Abstract
The aim of the study was to detect the infection level of honey bees with Nosema apis and/or Nosema ceranae using microscopic and molecular analysis from indigenous honeybee race of eight Saudi Arabian geographical regions. A detailed survey was conducted and fifty apiaries were chosen at random from these locations. Infection level was determined both by microscope and Multiplex-PCR and data were analyzed using bioinformatics tools and phylogenetic analysis. Result showed that N. ceranae was the only species infecting indigenous honeybee colonies in Saudi Arabia. As determined by microscope, Nosema spores were found to be in 20.59% of total samples colonies, while 58% of the samples evaluated by PCR were found to be positive for N. ceranae, with the highest prevalence in Al-Bahah, a tropical wet and dry climatic region, whereas low prevalence was found in the regions with hot arid climate. Honeybees from all eight locations surveyed were positive for N. ceranae. This is the first report about the N. ceranae detection, contamination level and distribution pattern in Saudi Arabia.
Keywords: Nosema ceranae, Prevalence, Molecular detection, Honeybee: Saudi Arabia
1. Introduction
Western honey bees (Apis mellifera) are a highly valued resource worldwide and are of great relevance for humans and the entire ecosystem, not only as a honey and wax producer but also as a pollinator of agricultural and horticultural crops and wild flora (vanEngelsdorp and Meixner, 2010). The total annual global economic worth of pollination amounts to 212 billion USD, representing 9.5% of the value of the global agricultural production (Gallai et al., 2009). However, very unfortunately, honeybee is facing enormous threat worldwide (USA, Europe, Middle East) (Crailsheim et al., 2009, vanEngelsdorp et al., 2009, vanEngelsdorp et al., 2010, Haddad et al., 2009, Soroker et al., 2009) including Saudi Arabia (Alattal and AlGhamdi, 2015).
Beekeeping is one of the long-standing practices in rural Saudi Arabia and is one of the most important economic activities for the communities (Al-Ghamdi and Nuru, 2013). Approximately 5000 beekeepers maintain more than one million honeybee colonies and produce approximately 9000 metric tons of honey annually (Al-Ghamdi, 2007). Apis mellifera jemenitica Ruttner (= yemenitica auctorum: vide Engel, 1999), the smallest race of A. mellifera, is the only race of A. mellifera naturally found in the country and has been used in apiculture at least 2000 BC. Traditional beekeeping is mostly practiced using this race, because it is well adapted to the semi-arid to semi-desert conditions of Saudi Arabia (Alqarni et al., 2011, Al-Ghamdi and Nuru, 2013). Also, honey produced by this native bee (A. m. jemenitica) is sold at 10–20 times high rates than imported honeys (Al-Ghamdi personal Comm.)
Despite the great potential and multiple opportunities for beekeeping in Saudi Arabia, the bee-keeping industry is steadily growing in the country with different opportunities and, of course, many challenge. The major challenge is occurrence and distribution of honeybee disease in the country (Al-Ghamdi, 1990, Al-Ghamdi, 2010, Alattal and AlGhamdi, 2015, Ansari et al., 2016, Ansari et al., 2017).
A mysterious decline in honeybee colonies has gained worldwide attention, including in Saudi Arabia. In the last decades, significant losses have been observed in indigenous honeybee colonies in Saudi Arabia (Alattal and AlGhamdi, 2015). Much attention has been given to Colony Collapse Disorder (CCD), which is a syndrome specifically defined as a dead colony with no adult bees and with no dead bee bodies but with a live queen, and usually honey and immature bees, still presents (vanEngelsdorp et al., 2009). Several causes of these large-scale losses have been reported, including honey bee parasites (Varroa destructor, Acarapis woodi); pathogens (Nosema spp. and bee viruses); pesticides, harsh environment, use of antibiotics, poor nutrition, and migratory beekeeping practices (Kevan et al., 2007, Higes et al., 2008, Naug, 2009, vanEngelsdorp et al., 2009, Bacandritsos et al., 2010, vanEngelsdorp and Meixner, 2010; Alattal and AlGhamdi, 2015).
Nosemosis is a fungal infection of honey bees caused by either Nosema apis or N. ceranae. N. apis was the historic species infecting European honey bees (Matheson, 1996). N. ceranae was previously isolated in naturally infected Apis cerana worker bees in China (Fries et al., 1996) and later has been described infecting Apis mellifera in Europe (Higes et al., 2006). Currently, this parasite is widespread all over the world and has shown the capacity of infection in other Hymenoptera different from the honeybees (Plischuk et al., 2009), and it is now a common infection of European honeybees and is highly virulent to its new host (Chen et al., 2009a). This is problematic for beekeepers because N. ceranae has a different seasonal phenology than N. apis, causing more significant problems for beekeepers in summer months and in warm climates (Bourgeois et al., 2010). Both Nosema spp. can co-infect honey bees (Chen et al., 2009b; Paxton et al., 2007, Forsgren and Fries, 2010). Although co-infections occur, N. ceranae has become the predominant species in many regions (Chen et al., 2009a, Klee et al., 2007, Williams et al., 2008, Razmaraii et al., 2013, Haddad, 2014).
Both Nosema spp. invade the midgut epithelial cells of adult honeybees (A. mellifera), i.e. worker bees, drones, and queens, and caused honey bee disease known as nosemiasis or nosemosis (Fries, 1988, Fries, 2010, Higes et al., 2007). This disease negatively affects productivity and survival of honeybee colonies, adult bee longevity, queen bees, brood rearing, bee biochemistry, pollen collection, and other bee behaviors (Botías et al., 2013, Huang, 2012). In contrast to the clinical symptoms of N. apis, such as crawling bees and dysentery (Liu, 1988), infection with N. ceranae is symptomless apart from reports of a massive colony depopulation and collapse (Huang et al., 2007, Paxton, 2010), reduced honey production (Higes et al., 2008). The impact of N. ceranae infection on colony survival is unclear and has been found in both healthy colonies (vanEngelsdorp et al., 2009, Cox-Foster et al., 2007, Gisder et al., 2010) and those undergoing sudden collapses (Higes et al., 2008, Higes et al., 2009; Bacandritsos et al., 2010, Martín-Hernández et al., 2007). Thus, investigation of Nosema species seems important.
Routine optical microscopy assessment can confirm infection with Nosema species, but it is impossible to distinguish between the species because the spores of the two Nosema species are very similar and can hardly be distinguished by light microscopy, so that in the absence of clear morphological characteristics for species recognition, other techniques using molecular markers may greatly assist in the diagnosis and identification of honeybee microsporidians Thus, it is necessary to use molecular diagnostic tools and identification methods (Gajger et al., 2010). The PCR technique provides a very sensitive test for detecting microsporidian infection because it enables detection of the parasite even at very low levels of infection.
In some bordered countries of Saudi Arabia (Egypt, Israel, Jordan, Iraq and Iran), Nosema infection in honeybee colonies has been reported previously (Alzubaidy and Ali, 1994, El-Shemy et al., 2012, Nabian et al., 2011, Razmaraii et al., 2013, Aroee et al., 2016, Soroker et al., 2009). However, even though some preliminary studies have been conducted on Nosemosis in honeybee in Saudi Arabia (Al-Ghamdi, 1990; Alattal and AlGhamdi, 2015; Abdel-Baki et al., 2016). Recently, in Riyadh region of Saudi Arabia, Nosemosis has been recognized by the presence of Nosema spores through light microscopy, assuming N. apis to be the causal agent (Abdel-Baki et al., 2016). These findings have led to a demand for PCR based research that determine, which species of Nosema have been present in A. m. jemenitica, the indigenous honeybee race of Saudi Arabia in a recent past using detailed survey and molecular characterization.
