutant failed even though we successfully used the same method to construct an adeB deletion mutant.[18] We will attempt to construct the soxR-deletion mutant using new approaches in the future. Only studying the regulatory effects of SoxR on efflux pump genes is another limitation of our study. Studying other regulatory elements could increase our understanding of the role of SoxR in antibiotic resistance and fitness in A. baumannii. Future transcriptome analyses and functional studies are needed to fully understand the detailed regulatory mechanism of SoxR.
This is the first time, to our knowledge, that SoxR has been associated with the regulation of fitness and antibiotic resistance in A. baumannii. We found a correlation between decreased antibiotic susceptibility and the frequency of clinical strains carrying the Gly39Ser mutation in soxR. SoxR overexpression was associated with a significant fitness cost in vitro, and soxR overexpression correlated with the decreased expression of the efflux pump genes abeS, abeM, adeJ, and adeG. These results reveal that SoxR is a negative regulator of efflux pump gene expression and affects antibiotic resistance and fitness in A. baumannii.
Acknowledgments
We thank Dr. Yunsong Yu for donating the plasmid pWH1266.
Footnotes
Abbreviations: CFU = colony-forming units, CI = competitive index, CRAB = carbapenem-resistant A. baumannii, CSAB = carbapenem-susceptible A. baumannii, LB = Luria–Bertani, MATE = multidrug and toxic compound extrusion, PCR = polymerase chain reaction, RND = resistance-nodulation-division, RT-PCR = reverse transcription PCR, SMR = small multidrug resistance.
This study was supported by the Beijing Natural Science Foundation (7153177) and the National Natural Science Foundation of China (31170125 and 81625014).
The authors have no conflicts of interest to disclose.
References
- [1].Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 2008;21:538–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science 2009;325:1089–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Koutsolioutsou A, Pena-Llopis S, Demple B. Constitutive soxR mutations contribute to multiple-antibiotic resistance in clinical Escherichia coli isolates. Antimicrob Agents Chemother 2005;49:2746–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Webber MA, Piddock LJ. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob Agents Chemother 2001;45:1550–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bialek-Davenet S, Marcon E, Leflon-Guibout V, et al. In vitro selection of ramR and soxR mutants overexpressing efflux systems by fluoroquinolones as well as cefoxitin in Klebsiella pneumoniae. Antimicrob Agents Chemother 2011;55:2795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li H, Liu F, Zhang Y, et al. Evolution of carbapenem-resistant Acinetobacter baumannii revealed through whole-genome sequencing and comparative genomic analysis. Antimicrob Agents Chemother 2015;59:1168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Coyne S, Guigon G, Courvalin P, et al. Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob Agents Chemother 2010;54:333–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Marchand I, Damier-Piolle L, Courvalin P, et al. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother 2004;48:3298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rumbo C, Gato E, Lopez M, et al. Contribution of efflux pumps, porins, and beta-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 2013;57:5247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hunger M, Schmucker R, Kishan V, et al. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 1990;87:45–51. [DOI] [PubMed] [Google Scholar]
- [11].Liu D, Liu ZS, Hu P, et al. Characterization of surface antigen protein 1 (SurA1) from Acinetobacter baumannii and its role in virulence and fitness. Vet Microbiol 2016;186:126–38. [DOI] [PubMed] [Google Scholar]
- [12].Pomposiello PJ, Demple B. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol 2001;19:109–14. [DOI] [PubMed] [Google Scholar]
- [13].Fabrega A, Martin RG, Rosner JL, et al. Constitutive SoxS expression in a fluoroquinolone-resistant strain with a truncated SoxR protein and identification of a new member of the marA-soxS-rob regulon, mdtG. Antimicrob Agents Chemother 2010;54:1218–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Palma M, Zurita J, Ferreras JA, et al. Pseudomonas aeruginosa SoxR does not conform to the archetypal paradigm for SoxR-dependent regulation of the bacterial oxidative stress adaptive response. Infect Immun 2005;73:2958–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Coyne S, Courvalin P, Perichon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 2011;55:947–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li XZ, Plesiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 2015;28:337–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Srinivasan VB, Rajamohan G, Gebreyes WA. Role of AbeS, a novel efflux pump of the SMR family of transporters, in resistance to antimicrobial agents in Acinetobacter baumannii. Antimicrob Agents Chemother 2009;53:5312–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li H, Wang X, Zhang Y, et al. The role of RND efflux pump and global regulators in tigecycline resistance in clinical Acinetobacter baumannii isolates. Future Microbiol 2015;10:337–46. [DOI] [PubMed] [Google Scholar]


