Supplemental Digital Content is available in the text
Keywords: MDR1, meta-analysis, nephrotic syndrome, polymorphisms
Abstract
Background:
Studies have investigated rs1128503, rs1045642, and rs2032582 in multidrug resistance protein 1 (MDR1) for association with susceptibility to idiopathic nephrotic syndrome (INS) and steroid resistance. However, because these findings were inconsistent, we performed a meta-analysis to determine whether there was evidence of a role of these MDR1 variants in INS.
Methods:
The PubMed, Embase, and Web of Science databases were systematically searched to identify studies that examined MDR1 polymorphisms with susceptibility to INS and/or to steroid resistance. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were calculated by a fixed-effects or random-effects model based on heterogeneity.
Results:
We selected 9 case-control studies that included 928 patients with INS, of which steroid resistance data were available for 724 (236 were steroid resistant and 488 were steroid sensitive), and 879 healthy controls. All subjects were children. No significant relationships between these polymorphisms and INS susceptibility were identified. Significantly increased risk of steroid resistance was observed with rs1128503 allelic (OR = 1.49, 95% CI = 1.20–1.86) and genotypic (OR = 1.97, 95% CI = 1.18–3.30; OR = 2.03, 95% CI = 1.43–2.88) comparisons, and with allelic (OR = 1.56, 95% CI = 1.05–2.31) and genotypic (OR = 2.85, 95% CI = 1.15–7.07; OR = 2.21, 95% CI = 1.01–4.8) comparisons to rs2032582 in Caucasian populations. However, this association between rs2032582 and steroid resistance was not robust enough to withstand corrections for multiple comparisons. Similarly, we found that the rs1128503T-rs2032582G-rs1045642C (T-G-C) haplotype was associated with an increased risk of steroid resistance (OR = 2.02, 95% CI = 1.13–3.59), while the wild-type C-G-C haplotype was associated with a decreased risk (OR = 0.32, 95% CI = 0.12–0.88) in Caucasians; however, these findings were not significant following adjustments for multiple comparisons.
Conclusions:
MDR1 rs1128503, rs1045642, and rs2032582 polymorphisms are not associated with INS susceptibility; however, there is evidence of an association between rs1128503 and increased risk of steroid resistance in children with INS, which indicates MDR1 may play a role in steroid resistance found in children with INS.
1. Introduction
Nephrotic syndrome (NS) is a common glomerular disease characterized by massive proteinuria and hypoalbuminemia at any age. The most frequent type is idiopathic nephrotic syndrome (INS), which clinically manifests with histological differences, such as minimal change nephropathy and focal segmental glomerulosclerosis.[1] Glucocorticoids (GCs) are recommended as a mainstay of therapy[2]; however, patients with INS show a varied response. Although most INS sufferers respond to steroids, some subjects remain steroid dependent or steroid resistant, especially in children.[3] Furthermore, a poor response to steroids indicates an unfavorable prognosis and high risk of end-stage renal disease.[4] Therefore, it is essential to identify risk factors contributing to failure of immunosuppressive treatment. Genetic backgrounds were reported as a prominent etiology of INS. Variants of genes that encode podocyte proteins result in structural defects of the glomerular filtration barrier and subsequent onset of NS as well as steroid resistance.[5–7] Additionally, factors that modulate response to therapeutic regimens such as P-glycoprotein (P-gp) are another risk.[8] P-gp is encoded by multidrug resistance protein 1 (MDR1), which is also known as ATP binding cassette subfamily B member 1 (ABCB1), and acts as a transmembrane efflux pump for numerous drugs involved in absorption, distribution, and elimination. GCs are transported by P-gp and induce MDR1 expression.[9] In addition, expression of MDR1 and P-gp activity are negatively correlated with steroid sensitivity in children with NS.[10–12] Furthermore, there is evidence that polymorphisms in MDR1 may influence disease occurrence and response to drug treatment through changes in gene expression and P-gp activity.[13]
During the past decade, there has been increasing interest whether there is an association between MDR1 polymorphisms and aspects of INS, including susceptibility to the disorder and steroid responsiveness. Several case-control studies have investigated whether INS risk and the therapeutic effect of GCs are associated with common single nucleotide polymorphisms (SNPs) found in the exons of MDR1, specifically, rs1128503 (C1236T), rs1045642 (C3435T), and rs2032582 (G2677T/A). However, these studies yielded inconsistent results. Youssef et al[14] reported that the T allele and TT genotype of rs1045642, and the T allele and GT, TT, and TT + AA genotypes of rs2032582 were significantly increased in patients with INS, but no significant evidence of association was found with any of these 3 SNPs in the cohort tested by Cizmarikova et al.[15] Jafar et al[16] showed that the frequencies of homozygous mutant TT/AA genotypes of rs2032582 were significantly increased in affected children with steroid resistance, whereas Chiou et al[17] observed a trend with the T allele of rs1128503 in their cohort.
To confirm genetic markers of INS risk and steroid resistance, and conforming to Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) checklists and guidelines,[18] we performed a meta-analysis to assess whether there is evidence of a correlation between these three MDR1 polymorphisms and increased risk of susceptibility to INS or to steroid resistance. Furthermore, our findings may show whether these polymorphisms are informative as potential biomarkers for INS risk and steroid responsiveness.
2. Methods
2.1. Ethics
Our meta-analysis used previously published studies, and therefore, ethical approval and informed consent for our study were not applicable. However, all studies that were included in our meta-analysis received ethics approval from the respective ethics committee and patient consent was obtained.
2.2. Search strategy
We searched for eligible studies using the following databases: PubMed, Embase, and Web of Science, using the key words or phrases “MDR1, ABCB1, multidrug resistance gene, multidrug resistance protein 1,” “polymorphism (s),” and “nephrotic syndrome.” There was no limitation of language and the last search update was performed on October 15, 2016. In addition, references from related studies and reviews were manually searched to identify additional studies.