2. Materials and methodology
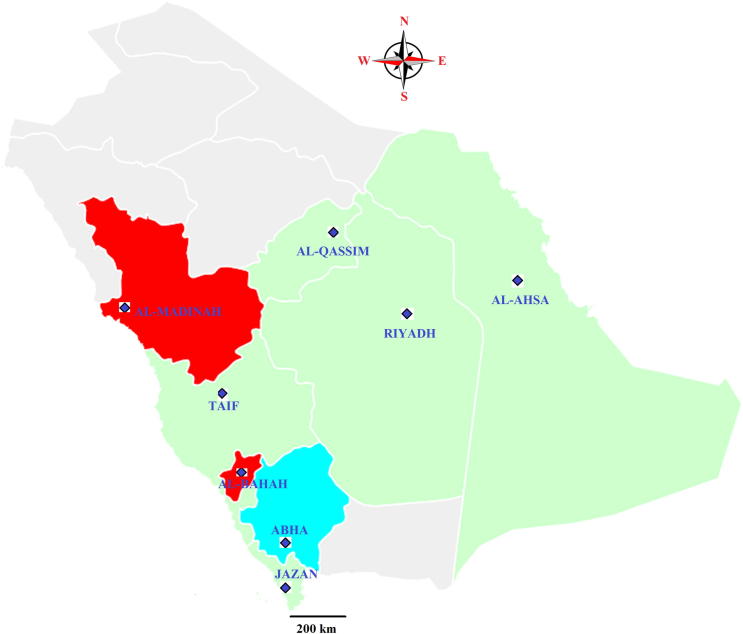
The presence of nosemosis in honeybee colonies was investigated in different beekeeping locations and eco-regions, during the spring season (March to April 2015), the active season for honeybees in Saudi Arabia. Eight different geographical localities, where beekeeping is common were included in this survey (Fig. 1): Al-Ahsa (25° 25′ 46″ N, 49° 37′ 19″ E), Abha (18° 13′ 24″ N, 42° 30′ 26″ E), Jazan (16° 53′ 21″ N, 42° 33′ 40″ E), Taif (21° 16′ 0″ N, 40° 25′ 0″ E), Al-Madinah (24° 28′ 0″ N, 39° 36′ 0″ E), Al-Bahah (20° 0′ 0″ N, 41° 30′ 0″ E), Al-Qassim (25° 49′ 19.72″ N, 42° 50′ 6.85″ E) and Riyadh (24°43′19.2″N 46°37′37.2″E). Total 50 random apiaries were visited in all locations, and 10 colonies in each apiary were inspected.
Fig. 1.
Sampling locations in Saudi Arabia with different eco-regions, from which A. mellifera were collected and tested for N. apis and N. ceranae (a) Red color: Tropical wet and dry or Tropical savanna (Aw) climate (b) Sky blue color: Cold semi-arid (BSk) climate (c) Green color: Hot arid (BWh) climate.
2.1. Sampling
A total of fifty seemingly healthy apiaries of A. m. jemenitica, indigenous race of Saudi Arabia, owned by different beekeepers were randomly selected following stratified randomization procedures (Moher et al., 2010). Samples were collected from local (A. m. jemenitica) bee races only (10 hives from each apiary) from March to April (major nectar flow period in Saudi Arabia) during the year 2015. The samples were collected from eight major beekeeping regions of Saudi Arabia, based on, the beekeeping management schedule and the categorization of geographical regions, For instance, Riyadh, Al-Qassim, Al-Ahsa, Taif and Jazan (Hot arid climate region), Al-Madinah and Al-Bahah (Tropical wet and dry climate), and Abha (Cold semi-arid climate). The sampling hives had not been treated against Nosema disease for at least 6 months. In each hive, approximately 100 worker bees were collected from outer honey frames of the brood chamber, placed in falcon tube containing preservative buffer (RNA Later®), transported to the laboratory of the Bee Research Unit (BRU) at the Department of Plant Protection of the Faculty of Food and Agriculture Sciences at King Saud University and stored at −20 °C until analyzed.
2.2. Data collection
The data collected from the survey included the following information for each inspected apiary: the date of inspection, the apiary location (to facilitate repeat visits), the name of the owner, the hive type (local or modern), the honeybee race (indigenous or imported), the number of honeybee colonies and the colonies having some unusual symptoms.
2.3. Microscopic examination of spores
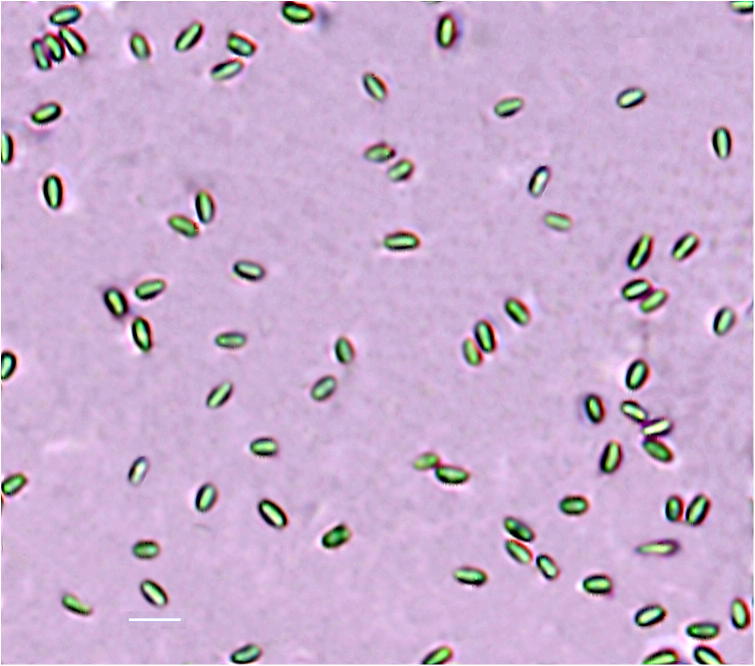
Samples were initially analyzed by phase contrast microscopy for presence or apparent absence of Nosema spp. spores. For each sample, the abdomens of 30 adult bees were macerated in 30 mLof ddH2O, the suspension was filtered and centrifuged for 5000 rpm for 10 min and the homogenate examined under a Phase Contrast Microscope (Olympus BX51, model BX51TF, Japan, equipped with an Olympus DP71 camera (Olympus, Japan) at 400 × magnification (Fig.2B), and photographed (OIE, 2008). Measurements are presented in micrometers and data are expressed as the mean followed by the range in parentheses. As morphological characteristics of N. ceranae and N. apis spores are similar and can hardly be distinguished by optical microscopy, all samples were also screened by multiplex 96 polymerase chain reaction (M-PCR) assay based on 16S rRNA-gene-targeted species specific primers to distinguish between N. ceranae and N. apis. Positive samples were also used for further molecular diagnosis as discussed below.
Fig. 2.
Spores of Nosema ceranae have an oval shape with a dark border under Phase contrast microscope at 400X magnification. Scalebar = 10 μm.
