Abstract
Two major DNA double-strand break repair pathways exist in all eukaryotes, nonhomologous DNA end joining (NHEJ) and homologous recombination (HR). Although both pathways can function throughout the cell cycle, NHEJ predominates in G0/G1 (when a replicated sister chromatid is unavailable), whereas HR makes a more substantial contribution in S and G2. How a cell chooses between these two important DNA repair pathways is largely unknown. DNA-dependent protein kinase (DNA-PK) is critical for NHEJ. Here, we describe two conserved splice variants of a catalytic subunit of DNA-PK (DNA-PKcs) that are expressed predominately in nondividing cells. Although both encode stable products, neither reverses the NHEJ defects in DNA-PKcs-deficient cells. In fact, cells expressing one of the DNA-PKcs variants are slightly more radiosensitive than cells completely deficient in DNA-PKcs. We investigated whether cells expressing the DNA-PKcs variants had any other DNA repair deficits and found that these cells are considerably more sensitive to both etoposide and mitomycin C than cells that express no DNA-PKcs at all. Because repair of DNA damage induced by these two agents requires intact HR, we tested whether the NHEJ-defective variants of DNA-PKcs inhibit double-strand break-induced HR in an integrated substrate. In cells expressing the NHEJ-defective variants, HR was markedly reduced. Because the splice variants are expressed highly only in nondividing cells, quiescent cells would be afforded a mechanism to inhibit repair by means of HR when sister chromatids are not available as templates for accurate repair with low risk of genome rearrangement, thereby enhancing genome stability.
Keywords: DNA repair, DNA-dependent protein kinase, nonhomologous DNA end joining pathway
In 1985, a protein kinase activity was described that depends on the presence of free DNA ends for activation, the DNA-dependent protein kinase (DNA-PK) (1). Later it was demonstrated that this nuclear, S/T protein kinase consists of two functional subunits: the Ku heterodimer (known as the regulatory subunit) directs a catalytic subunit (DNA-PKcs) to DNA ends (2, 3).
In higher eukaryotes, the nonhomologous DNA end joining pathway (NHEJ) is thought to be the primary pathway that repairs DNA double-strand breaks (DSBs) (reviewed in refs. 4 and 5). However, it is clear that a second DSB repair pathway, homologous recombination (HR), is also critical in higher eukaryotes to maintain DNA integrity after DSBs (6). The fact that both of these pathways function to maintain genomic stability in higher eukaryotes is underscored by reports demonstrating more severe genomic instability in animals with defects in both pathways (7, 8). How a cell chooses between these two important DNA repair pathways has not been completely elucidated.
Although it is clear that DNA-PK is required for both NHEJ and V(D)J recombination, its precise role in repair is only partially understood (9–13). Because DNA-PK's protein kinase activity is required for its functional role in NHEJ, it has been suggested that DNA-PK may activate downstream repair mediators (14, 15). To date, the only physiologically relevant DNA-PK target defined is DNA-PK itself (16). There is also evidence that DNA-PK functions as a molecular scaffold to target other repair factors to damaged DNA (reviewed in refs. 17 and 18). An emerging consensus is that DNA-PK acts as a “gatekeeper” to regulate access of DNA ends to other repair factors like XRCC4, DNA ligase IV, and Artemis (19–22).
Here, we analyze two DNA-PKcs splice variants. Splice variation occurs within the phosphatidylinositol 3-kinase (PI3-kinase) domain and is achieved by the following two mechanisms: (i) exon skipping where a single exon is differentially inserted into the transcript (as described in ref. 23) and (ii) intron retention. The two splice variants are only weakly expressed in cultured cell lines; however, expression of the splice variants was similar to full-length DNA-PKcs in all normal human tissues examined. Transfection studies demonstrate (14, 15) that the full-length DNA-PKcs transcript generates an active form of the protein, which fully complements the DNA repair defects in DNA-PKcs-deficient cell lines. In this article, we demonstrate that the two alternative isoforms lack protein kinase enzymatic activity and thus cannot function in NHEJ. The structure of the two alternative isoforms suggests a possible regulatory function, and we demonstrate that one of the alternative isoforms inhibits HR. We suggest that this DNA-PKcs isoform functions in normal, nondividing cells to inhibit HR and thus prevent loss of genetic material that can occur during HR in the absence of a sister chromatid.
Methods
Cell Lines. Cell lines used in this study were as follows: Ramos B cell lymphoma; HeLa, cervical carcinoma; LS, colon carcinoma; U937, monocytic leukemia; SBC, pre B cell lymphoma; A431, keratinocyte cell line; HepG2, hepatoma; Sf19, murine severe combined immunodeficient (SCID) fibroblasts; CHO, Chinese hamster ovary cells; V3, CHO DSB repair mutant cell line; XR-1, CHO DSB repair mutant cell line; and MSU1.1, normal human fibroblast cells. Peripheral blood T cells were isolated, cultured, and activated as described in ref. 24.
PCR. RNA isolated from cell lines and human tissue (purchased from Clontech) was converted to cDNA. PCR was performed by using Elongase according to the manufacturer's recommendations (GIBCO/BRL). Amplified transcripts were subcloned and sequenced. Amplification primers used to assess relative transcript levels were 5′-CTCTTCGAGGTCATGAAT and 5′-ATGTCTGTCTCCAATCCC. Total genomic DNA was isolated from the HeLa cells. Two overlapping DNA fragments spanning this region were amplified and sequenced.