2.3. Definition of NS/SS/SR
We defined NS in children according to the International Study of Kidney Disease in Children (ISKDC) as proteinuria of 40 mg/m2/h and hypoalbuminemia (serum albumin <2.5 g/dL). Steroid sensitive (SS) was defined as the disappearance of proteinuria within 4 weeks of initial standard steroid therapy, which was treatment with prednisone 2 mg/kg/d or 60 mg/m2 body surface area/day. Patients who had persistent proteinuria after 4 weeks of prednisone therapy were categorized as steroid resistant (SR).[19]
The diagnosis of NS in adults was heavy proteinuria (24-hour urinary protein excretion >3 g) and hypoalbuminemia (serum albumin <2.5 g/dL).[20] SS was assigned to affected adults with negative proteinuria after 16 weeks of standard steroid therapy (prednisone 1 mg/kg/d), while persistent proteinuria was considered SR.[21]
2.4. Inclusion and exclusion criteria
To identify eligible studies for our meta-analysis, we used the following inclusion criteria: (1) case-control study, (2) investigated whether there were associations between the MDR1 polymorphisms, rs1128503, rs1045642, and rs2032582, and susceptibility to INS and/or steroid responsiveness in INS, and (3) detailed genotype frequencies were available.
Studies were excluded based on the following criteria: (1) study included secondary NS, (2) detailed genotype data were not available, (3) meeting abstract, (4) family-based study of pedigrees, or (5) duplication of previous publications.
2.5. Data extraction
Two reviewers (Y-QX and X-CG) independently extracted the following data of each eligible study: author name, year of publication, country, ethnicity, subpopulation, diagnostic criteria of NS, the definition of SS and SR, genotyping method, Hardy–Weinberg equilibrium (HWE), sample size of cases, controls, SS, and SR groups, and genotype frequencies of all groups.
2.6. Assessment of methodological quality
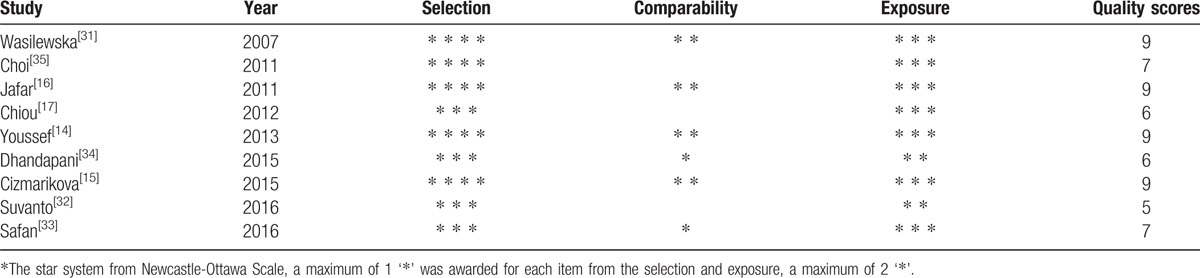
Two investigators (S-SH and YL) independently evaluated the quality of the included studies according to the Newcastle–Ottawa Scale (NOS) for a case-control study.[22] Three dimensions including selection, comparability, and exposure were categorized in NOS. A star system was used to semi-quantitatively assess study quality. A maximum of 1 star was awarded for each item from the selection and exposure categories, whereas a maximum of 2 stars could be assigned for comparability. In the context of our meta-analysis, studies with 7 or more stars were considered high quality, whereas studies between 4 and 6 stars were considered medium quality, and studies with less than 4 stars were considered poor quality.[23] Differences in scoring were resolved by discussion to achieve consensus, and confirmed by another reviewer (YW).
2.7. Statistical analysis
HWE of the control group was evaluated using an online tool (available at https://ihg.gsf.de/cgi-bin/hw/hwa1.pl) for each study.[24] The association between MDR1 polymorphisms and INS risk was assessed by determining the odds ratio (OR) and 95% confidence interval (CI). Pooled ORs were generated using the following genetic models: allelic comparisons (rs1128503 T vs. C, rs1045642 T vs. C, and rs2032582 T/A vs. G), homozygous model (rs1128503 TT vs. CC, rs1045642 TT vs. CC, and rs2032582 TT/AA vs. GG), heterozygous model (rs1128503 CT vs. CC, rs1045642 CT vs. CC, and rs2032582 GT/GA vs. GG), dominant model (rs1128503 TT + CT vs. CC, rs1045642 TT + CT vs. CC, and rs2032582 TT/AA + GT/GA vs. GG), and recessive model (rs1128503 TT vs. CC + CT, rs1045642 TT vs. CC + CT, and rs2032582 TT/AA vs. GG + GT/GA). P values were determined with the Z-test and P < .05 was considered statistically significant. The reported P was adjusted using Bonferroni–Holm correction (BON) and Benjamini–Hochberg False Discovery Rate (FDR) methods to control the overall false positive rate.[25,26] In addition, we performed haplotype analysis comparing each haplotype to all other haplotypes (i.e., N-N-N vs. non-N-N-N).
The Q test and I2 statistic were carried out to assess heterogeneity, which was considered significant with P < .10 or I2 > 50%.[27] Either a fixed-effects model or a random-effects model was adopted according to the heterogeneity.[28] Subgroup analysis was performed to explore heterogeneity stratified by ethnicity and subpopulation. We used sensitivity analysis to test the reliability of our results by singly omitting each study in turn, and studies that did not meet HWE in the control group were also removed from the sensitivity analysis. Publication bias was evaluated by Begg funnel plot and Egger linear regression test, and P < .05 was considered statistically significant.[29,30] All statistical analyses were performed by STATA version 12.0 (StataCorp, College Station, TX).