2.4. Genomic DNA isolation
For DNA extraction, the abdomen of the 30 individual honeybees was cleanly cut off with scissors, immediately put into a vial and homogenized in 100 μl Krebs Ringer solution pH 7.4 (123 mM NaCl, 1.3 mM CaCl2, 5 mM KCl, 100 mM HEPES, 5 mM D-glucose, 1.5% bovine serum albumin free fatty acid) with a pestle. DNA was extracted from pools using a CTAB buffer (100 mM Tris HCl, (pH 8.0); 20 mM EDTA, (pH 8.0); 1.4 M NaCl; 2% (w/v) cetyltrimethylammonium bromide; 0.2% (v/v) 2-mercaptoethanol) plus proteinase K, overnight incubation at 56 °C followed by a phenol/chloroform/isoamyl alcohol (25:24:1) extraction. and precipitated overnight at −20 °C with isopropanol, recovered by means of centrifugation, resuspended in TE 1×, and treated with RNase (100 μg/ml). The DNA (3–5 μg) was loaded onto 1.2% agarose gels, which were stained with ethidium bromide after migration.
2.5. Multiplex PCR (M-PCR) amplification using N. apis and N. ceranae-specific primers
Each DNA sample was subjected to molecular identification by PCR assay using the N. apis-specific primers (321APIS, FOR 5′-GGGGGCATGTCTTTGACGTACTATGTA-3; 321APIS REV 5′-GGGGGGCGTTTAAAATGTGAAACAACTATG-3′) and N. ceranae-specific primer (218MITOC FOR 5′-CGGCGACGATGTGATATGAAA-ATATTAA-3′; 218MITOC REV 5′-CCCGGTCATTCTCAAACAAAA-AACCG-3′) described by Martin-Hernandez et al. (2007). The PCR reactions were performed in a total volume of 50 μL containing 5 μl 10x PCR buffer (100 mM Tris–HCl, pH 8.3, 500 mM KCl, 4 mM MgCl2, 1% Triton X-100), 200 μM of each deoxynucleotide triphosphates, 2U Taq DNA polymerase enzyme (Promega, USA), 100 ng of each primer and 10 ng target DNA were added. The surface of the mixture was covered with 100 μl mineral oil. The following PCR conditions were used: initial denaturation at 95 °C (2 min), and 35 cycles of (95 °C for 30 s, 55 °C for 30 s and 72 °C for 60 s) and a final extension cycle at 72 °C (5 min) ended the PCR. Negative controls (from DNA extraction) were included in all PCR experiments. (OIE Terrestrial manual, 2008; Martin-hernandez et al., 2007). Samples of the amplicons were electrophoresed in 1.5% agarose gel. Approximate product size was determined using the 100-bp molecular size marker (Promega, USA). The PCR product was visualized and photographed using a Gel Doc EZ system (Bio-Rad, USA) with a band at 321 bp for N. apis and at 218-219 for N. ceranae.
3. 16S rRNA gene sequencing and phylogenetic analysis
PCR products were purified using GenElute PCR Clean-Up Kit (Sigma–Aldrich, India) and send to BGI Genomics Co., Ltd (Hong Kong, China) for both end sanger sequencing. The sequences obtained were manually edited using Sequencher 4.5 (Gene Codes Corp.) and were aligned using the Bioedit sequence editor software version 7.0.5.3. These sequences have been submitted to GenBank (accession number KY022481 and KY022482 for ksuNC4 and ksuNC6 respectively). Partial 16S rRNA gene sequences of the isolates were compared with 16S rRNA gene sequences available by the BLAST search (Altschul et al., 1990), in the National Centre for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/). Multiple sequence alignments were performed using CLUSTALW version 1.8 (Thompson et al., 1994). Phylogenetic tree was constructed by the neighbor-joining method (Saitou and Nei, 1987), and the reliability of the tree topology was evaluated by bootstrap analysis using MEGA 6.06 software (Tamura et al., 2013).
3.1. Data analysis
The corresponding 95% confidence intervals (95% CI) were calculated and differences among prevalence values were compared by Fisher’s exact test. P values <0.05 were considered significant. The Cohen's Kappa coefficient was used as a measure of agreement between microscopy and M-PCR. The following ranges were considered for interpretation of the Cohen's Kappa coefficient: poor agreement = less than 0.00, slight agreement = 0.00–0.20, fair agreement = 0.21–040, moderate agreement = 0.41–0.60, substantial agreement = 0.61–0.80, and almost perfect agreement = 0.80–1.00.
3.2. Statistical analysis
The prevalence of Nosema spp. contamination levels in honeybees from different geographical regions of Saudi Arabia was calculated by descriptive statistics and a confidence interval (CI) of 95%.
4. Results
4.1. Field survey
During survey presence of disease agents and parasite was monitored and results showed that Varroa mite is frequently infesting honeybee colonies in all the sampled locations. Presence of Nosema was recorded in all locations (Table 1). Majority of infection have been reported in Al-Bahah, Abha and Taif region of Saudi Arabia. These are the three major beekeeping regions in Saudi Arabia and most of the honey has been produced by these three areas. In comparison to Riyadh, Al-Qassim and Al-Ahsa, these three regions have high rainfall and moderate temperature (Table 1).
Table 1.
Prevalence and estimated contamination of N. ceranae in various eco-regions of Saudi Arabia based on altitude, average temperature, rainfall, relative humidity, climate, monitoring time.
| Sampling Area | Altitude (m) | Average Temperature (°C) |
Average rainfall (mm) |
Average Relative Humidity |
No. of inspected apiaries/ samples collections | Microscopic Positive (%) | PCR Positive (%) | No. of infected Apiaries | Climate* | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| March | April | March | April | March | April | |||||||
| Riyadh | 612 | 21 | 23 | 23 | 25 | 32 | 28 | 6/60 | 12 (20) | 19 (31.66) | 3 | Hot arid(BWh) climate |
| Al-Qassim | 753 | 17 | 22 | 26.7 | 28.8 | 35 | 33 | 4/40 | 09 (18.36) | 13 (32.5) | 2 | Hot arid(BWh) climate |
| Al-Ahsa | 179 | 20.9 | 25.8 | 16.2 | 10.7 | 44 | 38 | 2/20 | 3 (15) | 6 (30) | 1 | Hot arid(BWh) climate |
| Al-Madinah | 608 | 23.6 | 26.8 | 9.8 | 9.6 | 25 | 22 | 05/50 | 13 (26) | 17 (34) | 2 | Tropical wet and dry or Tropical savanna (Aw) climate |
| Taif | 1879 | 20 | 21.8 | 15.1 | 35.7 | 47 | 47 | 10/100 | 19 (19) | 22 (22) | 5 | Hot arid (BWh)climate |
| Al-Bahah | 2150 | 16.5 | 18.5 | 16.5 | 36.3 | 46 | 45 | 10/100 | 31 (31) | 39 (39) | 8 | Tropical wet and dry or Tropical savanna (Aw) climate |
| Abha | 2270 | 15.5 | 17.5 | 47.2 | 47.9 | 62 | 60 | 07/70 | 19 (27.14) | 25 (35.71) | 5 | Cold semi-arid (BSk) climate |
| Jazan | 5 | 28 | 30 | 5.7 | 14.6 | 66 | 64 | 6/60 | 07 (12.06) | 09 (15.51) | 3 | Hot arid(BWh) climate |
Köppen-Geiger climate classification.
4.2. Microscopic examination
Spores of Nosema spp. were detected at 400x magnification under Phase contrast microscope (Fig. 1B). Phase contrast microscopy of the midgut content revealed the presence of large numbers of Nosema spp. spores. Nosema spores were oval shaped, measuring 3.0–4.0 μm in width and 5.0–7.0 μm in length (n = 30) (Fig. 2). Nosema infection has been found in all locations of sample collections. As determined by microscope, Nosema spp. were found in 20.59% of total sampled colonies. In hot arid climatic eco-regions like Riyadh (20%), Al-Qassim (18.36%), Al-Ahsa (15%), Taif (19%) and Jazan (12.06%) of samples were found microscopically positive. Conversely, in Tropical savanna climate region like Al-Madinah (26%), Al-Bahah (31%) and Cold semi-arid climatic region, Abha (27.14%) infection was more than hot arid climatic sampling locations. A significant difference in the infection level was found in hot arid climatic eco-regions and Tropical savanna climate region together with cold semi-arid climatic region (P < 0.001) (Table 1).