Plasmid Construction and Transfections. Construction of the wild-type (WT) human DNA-PKcs expression vector is described in ref. 25. To generate the plasmid encoding isoform III, a PCR fragment that lacked exon 80 was subcloned into the parental plasmid. To make the plasmid encoding isoform II, an alternative oligonucleotide was used that introduced the stop codon present at the start of intron D just 3′ of exon 82 followed by a NotI site. For transfections into CHO cells, an N-terminal FLAG epitope was added to the expression vectors by subcloning an oligonucleotide encoding the FLAG epitope. Murine SCID cell lines and CHO cells expressing human DNA-PKcs were derived as described in refs. 14 and 16. V(D)J recombination assays were performed as described in ref. 16.
DNA-PK Microfractionation and Measurement of Protein Kinase Activity. DNA-PK activity was assessed as described in ref. 16 with the following exception. For the experiments presented in Fig. 4, the DNA-cellulose beads first were mixed with preswollen cellulose beads at a ratio of 1:4. Immunoblotting was performed as described in refs. 16 and 25.
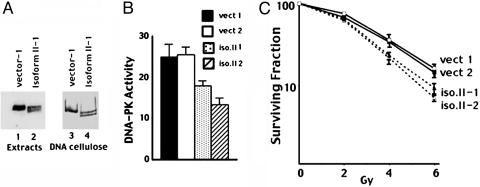
Fig. 4.
DNA-PKcs isoform II modestly inhibits DNA-PK activity and modestly radiosensitizes WT CHO cells. (A) Immunoblot analyses of WCE or DNA cellulose fractions from CHO cells stably transfected with vector or isoform II as indicated. (B) WCE (300 μg) prepared from CHO cells transfected with vector or isoform II were assayed for DNA-PK activity. Each cell extract was tested in duplicate, and six extracts were tested for each cell line. (Bars indicate SD of six experiments.) (C) Radiation resistance was assessed of CHO cells stably transfected with vector and isoform II. Data are presented as percent survival of unirradiated controls. (Bars represent SEM from four experiments.)
DNA Damage Sensitivity Assays. Cells (103) were exposed to various amounts of ionizing radiation by using a 60Co source and immediately seeded in complete medium containing 10% FBS. After 7 days, colonies were fixed with methanol, stained with crystal violet, and counted. Mitomycin C (MMC) and etoposide sensitivities were assayed by plating 200 cells in 100-mm dishes in complete medium. Four hours later, MMC or etoposide was added to the indicated concentrations, cultures were incubated for 7 days, and colonies were stained and counted as above.
HR Assays. V3 cells harboring the HR substrate (cell strain VD7) are described in detail in ref. 26. Expression constructs encoding WT DNA-PKcs, isoform II, or mutant ABCDE were stably integrated into the VD7 cell strain as described in ref. 16 except that pCDNA6 was cotransfected to provide blasticidin resistance. Assessment of DSB-induced HR was essentially as described in ref. 26.
Results
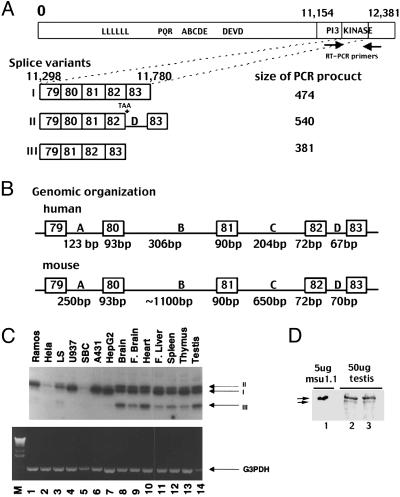
Splice Variation Within the PI3-Kinase Domain of Human DNA-PKcs. The PI3-kinase domain spans only the C-terminal 408 aa of the 4,126-aa protein (Fig. 1A). In our previous studies, the entire human DNA-PKcs cDNA and ≈95% of the equine DNA-PKcs cDNA were isolated by RT-PCR (25, 27). Splice variation was only consistently observed within the region encoding the PI3-kinase domain. A variety of different human transcripts were cloned that could be explained by two variably inserted sequences between nucleotides 11,298 and 11,780. The genomic organization of the human and murine DNA-PKcs genes shows that this region is encoded by five exons separated by four introns (designated A, B, C, and D; Fig. 1B). The murine DNA-PKcs gene has been studied previously (28); for consistency, we designated the exons here as 79–83.
Fig. 1.
Splice variants of DNA-PKcs. (A) The top portion is a representation of the DNA-PKcs transcript. Approximate positions of the leucine zipper motif (LLLLLL), the major autophosphorylation site cluster (ABCDE), a second autophosphorylation site cluster (PQR), the caspase cleavage site (DEVD), and the PI3-kinase domain are shown. The region between nucleotides 11,298 and 11,780 is encoded by five exons, 79–83, separated by four introns, A–D. Positions of PCR amplification primers are indicated with arrows. Structures of the three splice variants also are depicted. Exon 80 spans nucleotides 11,394–11,486; exon 81 spans nucleotides 11,487–11,577; and exon 82 spans nucleotides 11,578–11,660. (B) The genomic organization of this portion of human and murine DNA-PKcs. (C) Expression of DNA-PKcs splice variants was assessed by RT-PCR using RNA from human cells and tissues. Arrows denote the three major transcripts and the G3PDH amplified fragment. (D) DNA-PKcs expression was assessed in human tissue and cell extracts by immunoblotting. Extracts are loaded as follows: lane 1, 5 μg of MSU1.1; lanes 2 and 3, 50 μg of testis.