3. Results
3.1. Characteristics of studies included in the meta-analysis
Fifty-nine publications were identified using our search criteria from the PubMed, Embase, and Web of Science databases of which 9 studies were included in our meta-analysis according to the inclusion and exclusion criteria (Fig. 1).[14–17,31–35] All of the participants in these studies were children consisting of 928 patients with INS and 879 healthy controls; moreover, steroid resistance data were available for 724 patients comprising 488 patients with SS and 236 patients with SR, which were compared. The subjects in 4 studies were Asian[16,17,34,35] and those from the other 5 were Caucasian.[14,15,31–33] Seven studies reported genotype frequencies between patients with INS and healthy controls,[14–16,31,32,34,35] and 7 studies showed the genotype data of SS and SR groups.[14–17,32,33,35] Haplotype frequencies of both case and control cohorts were described in part in 4 studies.[14,15,32,35] SS and SR groups were also compared in 4 studies (details provided in Table 1).[14–16,35] Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) was used for genotyping in 6 studies,[14–16,31,34,35] the TaqMan genotyping assay was performed in 1 study,[17] and the 2 remaining studies used real-time PCR.[32,33] Blood samples were collected for genotyping in all studies. We tested the controls from all 9 studies for HWE and found that the controls in 1 study were not in HWE for rs1045642 and rs2032582 (P < .001).[34] The quality of the included studies for our meta-analysis was assessed by NOS and all studies were either of medium or high quality in terms of selection and exposure. However, we found that comparability was poor in some studies (Table 2).
Figure 1.
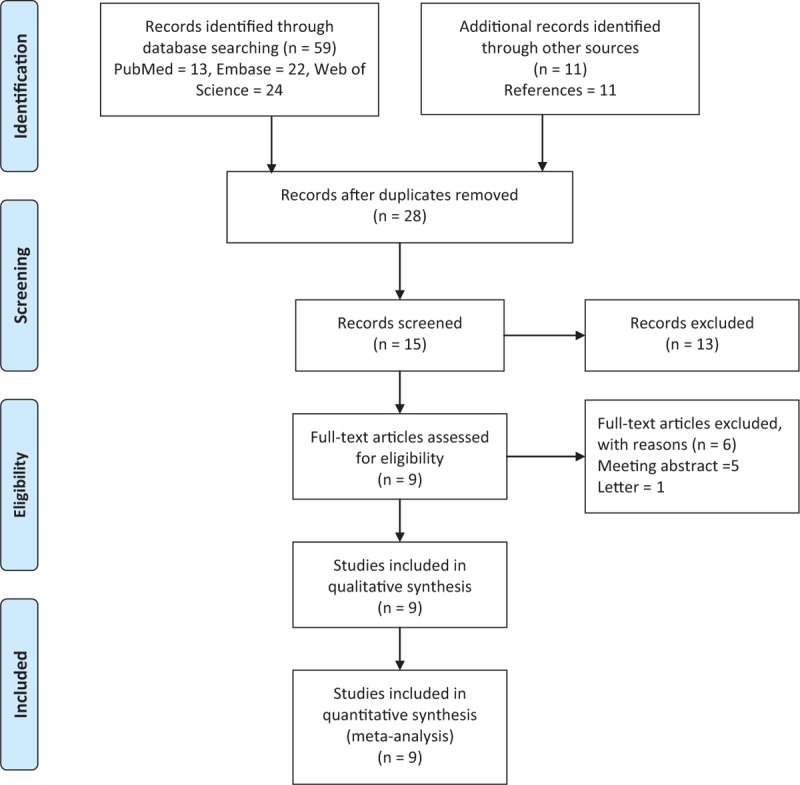
Flow chart of study selection.
Table 1.
Characteristics of the individual studies included in the meta-analysis.
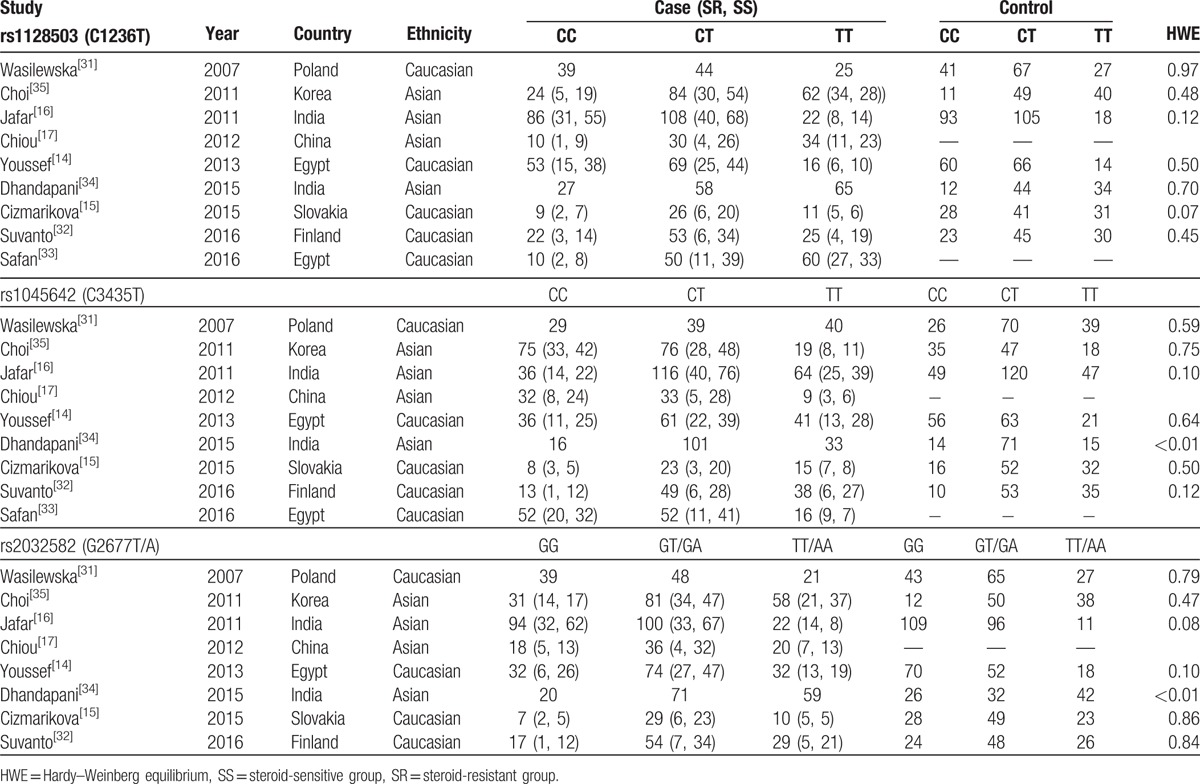
Table 2.