4.3. Molecular characterization of Nosema spp
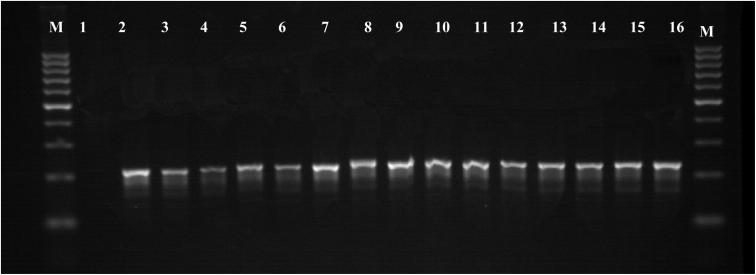
There is no previous record of presence of Nosema spp. using molecular identification in Saudi Arabia. Therefore, we initiate to confirm the Nosema spp. identification by M-PCR and DNA sequencing by collecting samples from geographically distinct locations throughout Saudi Arabia. Based on the PCR analysis, Nosema infections were detected in apiaries from all the eight beekeeping regions examined. Detection by PCR using N. apis and N. ceranae specific primers found that 29 out of 50 apiaries (58%) were positive for N. ceranae (Table 1). But no apiaries were found to be positive for N. apis (Fig. 3). Infection was found in the samples collected from Tropical savanna climatic regions and cold semi-arid climatic region in comparison to hot arid climatic region. Overall, a total of 29 out of 50 (29%, 95% CI: 41.96–58.04%) apiaries tested positive for Nosema infection by microscopy and M-PCR. Therefore, the Cohen's Kappa coefficient for the association between results of microscopy and results of M-PCR was 1, indicating that there was a perfect level of agreement between the two diagnostic methods in all the bee samples. In hot arid climatic eco-regions, 31.6%, 32.5%, 30%, 22% and 15.51% from Riyadh, Al-Qassim, Al-Ahsa, Taif, and Jazan respectively were found to be PCR positive. Conversely, in Tropical savanna climate region, 34% (Al-Madinah), 39% (Al-Bahah) and Cold semi-arid climatic region 35.71% (Abha) were PCR positive (Table 1).
Fig. 3.
Agarose gel showing amplification of the part of small subunit of ribosomal RNA by using primer 218MITOC and 321APIS. Lane M: DNA Marker 100 bp ladder: Lane 2: No amplification in negative control; Lane 1–14: Honeybee samples positive for N. ceranae; Lane 15: PCR product of positive control of 218 bp for N. ceranae.
4.4. BLAST and phylogenetic analysis
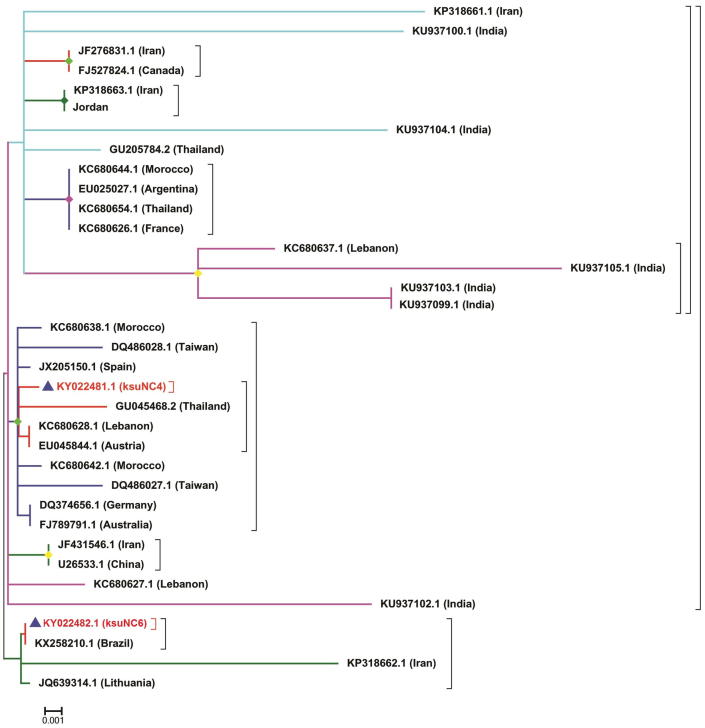
Similar to that observed in other countries (Medici et al., 2012), we observed some intraspecific variations in the 16S SSU of N. ceranae in Saudi Arabia. Comparative analysis of 16S rRNA gene sequences of ksuNC4 and ksuNC6 isolates showed 217/218 (99%) sequence identity. Over the entire sequence range analyzed (218 bp), only one position (18th bp) was polymorphic with one gap. The BLAST search of these sequences against GenBank Nucleotide database, the highest similarity (99%) was found with N. ceranae 16S rRNA suquences. A Nucleotide BLAST search showed that the DNA sequence obtained from the ksuNC4 (218 bp) isolate showed a 100% sequence identity with the 16S rDNA of some N. ceranae isolates (gb| KC680636.1, gb| KC680629.1 and gb| DQ329034.1). The first two close hit of ksuNC4 closely related to the DNA sequence of N. ceranae isolated from honeybee samples from Lebanon (Roudel et al., 2013), and the third close hit belongs to the sequence of N. ceranae isolated in Spain (Higes et al., 2007). Similarly, the DNA sequence obtained from the ksuNC6 isolate (417 bp) showed a 100% sequence identity with the 16S rDNA of other N. ceranae isolates (gb| KC680641.1, gb| KC680642.1 gb| KC680637.1 and gb| JF431546.1). First two closely related hits of ksuNC6 belongs to N. ceranae isolates from Moroccon Honeybee, third one from Lebanon (Roudel et al., 2013), and the forth close hit belongs to the sequence of N. ceranae isolated in Iran (Nabian et al., 2011). This indicated that Lebanon (northern bordered country of Saud Arabia) and Iran (eastern bordered country) the close neighbors of Saudi Arabia have same type of genotype as found in Saudi Arabia.
The evolutionary relationship between the two isolates and previously reported isolates were constructed using MEGA6.06 software (Tamura et al., 2013). The results illustrate the degree of evolutionary relatedness between the two Saudi Arabian isolates and other previously reported isolates (Fig. 4). From our study, the ksuNC4 isolate showed greater relatedness to three previously reported N. ceranae isolates {gb| KC680629.1 (Lebanon), gb| KC680636.1 (Lebanon) and gb| DQ329234.1 (Spain)}, while isolate ksuNC6 formed a separate clade by itself with gb| KC680637.1 (Lebanon), gb| JF431546.1 (Iran) and gb| KC680642.1 (Morocco). This indicates that the genotypes of the two isolates differ. The ksuNC4 genotype was isolated from the apiary located in Al-Bahah and the isolate ksuNC4 was isolated from Jazan, both are two different geographical regions of Saudi Arabia.
Fig. 4.
Neighbor-joining phylogenetic tree of the two N. ceranae isolates (ksuNC4 and ksuNC6) based on 16S rRNA gene sequence comparisons and closest NCBI (BLASTn) strains based on the 16S rRNA gene sequences (neighbor-joining tree method). The scale bar indicates 0.001 nucleotide substitutions per nucleotide position. The numbers at node show the bootstrap values obtained with 500 resampling analyses.