One splice variant (transcript III) lacks exon 80 (isoform III; Fig. 1A). In fact, exon 80 is absent in the first reported DNA-PKcs sequence (29). This exon encodes a 31-aa insertion at position 3797 within the PI3-kinase domain of DNA-PKcs. Transcript I (which includes exon 80, referred to hereafter as full-length or WT DNA-PKcs) is the predominant form expressed in cell lines, and this form encodes an active protein kinase that is fully functional in NHEJ (14, 15). The second major splice variant (transcript II) retains intron D (isoform II) inserting an additional 67 nt into the DNA-PKcs transcript. The first codon of intron D is a termination codon that would truncate the protein at amino acid 3883 (deleting the conserved protein kinase motifs). DNA-PKcs splice variation also was studied in other species (see Supporting Text, Table 2, and Fig. 7, which are published as supporting information on the PNAS web site). In summary, transcript II was observed in all species tested. In contrast, transcript III was detected in human and horse but was absent in mouse and rat in agreement with the findings of Blunt et al. (30). The fact that transcript II has been evolutionarily conserved suggests that it may be functionally relevant.
All Three DNA-PKcs Transcripts Are Present in Normal Human Tissues. Expression patterns of the three transcripts are markedly different when comparing normal tissues with transformed cell lines (Fig. 1C). Whereas all three transcripts are abundantly expressed in all normal human tissues, transcripts II and III are only weakly expressed in cultured cell lines. The predicted molecular weights of the three polypeptides encoded by the three DNA-PKcs transcripts are 469, 441, and 465 kDa. Because transcripts II and III are only expressed to a significant extent in normal tissues, we assessed DNA-PKcs protein expression in human tissue extracts (Fig. 1D). Moll et al. (31) examined DNA-PKcs expression in human tissues and malignancies and documented differential DNA-PKcs protein expression in certain tissues even though mRNA levels were generally similar. They concluded that the differential tissue expression was the result of posttranscriptional regulation. We assessed DNA-PKcs expression in normal human testis [shown by Moll et al. (31) to express abundant DNA-PKcs protein] and compared expression with that in a human fibroblast cell line (Fig. 1D). As can be seen, DNA-PKcs is readily detectable by using 50 μg of testis extract, although the signal is approximately one-fifth that observed in just 5 μg of MSU1.1 extract. In testis, two distinctly migrating species of DNA-PKcs could be detected: one with the same mobility as full-length DNA-PKcs and one with a faster mobility consistent with the predicted mass of isoform II. Our data are consistent with those of Moll et al. (31) in that we found much lower DNA-PKcs levels in normal human tissues as compared with cultured cell lines. We found no evidence of distinct 469- and 465-kDa species (the predicted mass of full-length and isoform III) in tissues, although it is unlikely that such large polypeptides with such similar mass could be discriminated by SDS/PAGE.
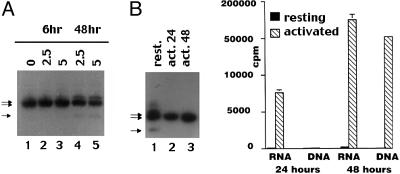
Expression of DNA-PKcs Splice Variants Is Altered by Ionizing Radiation and Cell Proliferation. We next assessed transcript expression in cells exposed to ionizing radiation and found that isoforms II and III are induced after ionizing radiation (Fig. 2). Transcript expression was not altered by UV irradiation, MMC, or etoposide in the cultured cell lines tested (data not shown). The altered ratios of full-length DNA-PKcs to transcripts II and III after irradiation mimics those found in normal tissues. We considered that the change induced by irradiation might reflect a difference in dividing vs. nondividing cells. Thus, we next assessed splice variation in peripheral blood T lymphocytes that were unstimulated or stimulated (and thus actively dividing) with phytohemagglutinin, anti-CD28, and phorbol 12-myristate 13-acetate. All three transcripts were detected in unstimulated peripheral blood T cells, which are largely nonproliferating as assessed by thymidine incorporation (Fig. 2B). In contrast, in activated, proliferating cells (either 24 or 48 h after stimulation), only full-length DNA-PKcs was detected. These data suggest that isoforms II and III are primarily expressed in nondividing cells. This conclusion is supported by the observation that in fetal tissues and thymus (tissues with high numbers of dividing cells), levels of transcripts II and III were decreased compared with other adult tissues (Fig. 1C).
Fig. 2.
DNA-PKcs splice variation is associated with cell proliferation. (A) Ramos cells were treated with 0–5 Gy, harvested 6 or 48 h later, and subjected to RT-PCR of DNA-PKcs. (B Left) Splice variation was analyzed in resting (unstimulated) peripheral blood T cells or T cells activated with a combination of phytohemagglutinin, anti-CD28 mAb, and phorbol 12-myristate 13-acetate for the times indicated. (Right) 3H thymidine and uridine incorporation of duplicate T cell cultures.