Quality assessment based on the Newcastle–Ottawa Scale.
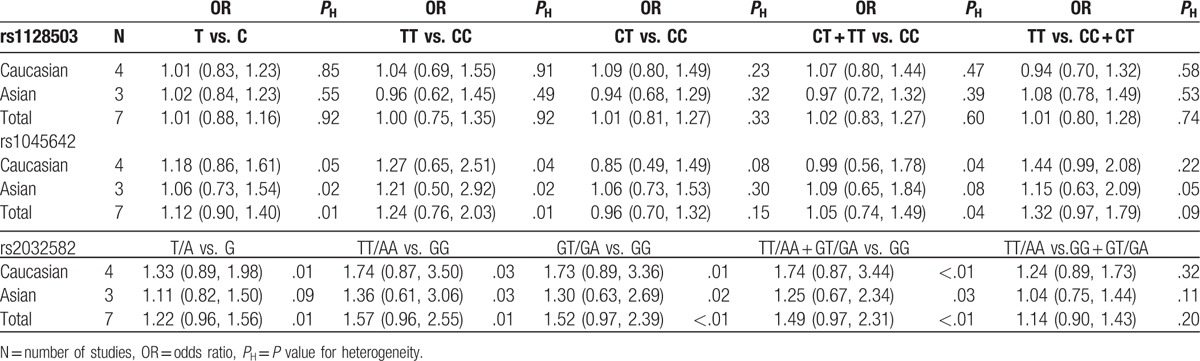
3.2. Associations between MDR1 polymorphisms and INS susceptibility
Because no significant heterogeneity was identified by Q test and I2 statistic in all genetic models of rs1128503, we used a fixed-effects model and found no evidence of an association in any comparison (Fig. 2). Using either a fixed-effects model or a random-effects model in our meta-analysis of rs1045642 and rs2032582, we found no significant correlation between these 2 SNPs and INS occurrence. Next, subgroup analysis was performed according to ethnicity, and we found no statistical evidence of an association for any of these 3 SNPs (Table 3). Because all subjects in these studies were children, subgroup analysis based on subpopulation was not performed.
Figure 2.
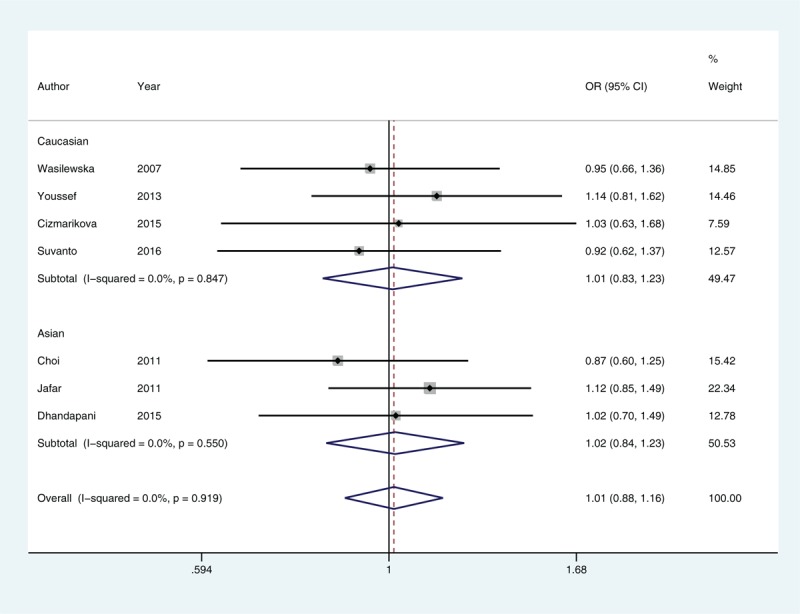
Forest plot of rs1128503 allelic comparisons in susceptibility to idiopathic nephrotic syndrome. CI = confidence interval, OR = odds ratio.
Table 3.
Meta-analysis of MDR1 polymorphisms and susceptibility to idiopathic nephrotic syndrome.