5. Discussion
During the last decade, an increase infection by Nosema parasite in the honeybee (Emsen et al., 2016) followed by increased numbers of honeybee colony death and decreased honey production has been reported worldwide. One of the main reasons that might explain these problems is the recent introduction of N. ceranae in honeybee in Europe (Higes et al., 2006) and appeared to be highly virulent (Paxton et al., 2007, Higes et al., 2007). Environmental conditions also strongly influence many parasitic relationships and, regardless of the effects of altitude, flora and colony management, in warm countries like Spain the influence of temperature on the consequences of N. ceranae has been observed (Martín-Hernández et al., 2012).
The prevalence of nosemosis disease has been proven to vary among regions and years (Mulholland et al., 2012). Although, N. apis has a world-wide distribution, it is not considered an important problem in tropical and sub-tropical regions (Wilson and Nunamaker, 1983). However, in temperate regions N. apis infections typically peak in the spring, decrease during the summer and then increase again in the fall before declining during the early winter months (Pickard and El-Shemy, 1989). N. ceranae was the most prevalent microsporidia found in A. mellifera in hotter regions (Mediterranean regions) and is reduced in colder climate (Fries, 2010, Gisder et al., 2010). Since N. ceranae infection appears to be more common in warmer climates and in specific geographical areas as N. ceranae spores are capable of surviving high temperature (60 °C) (Fenoy et al., 2009), this should be considered when importing bees from such areas (Fries, 2010).
In Sweden, 83% of colonies had N. apis only and 17% had both N. apis and N. ceranae (Fries, 2010). In Scotland, 70.4% of colonies revealed the presence of both N. ceranae and N. apis (Bollan et al., 2013). In east Azerbaijan province of Iran, an eastern bordered country of Saudi Arabia, 67.1% of colonies revealed the presence of N. ceranae (Razmaraii et al., 2013). Nabian et al. (2011) and Aroee et al. (2016), also reported the presence of N. ceranae in Iran. In Jordan, situated on northern border of Saudi Arabia, 23.9% of colonies found to be infected by N. ceranae and N. apis infection was not detected by PCR (Haddad, 2014). More evidences are available for prevalence of Nosema infection in some more cross bordered countries of Saudi Arabia, For instance, Iraq (Alzubaidy and Ali, 1994), Egypt (El-Shemy et al., 2012); Kuwait (OIE, 2004) and Oman (Matheson, 1993). Symptoms of nosemosis have been reported before among honey bees in Saudi Arabia (Al-Ghamdi, 1990; Matheson, 1993, Alattal and AlGhamdi, 2015). Recently, Abdel-Baki et al. (2016), observed the presence of Nosema spores using optical microscope from Honeybee in Riyadh region of Saudi Arabia.
The identification of Nosema in the Eight provinces surveyed in Saudi Arabia was expected, given that this Nosema species symptoms and microscopic examinations of spores has previously been reported in Saudi Arabia and cross border countries prevalence of Nosema infection. N. ceranae is recent fungal pathogen for Saudi Arabian honeybees as reported in this study. since Al-Ghamdi, 1990 and Abdel-Baki et al., 2016, suspects the symptoms of N. apis in honeybee of Saudi Arabia by morphological symptoms and microscopy. In this study, the high prevalence (58%) of N. ceranae together with the absence of N. apis infection in the present survey corroborates the findings of other authors that N. ceranae is definitely spread in Saudi Arabia and has basically replaced N. apis (Klee et al., 2007). High thermotolerance at 60 °C and 35 °C, resistance to desiccation, significant decrease in viability after freezing, and rapid degeneration of N. ceranae spores maintained at 4 °C were observed under experimental conditions (Fenoy et al., 2009). Therefore, it has been proposed that N. ceranae may be more prevalent in warmer climates (Fries, 2010) such as the typical middle east countries climate that occurs in the different study area. The present prevalence is close to values as high as 67% observed in Iran (Razmaraii et al., 2013) but considerably higher than, 23.9% and 49.2% as previously reported in Jordan (Haddad, 2014) and Thailand (Chupia et al., 2016), respectively. Different prevalence values reported in the literature may be due to differences in the number of apiaries examined, sampling methods, geographical areas, characteristics of honeybee population, diagnostic techniques, and other biotic and abiotic factors.
Based our findings, microscopy is still a valuable, relatively cheap and simple method to screen for the presence of Nosema infection in apiaries since a perfect agreement between microscopy and M-PCR was observed in the experiments. Unfortunately, very strong morphological similarities occur between N. apis and N. ceranae spores, resulting in a high risk of misdiagnosis. Both Nosema species spores are smooth and darkly outlined with elongated-elliptical shape and bright centre. N. apis spores end rounded and measure 6.0 μm in length and 3.0 μm in width. N. ceranae spores end sharper and measure 4.4 μm in length and 2.2 μm in width (Huang, 2012, Michalczyk et al., 2011). The main differences are noted with respect to the length of the polar filament, and they can be detected only under an electron microscope (Fries, 2010, Paxton, 2010).
In this scenario, molecular techniques such as M-PCR are needed for a reliable identification of Nosema to species level (Michalczyk et al., 2011). Indeed, the advent of new highly sensitive and specific molecular tools has played a key role for detection of N. ceranae in A. mellifera and for retrospective analyze of samples, showing that N. ceranae is not a new microsporidian agent in A. mellifera but it has infected this host during the last two decades (Guerrero-Molina et al., 2016). It is likely that the delay in a correct identification of N. ceranae in A. mellifera is attributable to the routine use of microscopy as a diagnostic technique for the identification of Nosema spores (Higes et al., 2010). Therefore, accurate identification of Nosema spores to species level by molecular tools should be especially useful for Saudi Arabian beekeepers.
A Nucleotide BLAST search showed that the DNA sequence obtained from the ksuNC4 (218 bp) isolate showed a 100% sequence identity with the with the other N. ceranae isolates from Lebanon (Roudel et al., 2013), whereas, ksuNC6 (217 bp) isolate showed a 100% sequence identity with N. ceranae isolates from Morocco and Lebanon. This indicated that Lebanon and Morocco, which are close neighbors of Saudi Arabia, have the same type of genotype as found in Saudi Arabia. It is unlikely that the primary infestation of N. ceranae in Saudi Arabia is due to human facilitated transportation of bee packages or bee products between both countries, but it seems that a third party is the main source of this type of infestation in both countries, which could be Egypt, where bee exportation is common to most of the African and middle east countries (Alattal and AlGhamdi, 2015).
Nosema spores are primarily spread to neighboring bees through fecal matter contaminating the environment (fecal-oral pathway) or, alternatively, they can also reach the crop and be regurgitated to other colony members during food exchange (oral-oral pathway) (Smith, 2012). Therefore, infections by both Nosema species can be transmitted among bees via ingestion of environmentally resistant mature spores from contaminated wax, combs, other hive interior surfaces, and water (OIE, 2013). Other potential routes of transmission include contamination of pollen, beekeeping material, and honey as well as cleaning activities and trophallaxis (Higes et al., 2010). Auto-infections can also occur (Higes et al., 2007). In our survey, we did not attempt to identify any source of infection. However, all these routes of spread and transmission of infective spores may have played a role in the presence of N. ceranae in the A. mellifera colonies investigated.