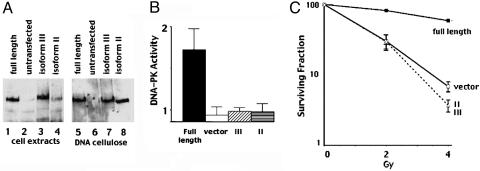
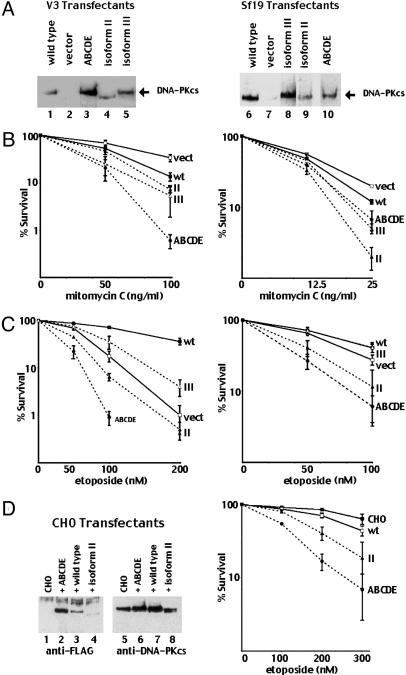
DNA-PKcs Isoforms II and III Do Not Complement the NHEJ Deficits in Murine SCID Cells. To assess the function of transcripts II and III, expression vectors encoding each were transfected into a murine SCID fibroblast cell line. The proteins encoded by transcripts II and III were stably expressed in Sf19 cells, and DNA cellulose binding of cell extracts demonstrated that isoforms II and III fractionate (with Ku, data not shown) onto DNA cellulose as efficiently as full-length DNA-PKcs (Fig. 3). However, neither isoform II that lacks the protein kinase motifs nor isoform III that contains conserved PI3-kinase motifs, had detectable DNA-PK activity (Fig. 3B). Consistent with previous reports (14, 15), complementation of the radiosensitivity of murine SCID cells was only observed in transfectants expressing the kinase competent form of DNA-PKcs (Fig. 3C). In fact, cells expressing isoform II consistently were slightly more radiosensitive than cells expressing no DNA-PKcs at all. Neither isoform II or III reverse the V(D)J recombination defects in Sf19 cells (Table 1). Whereas full-length DNA-PKcs efficiently complements both the coding joint and more modest signal joint deficit of the V3 cell line, neither isoform II nor III supported either coding or signal end joining. DNA-PKcs independent subpathway(s) of NHEJ have been reported (20). Residual signal end joining in DNA-PKcs deficient cells (dependent on Ku, XRCC4, and DNA ligase IV) may represent such a pathway. However isoforms II and III do not inhibit signal end joining. In sum, we conclude that isoforms II and III (when expressed in the absence of full-length DNA-PKcs) do not positively or negatively affect NHEJ.
Fig. 3.
DNA-PKcs isoforms II and III encode stable proteins that bind to DNA cellulose but lack protein kinase activity. (A) Immunoblot analyses of 250 μg of whole-cell extract (WCE; lanes 1–4) or DNA cellulose fractions (lanes 5–8) from Sf19 cells stably transfected with full-length DNA-PKcs, untransfected control, or isoform III or II. (B) WCE prepared from Sf19 cells transfected with full-length DNA-PKcs, vector, isoform II, or III were assayed for DNA-PK activity. Each cell extract was tested in duplicate, and at least three extracts were tested for each cell line. (Bars indicate SD.) (C) Radiation resistance of Sf19 fibroblasts stably transfected with full-length DNA-PKcs, vector, or isoform II or III was assessed. Data are presented as percent survival of unirradiated controls. (Bars represent SEM.)
Table 1. Isoforms II and III do not complement the V(D)J recombination deficit of DNA-PKcs-deficient cells.
| pJH290 (coding)
|
pJH201 (signal)
|
||||||
|---|---|---|---|---|---|---|---|
| Exp. | amp | cam | %R* | amp | cam | %R* | |
| No RAGS | 1 | 111,128 | 1 | 0.0009 | 88,044 | 0 | 0 |
| 2 | 40,368 | 1 | 0.002 | 85,840 | 0 | 0 | |
| 3 | 14,927 | 0 | 0 | 15,281 | 0 | 0 | |
| RAGS | 1 | 51,968 | 0 | 0 | 61,016 | 31 | 0.05 |
| 2 | 79,576 | 0 | 0 | 22,736 | 40 | 0.176 | |
| 3 | 708 | 0 | 0 | 5,546 | 6 | 0.108 | |
| RAGS plus full length | 1 | 58,348 | 402 | 0.69 | 31,436 | 496 | 1.58 |
| 2 | 49,300 | 345 | 0.7 | 9,976 | 230 | 2.306 | |
| 3 | 6,903 | 60 | 0.87 | 3,009 | 129 | 4.3 | |
| RAGS plus form II | 1 | 125,976 | 1 | 0.0008 | 78,068 | 70 | 0.09 |
| 2 | 70,064 | 0 | 0 | 17,632 | 46 | 0.261 | |
| 3 | 21,594 | 2 | 0.009 | 4,720 | 5 | 0.106 | |
| RAGS plus form III | 1 | 109,272 | 0 | 0 | 66,816 | 10 | 0.015 |
| 2 | 25,984 | 0 | 0 | 38,048 | 43 | 0.113 | |
| 3 | 11,682 | 0 | 0 | 6,844 | 11 | 0.161 | |
Transient V(D)J assays were performed as described in ref. 16. Briefly, RAG expression initiates recombination in V3 cells as assessed by plasmid substrates pJH290 (coding end joining) or pJH201 (signal end joining). RAGS, recombination-activating genes; Exp., experiment; amp, ampicillin; cam, chloramphenicol; form II, isoform II; form III, isoform III.