3.3. Associations between MDR1 polymorphisms and steroid responsiveness
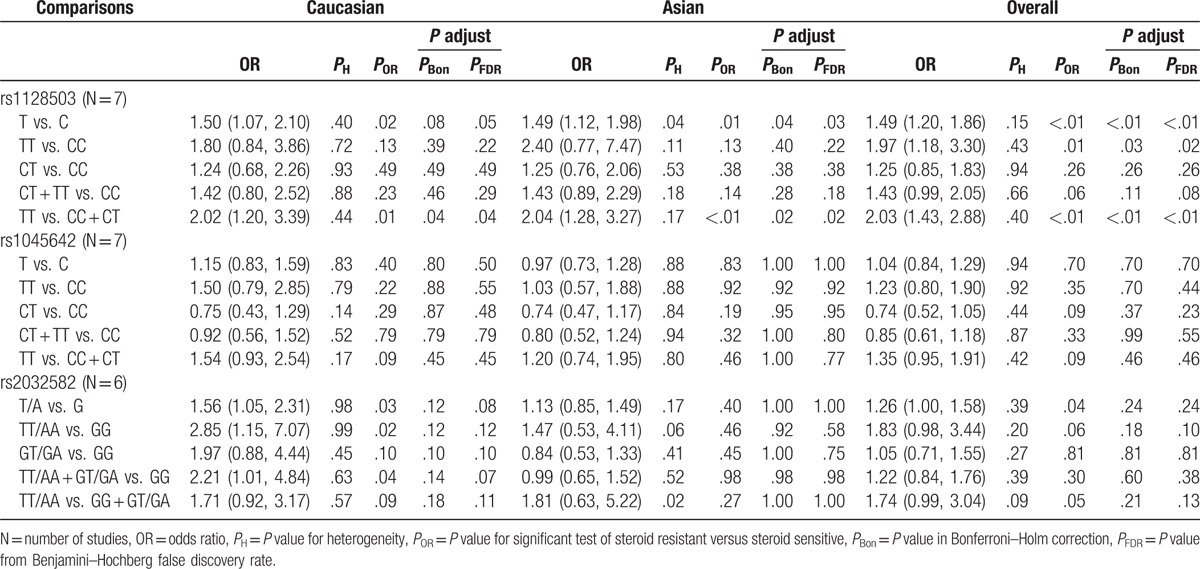
In our analysis to determine whether there was correlation between MDR1 polymorphisms and steroid responsiveness, and based on heterogeneity, the allelic comparison in rs1128503, and the homozygous model and recessive model in rs2032582 were pooled for testing in a random-effects model, while the remaining genetic models were performed in a fixed-effects model. We found a significantly increased risk of SR compared with SS with rs1128503 allelic comparisons (T vs. C, OR = 1.49, 95% CI = 1.20–1.86) and under both homozygous (TT vs. CC, OR = 1.97, 95% CI = 1.18–3.30) and recessive (TT vs. CC + CT, OR = 2.03, 95% CI = 1.43–2.88) models (Fig. 3). This association remained significant based on the adjusted P that was determined from Bonferroni–Holm correction and FDR methods. Our findings from subgroup analysis indicated that both Asian and Caucasian cohorts had an increased risk of SR in allelic comparisons and under a recessive mode of inheritance, but following corrections for multiple comparisons, the allelic comparison in Caucasian children showed no significant difference. In addition, no significant differences between rs1045642 and SR were found in any model tested as well as from subgroup analysis. However, we found from allelic comparisons that rs2032582 was associated with an increased risk of SR compared with SS (T/A vs. G, OR = 1.26, 95% CI = 1.00–1.58). Further subgroup analysis of rs2032582 indicated that Caucasians had an increased risk of SR compared with SS in an allelic comparison (T/A vs. G, OR = 1.56, 95% CI = 1.05–2.31) and under homozygous (TT/AA vs. GG, OR = 2.85, 95% CI = 1.15–7.07) and dominant (TT/AA + GT/GA vs. GG, OR = 2.21, 95% CI = 1.01–4.84) models. However, no statistical significance was detected in any genetic model tested and in subgroup analyses after P values were adjusted following FDR or Bonferroni–Holm correction for multiple testing (Table 4).
Figure 3.
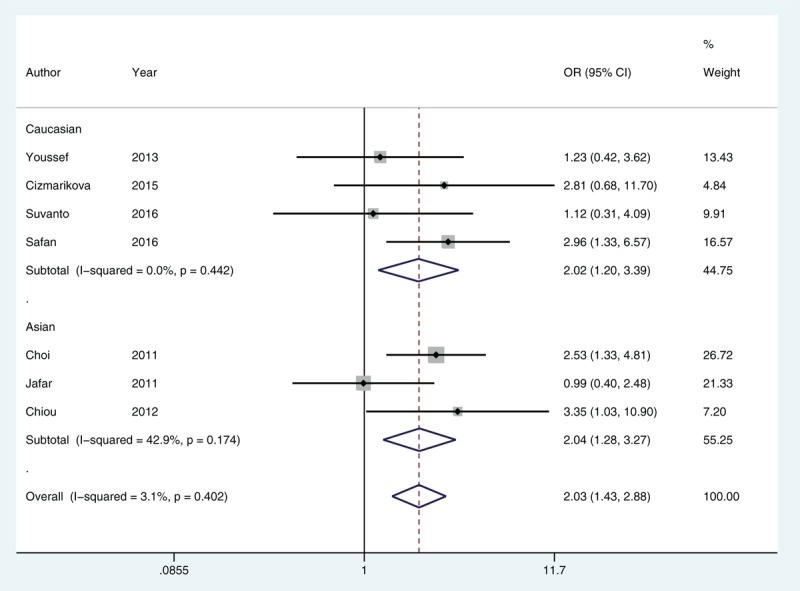
Forest plot of the recessive model for rs1128503 in steroid resistant versus steroid sensitive idiopathic nephrotic syndrome. CI = confidence interval, OR = odds ratio.
Table 4.
Meta-analysis of MDR1 polymorphisms and steroid responsiveness.