In contrast to nosemiasis caused by N. apis, N. ceranae affected bees that do not exhibit defecation near or inside the hive with evident dysentery but the main clinical symptom is dwindling, i.e. the progressive reduction in the number of bees in a colony with no apparent cause, until the point of collapse (Huang, 2012; Paxton, 2010). Sometimes dwindling may affect the whole apiary and other times only specific colonies may show symptoms. The disease sometimes occurs rapidly but may also occur over several months (vanEngelsdorp et al., 2009).
In a recent study in Argentina, an increasing gradient of infection and counts of Nosema spp. were observed from warmer to colder regions (Pacini et al., 2016). In accordance to these results of Pacini et al., 2016, in our study, maximum infection found in Al-Bahah (Tropical savanna (Aw) climate), Abha (Cold semi-arid (BSk) climate) and Al-Madinah (Tropical savanna (Aw) climate), whereas comparatively little infection has been observed in the samples collected from the apiaries situated in hot arid (BWh) climate (Riyadh, Al-Qassim, Al-Ahsa, Taif and Jazan) (Table 1). This uneven distribution pattern of Nosema spp. and infection level, may be caused by a diversity of yet unknown factors that have to be identified in future investigations as certain environmental conditions, beekeepers management practices and genetic background of honey bees that influences Nosema distribution. Alternatively, an ongoing displacement process of N. apis by N. ceranae may also result in the observed pattern yet such a chronological process can only be confirmed in a long-term study.
Our results show that N. ceranae is the only Nosema spp. found to infect honey bees in the different geographical regions of Saudi Arabia. None of the samples was infected with N. apis. From the available literature, we can understand that N. apis has been present in some Middle east countries (Al-Ghamdi, 1990; Alzubaidy and Ali, 1994; OIE, 2004; Matheson, 1993) as well as in the Europe and America (Paxton et al., 2007, Chen et al., 2009b) for the past decade. Recently N. ceranae emerging as new microspordian infection in middle east and north African countries as already reported in Europe, USA, Canada, and China (El-Shemy et al., 2012; Roudel et al., 2013, Haddad, 2014, Aroee et al., 2016, Williams et al., 2008, Higes et al., 2006), This indicates that N. ceranae is a new emerging pathogen for honey bees, and has presumably been transferred from its original host A. ceranae to A. mellifera (Klee et al., 2007) much earlier than previously recognized (Guerrero-Molina et al., 2016). The evidence, when N. ceranae appeared and started to parasitize Saudi Arabian bees is unknown, and it is difficult to investigate the past incidence because of a lack of bee samples. However total absence of N. apis might be due to better adaptation of N. ceranae to warm climate of Saudi Arabia (Forsgren and Fries, 2013; Martín-Hernández et al., 2012).
This is the first report of molecular detection of N. ceranae in Saudi Arabia. Further research and analysis of more colonies are needed to determine the actual prevalence of this new agent in the country. Intensive survey and further research are thus necessary to determine the distribution and prevalence of Nosema spp. in the Kingdom of Saudi Arabia and their Preventive measure. This report is an alarm for beekeeping industry of Saudi Arabia and protection from honeybee pathogens. Beekeepers must pay attention when moving their colonies in different season to void the pathogens including Nosema.
6. Conclusions
Overall, our results provide evidence that N. ceranae infection occurs frequently in the cohort of apiaries examined despite the lack of clinical signs. This suggests that colony disease outbreaks might probably be caused by other factors, both known and unknown, that singly or in combination may lead to higher susceptibility of honeybees to N. ceranae. The results confirmed the colonization of N. ceranae in Saudi Arabia and need further molecular study at a more extensive monitoring level in order to elucidate possible links between infection by N. ceranae and colony losses in Saudi Arabia.
Conflict of interest statement
None of the authors of this article has any conflict of interest.
Acknowledgements
The project was financially supported by King Saud University, Vice Deanship of Research Chairs. We are grateful to the beekeepers who allowed us to sample their hives.
Footnotes
Peer review under responsibility of King Saud University.
References
- Abdel-Baki A.A.S., Mares M.M., Dkhil M.A., Al-Quraishy S. First detection of Nosema sp., microsporidian parasites of honeybees (Apis mellifera) in Riyadh city, Saudi Arabia. J. King Saud Univ. Sci. 2016 [Google Scholar]
- Alattal Y., AlGhamdi A. Impact of temperature extremes on survival of indigenous and exotic honey bee subspecies, Apis mellifera, under desert and semiarid climates. B. Insectol. 2015;68(2):219–222. [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alzubaidy M.M., Ali A.B.M.H. First record of Nosema apis Zander on honey bees Apis mellifera L. Dirasat. Pure Appl. Sci. 1994;21(1):146–150. [Google Scholar]
- Ansari M.J., Al-Ghamdi A., Nuru A., Ahmed A.M., Ayaad T.H., Al-Qarni A., Alattal Y., Al-Waili N. Survey and molecular detection of Melissococcus plutonius, the causative agent of European Foulbrood in honeybees in Saudi Arabia. Saudi J. Biol. Sci. 2016 doi: 10.1016/j.sjbs.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari M.J., Al-Ghamdi A., Usmani S., Khan K.A., Alqarni A.S., Kaur M., Al-Waili N. In vitro evaluation of the effects of some plant essential oils on Ascosphaera apis, the causative agent of Chalkbrood disease. Saudi J. Biol. Sci. 2017;24(5):1004–1011. doi: 10.1016/j.sjbs.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ghamdi A.A. University of Wales; Cardiff, United Kingdom: 1990. Survey of Honeybee Diseases, Pests and Predators in Saudi Arabia. MPhil Thesis. [xvii] + 127 pp. [Google Scholar]
- Al-Ghamdi, A.A., 2007. Saudi beekeeping industry. In: Fifth International Arab Apicultural Conference, November 25–28, Tripoli.
- Al-Ghamdi A.A. King Saud University, College of Agriculture, Bee Research Unit; Riyadh: 2010. Comprehensive Study for Current Beekeeping Industry of Imported and Native Honeybee in Kingdom of Saudi Arabia. [Google Scholar]
- Al-Ghamdi A., Nuru A. Beekeeping in the Kingdom of Saudi Arabia: past and present practices. Bee World. 2013;90(2):26–29. [Google Scholar]
- Alqarni A.S., Hannan M.A., Owayss A.A., Engel M.S. The indigenous honey bees of Saudi Arabia (Hymenoptera, Apidae, Apis mellifera jemenitica Ruttner): their natural history and role in beekeeping. ZooKeys. 2011;134:83–98. doi: 10.3897/zookeys.134.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroee F., Azizi H., Shiran B., Kheirabadi K.P. Molecular identification of Nosema species in provinces of Fars, Chaharmahal and Bakhtiari and Isfahan (Southwestern Iran) Asian Pac. J. Trop. Biomed. 2016 [Google Scholar]
- Bacandritsos N., Granato A., Budge G., Papanastasiou I., Roinioti E., Caldon M., Falcaro C., Gallina A., Mutinelli F. Sudden deaths and colony population decline in Greek honey bee colonies. J. Invertebr. Pathol. 2010;105(3):335–340. doi: 10.1016/j.jip.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Bollan K.A., Hothersall J.D., Moffat C., Durkacz J., Saranzewa N., Wright G.A., Raine N.E., Highet F., Connolly C.N. The microsporidian parasites Nosema ceranae and Nosema apis are widespread in honeybee (Apis mellifera) colonies across Scotland. Parasitol. Res. 2013;112(2):751–759. doi: 10.1007/s00436-012-3195-0. [DOI] [PubMed] [Google Scholar]
- Botías C., Martín-Hernández R., Barrios L., Meana A., Higes M. Nosema spp. infection and its negative effects on honey bees (Apis mellifera iberiensis) at the colony level. Vet. Res. 2013;44(25):1–14. doi: 10.1186/1297-9716-44-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois A.L., Rinderer T.E., Beaman L.D., Danka R.G. Genetic detection and quantification of Nosema apis and N. ceranae in the honey bee. J. Invertebr. Pathol. 2010;103(1):53–58. doi: 10.1016/j.jip.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Chen Y.P., Evans J.D., Murphy C., Gutell R., Zuker M., Gundensen-Rindal D.A.W.N., Pettis J.S. Morphological, molecular, and phylogenetic characterization of Nosema ceranae, a microsporidian parasite isolated from the European honey bee, Apis mellifera. J. Eukaryot. Microbiol. 2009;56(2):142–147. doi: 10.1111/j.1550-7408.2008.00374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Evans J.D., Zhou L., Boncristiani H., Kimura K., Xiao T., Litkowski A.M., Pettis J.S. Asymmetrical coexistence of Nosema ceranae and Nosema apis in honey bees. J. Invertebr. Pathol. 2009;101(3):204–209. doi: 10.1016/j.jip.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Chupia V., Pikulkaew S., Krutmuang P., Mekchay S., Patchanee P. Molecular epidemiology and geographical distribution of Nosema ceranae in honeybees, Northern Thailand. Asian Pac. J. Trop. Dis. 2016;6(1):27–31. [Google Scholar]
- Cox-Foster D.L., Conlan S., Holmes E.C., Palacios G., Evans J.D., Moran N.A., Quan P.L., Briese T., Hornig M., Geiser D.M., Martinson V. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318(5848):283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- Crailsheim, K; Brodschneider, R; Neumann, P., 2009. The COLOSS puzzle: filling in the gaps. In: Proceedings of the 4th COLOSS Conference, 3-4 March 2009. Zagreb, Croatia, p. 46-47.