%R is calculated as the number of cam-resistant colonies divided by amp-resistant colonies × 100
Coexpression of Isoform II with Full-Length DNA-PKcs Weakly Inhibits DNA-PK Kinase Activity. Transcripts II and III were never observed in the absence of full-length DNA-PKcs (Fig. 1C). If isoforms II and III have a functional role in living organisms, it is likely that this function would be exerted in the presence of full-length DNA-PKcs. To test the capacity of isoform II to inhibit NHEJ in cells expressing full-length DNA-PKcs, FLAG-tagged isoform II was stably expressed in WT CHO cells.
We first tested DNA-PK activity in two clones expressing roughly equivalent levels of isoform II and endogenous DNA-PKcs (Fig. 4A) with a standard DNA-PK pulldown assay, but the results were inconsistent. We reasoned that because the number of Ku molecules in most cells vastly outnumbered DNA-PKcs molecules and if isoform II competes with full-length DNA-PKcs, then detecting that inhibition would require either limiting Ku or limiting DNA ends. We modified the DNA-PK pulldown assay to reduce free DNA ends by diluting the DNA cellulose with uncoupled cellulose beads. Under these conditions, a modest reduction of DNA-PK activity was detected in both clones coexpressing isoform II as compared with the control clones (Fig. 4B). However, expression of isoform II only modestly radiosensitized the transfected cells (Fig. 4C). These data suggest that isoform II only weakly competes with WT DNA-PKcs. In cultured mammalian cell lines, ionizing radiation-induced breaks are repaired predominately by NHEJ, with a minor contribution by HR (reviewed in ref. 32). Thus, if NHEJ were significantly reduced in these transfectants, one might expect more significant radiosensitivity. Modest radiosensitivity is more consistent with a deficiency in HR (see below).
NHEJ-Defective Forms of DNA-PKcs Sensitize Cells to MMC and Etoposide. Cells with HR defects are hypersensitive to agents that induce interstrand crosslinks (reviewed in ref. 32). Thus, to appraise the integrity of HR, we first assessed MMC sensitivity.
The modestly increased radiosensitivity of SCID fibroblasts expressing isoform II as compared with no DNA-PKcs at all (Fig. 2C) was similar to our studies of DNA-PKcs autophosphorylation site mutants that are nonfunctional in NHEJ because of an apparent block in DNA end processing (16). The prior studies were performed in the DNA-PKcs-deficient cell line V3, and, to complement those studies, we generated V3 transfectants expressing isoforms II and III as well as Sf19 transfectants expressing the DNA-PKcs autophosphorylation site mutant. Although we present data for only single clones, in most cases two or more clones (of each transfectant type) were tested and gave comparable results. Expression of WT DNA-PKcs rendered both V3 cells and Sf19 cells slightly more sensitive to MMC as compared with the vector only controls (Fig. 5B). This phenomenon has been observed by other investigators studying other DNA-PKcs-deficient cells (7) and likely relates to inefficient kinase activation by DNA-PK bound to interstrand crosslinks (ref. 33 and see Discussion). However, V3 and Sf19 transfectants expressing isoform II or III or the ABCDE mutant were considerably more sensitive to MMC than are cells expressing either full-length DNA-PKcs or no DNA-PKcs at all. These data suggest that NHEJ-defective variants of DNA-PK inhibit repair of interstrand crosslinks.
Fig. 5.
Expression of NHEJ-defective DNA-PKcs induces sensitivity to MMC and etoposide. (A) Immunoblot analyses of WCE from V3 or Sf19 cells stably transfected WT DNA-PKcs, vector, DNA-PKcs autophosphorylation site mutant ABCDE, or isoform II or III as indicated. (B) MMC sensitivity of V3 (Left) or Sf19 cells (Right) transfected with WT DNA-PKcs, vector, isoform II or III, or mutant ABCDE. (C) Etoposide sensitivity of V3 (Left) or Sf19 (Right) cells transfected with WT DNA-PKcs, vector alone, isoform II or III, or mutant ABCDE. (D Left) Immunoblot analyses of WCE from untransfected CHO cells (CHO) or CHO cells stably transfected with mutant ABCDE, WT DNA-PKcs, or isoform II. Anti-Flag, or anti-DNA-PKcs antibodies were used to detect DNA-PKcs. (D Right) Etoposide sensitivity of untransfected CHO cells (CHO) or CHO cells stably transfected with WT Flag-tagged DNA-PKcs, Flag-tagged isoform II, or ABCDE mutant. In B–D, data are presented as percent survival of untreated controls. (Error bars represent SEM of three experiments.)