3.4. Meta-analysis of haplotypes
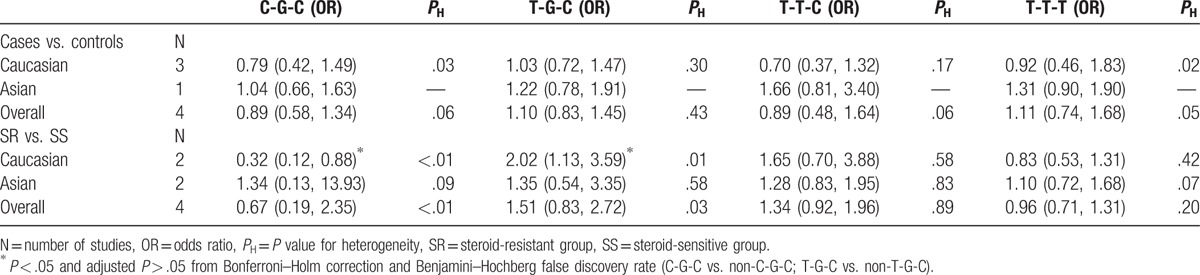
Using available data from these studies, we analyzed the 4 most frequent haplotypes of these SNPs between case and control groups (n = 436 and n = 438, respectively) as well as between SR and SS groups (n = 207 and n = 363, respectively). In our meta-analysis, we found no significant correlation between the MDR1 rs1128503-rs2032582-rs1045642 haplotypes, C-G-C, T-G-C, T-T-C, and T-T-T, with INS susceptibility. However, we found in Caucasian cohorts that the T-G-C haplotype was associated with an increased risk of SR compared with SS (T-G-C vs. non-T-G-C, OR = 2.02, 95% CI = 1.13–3.59), while in contrast we found that the wild-type C-G-C haplotype was associated with a decreased risk of SR compared with SS (C-G-C vs. non-C-G-C, OR = 0.32, 95% CI = 0.12–0.88). It should be noted that these findings were not significant following adjustments for multiple comparisons. No evidence of an association was found in studies of Asian cohorts (Table 5).
Table 5.
Meta-analysis of MDR1 rs1128503-rs2032582-rs1045642 haplotypes.
3.5. Sensitivity analysis
Sensitivity analyses were performed to identify the influence of each individual study on the pooled results. We re-tested all genetic models excluding each study in turn as well as excluding the study that deviated from HWE in the control group, and found that the pooled estimates did not change, which demonstrates the results of our meta-analysis were stable (Supplemental Figures S1 and S2).
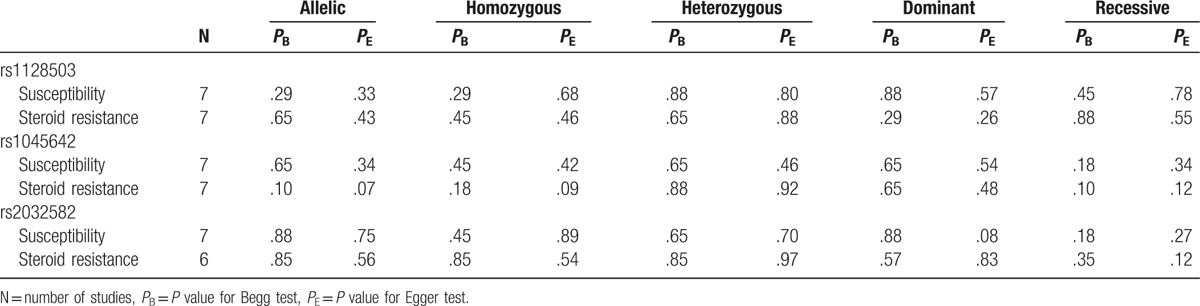
3.6. Publication bias
As shown in Table 6, no potential publication bias was found in any genetic model according to Begg funnel plot and Egger regression test.
Table 6.
Results of publication bias.
4. Discussion
Previous studies have identified over 50 SNPs in the coding region of MDR1, of which rs1128503 (C1236T), rs1045642 (C3435T), and rs2032582 (G2677T/A) have been of particular interest. These 3 SNPs showed the highest frequencies in Asian and Caucasian populations and the lowest in African populations.[36–38]MDR1 variants may affect the expression and function of P-gp that has a crucial role in the pharmacokinetics of several therapeutic drugs including glucocorticoids.[39] Significantly higher expression of MDR1 and P-gp activity were detected in steroid- and cyclosporine-resistant patients with INS,[11] and a positive correlation between P-gp expression and total prednisone dose has also been demonstrated in children with INS.[40] Therefore, the interindividual variability of steroid response found in patients with INS may be because of functional polymorphisms at MDR1.[10,41]
This is the first meta-analysis of MDR1 polymorphisms and INS, which consisted of 9 case-control studies including 928 patients with INS and 879 healthy controls to assess disease susceptibility, and 724 affected individuals consisting of 236 patients with SR and 488 patients with SS to evaluate steroid responsiveness. Our meta-analysis showed no evidence of a correlation between the 3 polymorphisms investigated and susceptibility to INS, which was consistent with the findings from the studies by Choi et al,[35] Cizmarikova et al,[15] and Suvanto et al.[32] Because P-gp affects the pharmacokinetics of GCs, the occurrence of idiopathic NS is unlikely to be related to MDR1 variants, which is in accordance with our findings. However, because Youssef et al[14] and Jafar et al[16] reported that rs1045642 and rs2032582 were associated with INS susceptibility, additional studies are warranted. It was previously determined that rs1128503 and rs1045642 were synonymous SNPs predicted to not cause amino acid changes.[42] Although we found no significant differences between variants of rs1045642 and patients with SR or SS in our meta-analysis, we found that the rs1128503 polymorphism was robustly associated with an increased risk of SR. Although rs1128503 is a silent SNP, it may still have an effect on protein translation rates thereby influencing protein folding and activity, which contributes to altered substrate specificity of P-gp.[43] In addition, rs1128503 exhibits strong linkage disequilibrium with other functional polymorphisms such as rs2032582.[44] rs2032582 is a non-synonymous variant that causes an amino acid substitution from alanine to serine or threonine that may lead to increased efficiency of the mutated protein by efflux of glucocorticoid or its active metabolites, which results in steroid resistance.[16,45] We found that rs2032582 was associated with a significantly increased risk of SR in Caucasian children with INS. However, the statistical significance of this association between rs2032582 and SR did not remain following FDR adjustments or Bonferroni–Holm corrections for multiple comparisons. Hence, the association between SR and this polymorphism may be a false positive result caused by type I error because of testing multiple hypotheses. Alternatively, we can speculate that this association may be a true positive result reflecting a molecular basis in SR that is not statistically significant in this analysis because of the limited sample sizes of the included studies or other confounding factors such as ethnic- or population-specific differences. Given the results of the association between rs2032582 and steroid resistance of INS were not sufficiently robust to withstand correction for multiple comparisons, further studies are required to verify whether there is a true association. Our findings from haplotype analysis indicated that the T-G-C haplotype was associated with an increased risk of SR with the contrasting finding that the wild-type C-G-C haplotype was associated with decreased SR risk. Our findings validated previous findings of an association between rs1128503 (C1236T) and an increased risk of SR; however, considering adjustments from correcting for multiple comparisons, the limited number of studies evaluated, and heterogeneity, the result should be considered with caution.