- El-Shemy A.A.H., Ibrahim Y.Y., El-kinani D.D. Detection of Nosema cerana Fries in Egypt and its seasonal fluctuations. Bull. Entomol. Soc. Egypt. 2012;1(89):25–37. [Google Scholar]
- Emsen B., Guzman-Novoa E., Hamiduzzaman M.M., Eccles L., Lacey B., Ruiz-Pérez R.A., Nasr M. Higher prevalence and levels of Nosema ceranae than Nosema apis infections in Canadian honey bee colonies. Parasitol. Res. 2016;115(1):175–181. doi: 10.1007/s00436-015-4733-3. [DOI] [PubMed] [Google Scholar]
- Engel M.S. The taxonomy of recent and fossil honey bees (Hymenoptera: Apidae; Apis) J. Hymenopt. Res. 1999;8:165–196. [Google Scholar]
- Fenoy S., Rueda C., Higes M., Martín-Hernández R., Del Aguila C. High-level resistance of Nosema ceranae, a parasite of the honeybee, to temperature and desiccation. Appl. Environ. Microbiol. 2009;75(21):6886–6889. doi: 10.1128/AEM.01025-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren E., Fries I. Comparative virulence of Nosema ceranae and Nosema apis in individual European honey bees. Vet. Parasitol. 2010;170(3):212–217. doi: 10.1016/j.vetpar.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Forsgren E., Fries I. Temporal study of Nosema spp. in a cold climate. Environ. Microbiol. Rep. 2013;5(1):78–82. doi: 10.1111/j.1758-2229.2012.00386.x. [DOI] [PubMed] [Google Scholar]
- Fries I. Infectivity and multiplication of Nosema apis Z. in the ventriculus of the honey bee. Apidologie. 1988;19(3):319–328. [Google Scholar]
- Fries I. Nosema ceranae in European honey bees (Apis mellifera) J. Invertebr. Pathol. 2010;103:73–79. doi: 10.1016/j.jip.2009.06.017. [DOI] [PubMed] [Google Scholar]
- Fries I., Feng F., da Silva A., Slemenda S.B., Pieniazek J. Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian Honey bee Apis cerana (Hymenoptera, Apidae) Eur. J. Protistol. 1996;32:356–365. [Google Scholar]
- Gajger I.T., Tomljanović Z., Petrinec Z. Monitoring health status of Croatian honey bee colonies and possible reasons for winter losses. J. Apic. Res. 2010;49(1):107–108. [Google Scholar]
- Gallai N., Salles J.-M., Settele J., Vaissie’re B.E. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econom. 2009;68(13):810–821. [Google Scholar]
- Gisder S., Hedtke K., Möckel N., Frielitz M.C., Linde A., Genersch E. Five-year cohort study of Nosema spp. in Germany: does climate shape virulence and assertiveness of Nosema ceranae? Appl. Environ. Microbiol. 2010;76(9):3032–3038. doi: 10.1128/AEM.03097-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Molina C., Correa-Benítez A., Hamiduzzaman M.M., Guzman-Novoa E. Nosema ceranae is an old resident of honey bee (Apis mellifera) colonies in Mexico, causing infection levels of one million spores per bee or higher during summer and fall. J. Invertebr. Pathol. 2016;141:38–40. doi: 10.1016/j.jip.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Haddad, N., Bataeneh, A., Albaba, I., Obeid, D., Abdulrahman, S., 2009. Status of colony losses in the Middle East. In: Proceedings of the 41st Apimondia Congress, Mointpellier, France. p. 36.
- Haddad N.J. First detection of Nosema ceranae in Jordan. Eur. Sci. J. 2014;10(33):91–96. [Google Scholar]
- Higes M., Martín R., Meana A. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J. Invertebr. Pathol. 2006;92(2):93–95. doi: 10.1016/j.jip.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Higes M., García-Palencia P., Martín-Hernández R., Meana A. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia) J. Invertebr. Pathol. 2007;94(3):211–217. doi: 10.1016/j.jip.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Higes M., Martín-Hernández R., Botías C., Bailón E.G., González-Porto A.V., Barrios L., Del Nozal M.J., Bernal J.L., Jiménez J.J., Palencia P.G., Meana A. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 2008;10(10):2659–2669. doi: 10.1111/j.1462-2920.2008.01687.x. [DOI] [PubMed] [Google Scholar]
- Higes M., Martín-Hernández R., Garrido-Bailón E., González-Porto A.V., García-Palencia P., Meana A., Del Nozal M.J., Mayo R., Bernal J.L. Honeybee colony collapse due to Nosema ceranae in professional apiaries. Environ. Microbiol. Rep. 2009;1(2):110–113. doi: 10.1111/j.1758-2229.2009.00014.x. [DOI] [PubMed] [Google Scholar]
- Higes M., Martín-Hernández R., Martínez-Salvador A., Garrido-Bailón E., González-Porto A.V., Meana A., Bernal J.L., Nozal M.J.d., Bernal J. A preliminary study of the epidemiological factors related to honey bee colony loss in Spain. Environ. Microbiol. Rep. 2010;2:243–250. doi: 10.1111/j.1758-2229.2009.00099.x. [DOI] [PubMed] [Google Scholar]
- Huang W.F., Jiang J.H., Chen Y.W., Wang C.H. A Nosema ceranae isolate from the honeybee Apis mellifera. Apidologie. 2007;38(1):30–37. [Google Scholar]
- Huang Z. Pollen nutrition affects honey bee stress resistance. Terr. Arthropod Rev. 2012;5(2):175–189. [Google Scholar]
- Kevan P.G., Guzmán-Novoa E., Skinner A., van Englesdorp D. Colony collapse disorder in Canada: do we have a problem? Hive Lights. 2007;20:14–16. [Google Scholar]
- Klee J., Besana A.M., Genersch E., Gisder S., Nanetti A., Tam D.Q., Chinh T.X., Puerta F., Ruz J.M., Kryger P., Message D. Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J. Invertebr. Pathol. 2007;96(1):1–10. doi: 10.1016/j.jip.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Liu T.P. Ultrastructural changes in Nosema apis in the midgut of the honeybee treated with thimerosal in vitro. Parasitol. Res. 1988;74:492–494. [Google Scholar]
- Martín-Hernández R., Meana A., Prieto L., Salvador A.M., Garrido-Bailón E., Higes M. Outcome of colonization of Apis mellifera by Nosema ceranae. Appl. Environ. Microbiol. 2007;73(20):6331–6338. doi: 10.1128/AEM.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Hernández R., Botías C., Bailón E.G., Martínez-Salvador A., Prieto L., Meana A., Higes M. Microsporidia infecting Apis mellifera: coexistence or competition. Is Nosema ceranae replacing Nosema apis? Environ. Microbiol. 2012;14(8):2127–2138. doi: 10.1111/j.1462-2920.2011.02645.x. [DOI] [PubMed] [Google Scholar]