Repair of DNA damage induced by topoisomerase poisons is thought to be resolved by either HR or NHEJ (34). It has been reported previously that mouse SCID fibroblasts are as sensitive to etoposide as WT controls (35), whereas V3 cells are more sensitive to etoposide than the parental strain (36). We found that both the Sf19 transfectants and V3 transfectants expressing WT DNA-PKcs were more resistant to etoposide than are vector controls, supporting previous conclusions that both NHEJ and HR can facilitate repair of etoposide damage. However, V3 and Sf19 transfectants expressing either isoform II or the ABCDE mutant were markedly more sensitive to etoposide than were cells expressing either WT DNA-PKcs or no DNA-PKcs at all, suggesting that these NHEJ-defective variants may inhibit HR (Fig. 5C).
Surprisingly, expression of isoform III in either V3 cells or Sf19 fibroblasts did not induce sensitivity to etoposide. In fact, in both cell strains, transfectants expressing isoform III were more resistant to etoposide than cells expressing no DNA-PKcs. These data suggest a possible functional role for isoform III in facilitating repair of etoposide-induced DNA damage.
To determine whether the NHEJ-defective variants could inhibit alternative DNA repair pathways in the presence of full-length DNA-PK, we assessed MMC or etoposide sensitivity of the WT CHO transfectants (Fig. 5D). For this experiment, we generated a CHO transfectant expressing FLAG tagged full-length DNA-PKcs, to rule out the possibility that our results might be an artifact of DNA-PKcs overexpression; additionally, we generated a CHO cell transfectant expressing FLAG tagged mutant ABCDE. We first tested MMC sensitivity and found all cell strains to be similarly sensitive to the crosslinking agent (data not shown). However, cells coexpressing either isoform II or ABCDE were considerably more sensitive to etoposide than were either untransfected CHO cells or CHO cells expressing flag-tagged WT human DNA-PKcs. This effect was more dramatic in the cells expressing ABCDE, probably reflecting the considerably higher expression level of ABCDE compared with isoform II (Fig. 5A). Thus, we conclude that isoform II and mutant ABCDE can inhibit the repair of etoposide damage, even in the presence of WT DNA-PKcs.
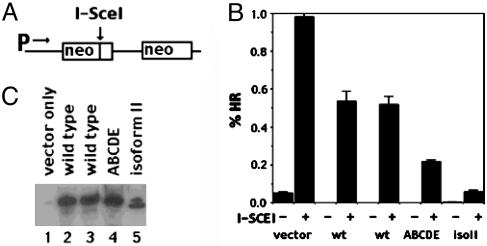
DNA-PKcs Isoform II and ABCDE Mutant Inhibit HR. The data presented thus far are consistent with a model whereby binding of functionally defective DNA-PKcs inhibits repair of DSBs by HR. However, because this conclusion is based entirely on cell survival after genotoxic stress, it is possible that the decreased cell survival reflects indirect effects (i.e., on checkpoint pathways) or effects on other DNA repair pathways. To test whether isoform II and the ABCDE mutant directly inhibit HR, we generated another panel of DNA-PKcs transfectants by using a V3-derived clone that harbors one copy of an integrated HR substrate (26). This substrate (diagramed in Fig. 6A) contains two nonfunctional neomycin resistance genes. The first is nonfunctional because of a frameshift mutation that is coincident with the restriction site for the homing endonuclease, I-SceI. The second is nonfunctional because it lacks a promoter. A DSB can be introduced in the first neo gene by transient expression of I-SceI. It has been shown previously that I-Sce-induced DSBs of this substrate are repaired by HR. In most instances, repair occurs by gene conversion in which both copies of the neo gene are retained (26). Production of G418-resistant colonies is a direct measure of successful DSB repair by HR. I-SceI was transiently expressed in the clonal transfectants, and G418-resistant clones were selected (Fig. 6C). Consistent with previous reports, repair by means of HR is much more robust in cells completely deficient in DNA-PKcs (vector controls) as compared with cells expressing DNA-PKcs (26). However, HR was markedly reduced in cells expressing either isoform II or the ABCDE mutant. Thus, we conclude that NHEJ-defective variants of DNA-PKcs directly inhibit HR.
Fig. 6.
Isoform II and mutant ABCDE inhibit DSB repair by HR. (A) Schematic of HR substrate utilized. (B) DSB-induced HR is shown. (C) Immunoblot analyses of WCE from VD7 cells stably transfected with vector only, WT DNA-PKcs, mutant ABCDE, or isoform II.
Discussion
Several recent reports suggest that DNA-PKcs may inhibit alternative DNA repair pathways by regulating access of DNA ends to repair factors. Dynan and colleagues (20) and Iliakis and colleagues (37) used in vitro end joining systems to demonstrate that kinase inactive DNA-PKcs can inhibit DNA-PKcs-independent subpathway(s) of NHEJ. Additionally, several reports using cellular systems have shown that inactivating DNA-PKcs (by kinase inhibitors) blocks other DNA repair and metabolism pathways. Thus, blocking DNA-PK's kinase activity inhibits both DSB-induced and spontaneous HR (26); inhibits telomere maintenance (38); and inhibits the repair of damage from topoisomerase II poisons (39). Emerging data suggest that DNA-PK that cannot undergo autophosphorylation (either by abrogating its kinase activity or by abrogating its autophosphorylation sites) may remain associated with DNA ends, thus preventing access to other repair factors (16, 18, 19, 40, 41). This hypothesis is a plausible explanation for how inactive DNA-PK inhibits other repair pathways. The genetic approach used here substantiates the idea that DNA-PK that cannot autophosphorylate inhibits alternative DNA repair pathways. However, DNA-PK can bind a variety of DNA discontinuities; thus, it is possible that DNA-PK's inhibitory capacity may vary depending on the DNA lesion involved.