Certain limitations should be considered with the interpretation of the findings of our meta-analysis. First, all participants were children. Because an evaluation of these SNPs in MDR1 in adults with INS has never been reported, the potential effect in adults is not known. Second, heterogeneity was significant in most comparisons of rs1045642 and rs2032582 for susceptibility analysis and subgroup analysis did not account for all the observed effects. These differences may be attributed to different genetic backgrounds, environments, and lifestyles. Third, homogeneity of baseline characteristics was poor when comparing SR to SS in some studies. Overall, well-designed studies including more ethnic groups and larger simple sizes are needed to validate the results of the current meta-analysis.
In conclusion, the present study found no evidence of an association between the MDR1 polymorphisms, rs1128503, rs1045642, and rs2032582, with susceptibility to INS in children. However, we found robust evidence that the rs1128503 polymorphism is associated with an increased risk of SR in Asian and Caucasian cohorts, and that the rs2032582 polymorphism may be associated with an increased risk of SR in Caucasian children. The findings from our meta-analysis may provide predictive genetic markers to determine glucocorticoid resistance in patients with INS.
Supplementary Material
Footnotes
Abbreviations: ABCB1 = ATP binding cassette subfamily B member 1, BON = Bonferroni–Holm correction, CIs = confidence intervals, FDR = Benjamini–Hochberg false discovery rate, GCs = glucocorticoids, HWE = Hardy–Weinberg equilibrium, INS = idiopathic nephrotic syndrome, ISKDC = International Study of Kidney Disease in Children, MDR1 = multidrug resistance protein 1, NOS = Newcastle–Ottawa Scale, NS = nephrotic syndrome, ORs = odds ratios, PCR-RFLP = polymerase chain reaction-restriction fragment length polymorphism, P-gp = P-glycoprotein, PRISMA = preferred reporting items for systematic reviews and meta-analyses, SNP = single nucleotide polymorphism, SR = steroid resistant, SS = steroid sensitive.
This study was supported by a grant from the National Natural Science Foundation of China (No. 81403361).
The authors report no conflicts of interest.
Supplemental digital content is available for this article.
References
- [1].Rheault MN. Nephrotic syndrome: updates and approaches to treatment. Curr Treat Options Peds 2016;2:94–103. [Google Scholar]
- [2].Lombel RM, Gipson DS, Hodson EM. Treatment of steroid-sensitive nephrotic syndrome: new guidelines from KDIGO. Pediatr Nephrol 2013;28:415–26. [DOI] [PubMed] [Google Scholar]
- [3].Teeninga N, Kistvan Holthe JE, Van Rijswijk N, et al. Extending prednisolone treatment does not reduce relapses in childhood nephrotic syndrome. J Am Soc Nephrol 2013;24:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Beins NT, Dell KM. Long-term outcomes in children with steroid-resistant nephrotic syndrome treated with calcineurin inhibitors. Front Pediatr 2015;27:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cil O, Besbas N, Duzova A, et al. Genetic abnormalities and prognosis in patients with congenital and infantile nephrotic syndrome. Pediatr Nephrol 2015;30:1279–87. [DOI] [PubMed] [Google Scholar]
- [6].Lovric S, Ashraf S, Tan W, et al. Genetic testing in steroid-resistant nephrotic syndrome: when and how. Nephrol Dial Transplant 2016;31:1802–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Becherucci F, Mazzinghi B, Provenzano A, et al. Lessons from genetics: is it time to revise the therapeutic approach to children with steroid-resistant nephrotic syndrome? J Nephrol 2016;29:543–50. [DOI] [PubMed] [Google Scholar]
- [8].Swierczewska M, Ostalska-Nowicka D, Kempisty B, et al. Molecular basis of mechanisms of steroid resistance in children with nephrotic syndrome. Acta Biochim Pol 2013;60:339–44. [PubMed] [Google Scholar]
- [9].Badr HS, El-Hawy MA, Helwa MA. P-glycoprotein activity in steroid-responsive vs. steroid-resistant nephrotic syndrome. Indian J Pediatr 2016;83:1222–6. [DOI] [PubMed] [Google Scholar]
- [10].Wasilewska A, Zoch-Zwierz W, Pietruczuk M, et al. Expression of P-glycoprotein in lymphocytes from children with nephrotic syndrome, depending on their steroid response. Pediatr Nephrol 2006;21:1274–80. [DOI] [PubMed] [Google Scholar]
- [11].Stachowski J, Zanker CB, Runowski D, et al. Resistance to therapy in primary nephrotic syndrome: effect of MDR1 gene activity [Polish]. Pol Merkur Lekarski 2000;8:218–21. [PubMed] [Google Scholar]
- [12].Youssef DM, Elbehidy RM, Abdelhalim HS, et al. Soluble interleukine-2 receptor and MDR1 gene expression levels as inflammatory biomarkers for prediction of steroid response in children with nephrotic syndrome. Iran J Kidney Dis 2011;5:154–61. [PubMed] [Google Scholar]
- [13].Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci USA 2000;97:3473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Youssef DM, Attia TA, El-Shal AS, et al. Multi-drug resistance-1 gene polymorphisms in nephrotic syndrome: impact on susceptibility and response to steroids. Gene 2013;530:201–7. [DOI] [PubMed] [Google Scholar]