- Matheson A. World bee health report. Bee World. 1993;74(4):176–212. [Google Scholar]
- Matheson A. World bee health update 1996. Bee world. 1996;77(1):45–51. [Google Scholar]
- Medici S.K., Sarlo E.G., Porrini M.P., Braunstein M., Eguaras M.J. Genetic variation and widespread dispersal of Nosema ceranae in Apis mellifera apiaries from Argentina. Parasitol. Res. 2012;110(2):859–864. doi: 10.1007/s00436-011-2566-2. [DOI] [PubMed] [Google Scholar]
- Michalczyk M., Sokół R., Szczerba-Turek A., Bancerz-Kisiel A. A comparison of the effectiveness of the microscopic method and the multiplex PCR method in identifying and discriminating the species of Nosema spp. spores in worker bees (Apis mellifera) from winter hive debris. Pol. J. Vet. Sci. 2011;14(3):385–391. doi: 10.2478/v10181-011-0058-z. [DOI] [PubMed] [Google Scholar]
- Moher D., Hopewell S., Schulz K.F., Montori V., Gøtzsche P.C., Devereaux P.J., Elbourne D., Egger M., Altman D.G. CONSORT 2010 explanation and elaboration: updated guidelines for reporting parallel group randomised trials. J. Clin. Epidemiol. 2010;63(8):e1–37. doi: 10.1016/j.jclinepi.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Mulholland G.E., Traver B.E., Johnson N.G., Fell R.D. Individual variability of Nosema ceranae infections in Apis mellifera colonies. Insects. 2012;3(4):1143–1155. doi: 10.3390/insects3041143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabian S., Ahmadi K., Shirazi M.N., Sadeghian A.G. First detection of Nosema ceranae, a microsporidian protozoa of European honeybees (Apis mellifera) in Iran. Iran J. Parasitol. 2011;6(3):89–95. [PMC free article] [PubMed] [Google Scholar]
- Naug D. Nutritional stress due to habitat loss may explain recent honeybee colony collapses. Biol. Cons. 2009;142(10):2369–2372. [Google Scholar]
- Office International des Épizooties (OIE) Animal health status and disease control methods. Part 2. Tables. Office International des Épizooties; Paris, France: 2004. World animal health 2004. [Google Scholar]
- Office International des Épizooties (OIE) OIE; Paris, France: 2008. Manual for diagnostic tests and vaccines for terrestrial animals, Chapter 2.2.4, Nosemosis of honey bees; p. 23. [Google Scholar]
- Office International des Épizooties (OIE), 2013. Listed diseases, infections and infestations. URL http://www.oie.int/en/animal-health-in-the-world/oie-listeddiseases-2013.
- Pacini A., Mira A., Molineri A., Giacobino A., Cagnolo N.B., Aignasse A., Zago L., Izaguirre M., Merke J., Orellano E., Bertozzi E. Distribution and prevalence of Nosema apis and N. ceranae in temperate and subtropical eco-regions of Argentina. J. Invertebr. Pathol. 2016;141:34–37. doi: 10.1016/j.jip.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Paxton R.J., Klee J., Korpela S., Fries I. Nosema ceranae has infected Apis mellifera in Europe since at least 1998 and may be more virulent than Nosema apis. Apidologie. 2007;38(6):558–565. [Google Scholar]
- Paxton R.J. Does infection by Nosema ceranae cause “Colony Collapse Disorder” in honey bees (Apis mellifera)? J. Apic. Res. 2010;49(1):80–84. [Google Scholar]
- Pickard R.S., El-Shemy A.A.M. Seasonal variation in the infection of honeybee colonies with Nosema apis Zander. J. Apic. Res. 1989;28:93–100. [Google Scholar]
- Plischuk S., Martín-Hernández R., Prieto L., Lucía M., Botías C., Meana A., Abrahamovich A.H., Lange C., Higes M. South American native bumblebees (Hymenoptera: Apidae) infected by Nosema ceranae (Microsporidia), an emerging pathogen of honeybees (Apis mellifera) Environ. Microbiol. Rep. 2009;1(2):131–135. doi: 10.1111/j.1758-2229.2009.00018.x. [DOI] [PubMed] [Google Scholar]
- Razmaraii N., Sadegh-Eteghad S., Babaei H., Paykari H., Esmaeilnia K., Froghy L. Molecular identification of Nosema species in East Azerbaijan province. Iran. Arch. Razi Inst. 2013;68(1):23–27. [Google Scholar]
- Roudel M., Aufauvre J., Corbara B., Delbac F., Blot N. New insights on the genetic diversity of the honeybee parasite Nosema ceranae based on multilocus sequence analysis. Parasitology. 2013;140(11):1346–1356. doi: 10.1017/S0031182013001133. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Smith M.L. The honey bee parasite Nosema ceranae: transmissible via food exchange? PLoS One. 2012;7(8):e43319. doi: 10.1371/journal.pone.0043319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroker, V., Hetzroni, A., Yacobson, B., Voet, H., Slabezki, S., Efrat, H., Chejanovsky, N., 2009. Colony losses in Israel: incidence of viral infection and beehive populations. In: Proceedings of the 41st Apimondia Congress, Mointpellier, France. p. 38.
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vanEngelsdorp D., Evans J.D., Saegerman C., Mullin C., Haubruge E., Nguyen B.K., Frazier M., Frazier J., CoxFoster D., Chen Y., Underwood R., Tarpy D.R., Pettis J.S. Colony collapse disorder: a descriptive study. PloS ONE. 2009;4:e6481. doi: 10.1371/journal.pone.0006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vanEngelsdorp D., Meixner M.D. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J. Invertebr. Pathol. 2010;103:80–95. doi: 10.1016/j.jip.2009.06.011. [DOI] [PubMed] [Google Scholar]
- vanEngelsdorp D., Hayes J., Jr, Underwood R.M., Pettis J.S. A survey of honey bee colony losses in the United States, fall 2008 to spring 2009. J. Apicult. Res. 2010;49(1):7–14. [Google Scholar]
- Williams G.R., Shafer A.B., Rogers R.E., Shutler D., Stewart D.T. First detection of Nosema ceranae, a microsporidian parasite of European honey bees (Apis mellifera), in Canada and central USA. J. Invertebr. Pathol. 2008;97(2):189–192. doi: 10.1016/j.jip.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Wilson W.T., Nunamaker R.A. The incidence of Nosema apis in honeybees in Mexico. Bee World. 1983;64(3):132–136. [Google Scholar]