MMC introduces DNA interstrand crosslinks. Turchi et al. (33) have demonstrated that although DNA-PK is able to efficiently bind crosslinked DNA, DNA-PK's enzymatic activity is only weakly induced. Thus, it is possible that once DNA-PK is bound to crosslinked DNA, it may be trapped. This mechanism has been proposed as the explanation for how cis-platinum (another agent that causes interstrand crosslinks) induces radiosensitization (33); furthermore, this mechanism likely explains why cells expressing DNA-PKcs are more sensitive to MMC than are cells lacking DNA-PKcs (Fig. 3B and ref. 7). NHEJ-defective DNA-PKcs variants render cells even more sensitive to MMC than WT DNA-PKcs. Thus, it seems likely that mutant DNA-PKcs is released substantially slower from DNA crosslinks than WT DNA-PKcs, either because of a lack of kinase activity and subsequent autophosphorylation (with isoform II and III) or because of the lack of autophosphorylation sites (with the ABCDE mutant); in either case, defective DNA-PKcs would block access to DNA crosslinks to other repair factors.
Etoposide inhibits the reversion of topoisomerase II DNA covalent intermediates. Repair of this damage occurs by conversion of the lesion to a DSB; either NHEJ or HR can repair these DSBs (reviewed in ref. 34). Mutant ABCDE and isoform II substantially potentiate etoposide sensitivity presumably because they inhibit repair of etoposide damage. We considered that phosphorylation of DNA-PK targets might inhibit the activity of proteins that facilitate the repair of etoposide damage. However, because isoform II (which lacks protein kinase activity) sensitizes cells as effectively as mutant ABCDE (which maintains catalytic activity), this hypothesis seems unlikely. Our current model is that both isoform II and ABCDE physically block access to the etoposide-induced lesions or repair intermediates. To date, it is not known whether DNA-PK binds to covalent topoisomerase II/DNA intermediates. However, it has recently been demonstrated that DNA-PK binds Holliday junctions (42). Thus, it is possible that the NHEJ-defective DNA-PK variants inhibit repair by means of several mechanisms: (i) by binding to the blocked intermediate, (ii) by binding to the converted DSB, or (iii) by binding directly to Holliday junctions during HR. Inhibition of I-SceI-induced DSBs could result either from direct binding to DNA ends, by binding Holliday junctions during HR, or both. Deciphering how NHEJ-defective DNA-PKcs variants inhibit other DNA repair pathways will be of considerable interest.
In haploid yeast, NHEJ is up-regulated and HR is down-regulated, and the opposite is true in diploid yeast (43). Similarly, in mammalian cells, HR is down-regulated in G1 and up-regulated during S/G2 phases (44). These forms of regulation ensure that HR is maximal when appropriate homologous templates (especially sister chromatids) are available for repair and that NHEJ is maximal when such templates are not available. The fact that splice variation leading to the expression of DNA-PKcs isoform II has been conserved in all species analyzed to date suggests that its expression is functionally relevant. Clearly, neither isoform II nor isoform III functions in NHEJ, and neither isoform substantially inhibits NHEJ in the presence of WT DNA-PKcs, but the inhibition of HR by isoform II suggests a possible negative regulatory role. We propose that the high-level expression of isoform II in nondividing cells would afford quiescent cells a mechanism to inhibit repair by HR and thus reduce the potential for loss of genetic information or rearrangements by HR when sister chromatids are not available as repair templates.
Supplementary Material
Acknowledgments
We thank Dr. Martin Gellert for helpful suggestions and insight during manuscript preparation. This work was supported by National Institutes of Health Public Health Service Grants R01A1048758 (to K.M.) and R01 CA100862 (to J.A.N.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NHEJ, nonhomologous end joining; HR, homologous recombination; DNA-PK, DNA-dependent protein kinase; DNA-PKcs, catalytic subunit of DNA-PK; DSB, double-strand break; MMC, mitomycin C; PI3-kinase, phosphatidylinositol 3-kinase; SCID, severe combined immunodeficient; WCE, whole-cell extract.