- [15].Cizmarikova M, Podracka L, Klimcakova L, et al. MDR1 polymorphisms and idiopathic nephrotic syndrome in Slovak children: preliminary results. Med Sci Monit 2015;21:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jafar T, Prasad N, Agarwal V, et al. MDR-1 gene polymorphisms in steroid-responsive versus steroid-resistant nephrotic syndrome in children. Nephrol Dial Transplant 2011;26:3968–74. [DOI] [PubMed] [Google Scholar]
- [17].Chiou YH, Wang LY, Wang TH, et al. Genetic polymorphisms influence the steroid treatment of children with idiopathic nephrotic syndrome. Pediatr Nephrol 2012;27:1511–7. [DOI] [PubMed] [Google Scholar]
- [18].Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].The primary nephrotic syndrome in children. Identification of patients with minimal change nephrotic syndrome from initial response to prednisone. A report of the International Study of Kidney Disease in Children. J Pediatr 1981;98:561–4. [DOI] [PubMed] [Google Scholar]
- [20].Kodner C. Diagnosis and management of nephrotic syndrome in adults. Am Fam Physician 2016;93:479–85. [PubMed] [Google Scholar]
- [21].Hogan J, Radhakrishnan J. The treatment of minimal change disease in adults. J Am Soc Nephrol 2013;24:702–11. [DOI] [PubMed] [Google Scholar]
- [22].Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Available at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.htm. Accessed October 17, 2016. [Google Scholar]
- [23].Dou H, Ma E, Yin L, et al. The association between gene polymorphism of TCF7L2 and type 2 diabetes in Chinese Han population: a meta-analysis. PLoS One 2013;8:e59495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang F, Ma YL, Zhang P, et al. A genetic variant in microRNA-196a2 is associated with increased cancer risk: a meta-analysis. Mol Biol Rep 2012;39:269–75. [DOI] [PubMed] [Google Scholar]
- [25].Holm S. A simple sequentially rejective multiple test procedure. Scan J Stat 1979;6:65–70. [Google Scholar]
- [26].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 1995;57:289–300. [Google Scholar]
- [27].Zintzaras E, Ioannidis JP. Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol 2005;28:123–37. [DOI] [PubMed] [Google Scholar]
- [28].DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177–88. [DOI] [PubMed] [Google Scholar]
- [29].Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50:1088–101. [PubMed] [Google Scholar]
- [30].Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wasilewska A, Zalewski G, Chyczewski L, et al. MDR-1 gene polymorphisms and clinical course of steroid-responsive nephrotic syndrome in children. Pediatr Nephrol 2007;22:44–51. [DOI] [PubMed] [Google Scholar]
- [32].Suvanto M, Jahnukainen T, Kestilä M, et al. Single nucleotide polymorphisms in pediatric idiopathic nephrotic syndrome. Int J Nephrol 2016;2016:1417456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Safan MA, Elhelbawy NG, Midan DA, et al. ABCB1 polymorphisms and steroid treatment in children with idiopathic nephrotic syndrome. Br J Biomed Sci 2016;10:1–6. [DOI] [PubMed] [Google Scholar]
- [34].Dhandapani MC, Venkatesan V, Rengaswamy NB, et al. Association of ACE and MDR1 gene polymorphisms with steroid resistance in children with idiopathic nephrotic syndrome. Genet Test Mol Biomarkers 2015;19:454–6. [DOI] [PubMed] [Google Scholar]
- [35].Choi HJ, Cho HY, Ro H, et al. Polymorphisms of the MDR1 and MIF genes in children with nephrotic syndrome. Pediatr Nephrol 2011;26:1981–8. [DOI] [PubMed] [Google Scholar]
- [36].Fung KL, Pan J, Ohnuma S, et al. MDR1 synonymous polymorphisms alter transporter specificity and protein stability in a stable epithelial monolayer. Cancer Res 2014;74:598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Marzolini C, Paus E, Buclin T, et al. Polymorphisms in human MDR1 (P-glycoprotein): recent advances and clinical relevance. Clin Pharmacol Ther 2004;75:13–33. [DOI] [PubMed] [Google Scholar]
- [38].Brambila-Tapia AJ. MDR1 (ABCB1) polymorphisms: functional effects and clinical implications. Rev Invest Clin 2013;65:445–54. [PubMed] [Google Scholar]
- [39].Wolking S, Schaeffeler E, Lerche H, et al. Impact of genetic polymorphisms of ABCB1 (MDR1, P-Glycoprotein) on drug disposition and potential clinical implications: update of the literature. Clin Pharmacokinet 2015;54:709–35. [DOI] [PubMed] [Google Scholar]
- [40].Wasilewska AM, Zoch-Zwierz WM, Pietruczuk M. Expression of P-glycoprotein in lymphocytes of children with nephrotic syndrome treated with glucocorticoids. Eur J Pediatr 2006;165:839–44. [DOI] [PubMed] [Google Scholar]
- [41].Funaki S, Takahashi S, Wada N, et al. Multiple drug-resistant gene 1 in children with steroid-sensitive nephrotic syndrome. Pediatr Int 2008;50:159–61. [DOI] [PubMed] [Google Scholar]
- [42].Kimchi-Sarfaty C, Oh JM, Kim IW, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 2007;315:525–8. [DOI] [PubMed] [Google Scholar]
- [43].Komar AA. Silent SNPs: impact on gene function and phenotype. Pharmacogenomics 2007;8:1075–80. [DOI] [PubMed] [Google Scholar]
- [44].Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, et al. Silent polymorphisms speak: how they affect pharmacogenomics and the treatment of cancer. Cancer Res 2007;60:9609–12. [DOI] [PubMed] [Google Scholar]
- [45].Kim RB, Leake BF, Choo EF, et al. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther 2001;70:189–99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