References
- 1.Walker, A. I., Hunt, T., Jackson, R. J. & Anderson, C. W. (1985) EMBO J. 4, 139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottlieb, T. M. & Jackson, S. P. (1993) Cell 72, 131-142. [DOI] [PubMed] [Google Scholar]
- 3.Dvir, A., Peterson, S. R., Knuth, M. W., Lu, H. & Dynan, W. S. (1992) Proc. Natl. Acad. Sci. USA. 89, 11920-11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Critchlow, S. E. & Jackson, S. P. (1998) Trends Biochem. Sci. 23, 394-398. [DOI] [PubMed] [Google Scholar]
- 5.Pastink, A., Eeken, J. C. & Lohman, P. H. (2001) Mutat. Res. 480–481, 37-50. [DOI] [PubMed] [Google Scholar]
- 6.Pierce, A. J., Stark, J. M., Araujo, F. D., Moynahan, M. E., Berwick, M. & Jasin, M. (2001) Trends. Cell Biol. 11, 52-59. [DOI] [PubMed] [Google Scholar]
- 7.Pluth, J. M., Fried, L. M. & Kirchgessner, C. U. (2001) Cancer Res. 61, 2649-2655. [PubMed] [Google Scholar]
- 8.Mills, K. D., Ferguson, D. O., Essers, J., Eckersdorff, M., Kanaar, R. & Alt, F. W. (2004) Genes Dev. 18, 1283-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dynan, W. S. & Yoo, S. (1998) Nucleic Acids Res. 26, 1551-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeggo, P. A. (1998) Adv. Genet. 38, 185-218. [DOI] [PubMed] [Google Scholar]
- 11.Jackson, S. P. (1997) Int. J. Biochem. Cell Biol. 29, 935-938. [DOI] [PubMed] [Google Scholar]
- 12.Anderson, C. W. & Carter, T. H. (1996) Curr. Top. Microbiol. Immunol. 217, 91-111. [DOI] [PubMed] [Google Scholar]
- 13.Gellert, M. (1997) Adv. Immunol. 64, 39-64. [DOI] [PubMed] [Google Scholar]
- 14.Kienker, L. J., Shin, E. K. & Meek, K. (2000) Nucleic Acids Res. 28, 2752-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurimasa, A., Kumano, S., Boubnov, N. V., Story, M. D., Tung, C. S., Peterson, S. R. & Chen, D. J. (1999) Mol. Cell. Biol. 19, 3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding, Q., Reddy, Y. V., Wang, W., Woods, T., Douglas, P., Ramsden, D. A., Lees-Miller, S. P. & Meek, K. (2003) Mol. Cell. Biol 23, 5836-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lees-Miller, S. P. & Meek, K. (2003) Biochemie 85, 1161-1173. [DOI] [PubMed] [Google Scholar]
- 18.Meek, K., Gupta, S., Ramsden, D. A. & Lees-Miller, S. P. (2004) Immunol. Rev. 200, 132-141. [DOI] [PubMed] [Google Scholar]
- 19.Weterings, E., Verkaik, N. S., Bruggenwirth, H. T., Hoeijmakers, J. H. J. & vanGent, D. C. (2003) Nucleic Acids Res. 31, 7238-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Udayakumar, D., Bladen, C. L., Hudson, F. Z. & Dynan, W. S. (2003) J. Biol. Chem. 278, 41631-41635. [DOI] [PubMed] [Google Scholar]
- 21.Li, Z., Otevrel, T., Gao, Y., Cheng, H. L., Seed, B., Stamato, T. D., Taccioli, G. E. & Alt, F. W. (1995) Cell 83, 1079-1089. [DOI] [PubMed] [Google Scholar]
- 22.Frank, K. M., Sekiguchi, J. M., Seidl, K. J., Swat, W., Rathbun, G. A., Cheng, H. L., Davidson, L., Kangaloo, L. & Alt, F. W. (1998) Nature 396, 173-177. [DOI] [PubMed] [Google Scholar]
- 23.Connelly, M. A., Zhang, H., Kieleczawa, J. & Anderson, C. W. (1996) Gene 175, 271-273. [DOI] [PubMed] [Google Scholar]
- 24.Owaki, H., Varma, R., Gillis, B., Bruder, J. T., Rapp, U. R., Davis, L. S. & Geppert, T. D. (1993) EMBO J. 12, 4367-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin, E. K., Rijkers, T., Pastink, A. & Meek, K. (2000) J. Immunol. 164, 1416-1424. [DOI] [PubMed] [Google Scholar]
- 26.Allen, C., Halbrook, J. & Nickoloff, J. A. (2003) Mol. Cancer Res. 1, 913-920. [PubMed] [Google Scholar]
- 27.Shin, E. K., Perryman, L. & Meek, K. (1997) J. Immunol. 158, 3565-3569. [PubMed] [Google Scholar]
- 28.Fujimori, A., Araki, R., Fukumura, R., Saito, T., Mori, M., Mita, K., Tatsumi, K. & Abe, M. (1997) Genomics 45, 194-199. [DOI] [PubMed] [Google Scholar]
- 29.Hartley, K. O., Gell, D., Smith, G. C., Zhang, H., Divecha, N., Connelly, M. A., Admon, A., Lees-Miller, S. P., Anderson, C. W. & Jackson, S. P. (1995) Cell 82, 849-856. [DOI] [PubMed] [Google Scholar]
- 30.Blunt, T., Gell, D., Fox, M., Taccioli, G. E., Lehmann, A. R., Jackson, S. P. & Jeggo, P. A. (1996) Proc. Natl. Acad. Sci. USA 93, 10285-10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moll, U., Lau, R., Sypes, M. A., Gupta, M. M. & Anderson, C. W. (1999) Oncogene 18, 3114-3126. [DOI] [PubMed] [Google Scholar]
- 32.Thacker, J. & Zdzienicka, M. Z. (2003) DNA Repair 2, 655-672. [DOI] [PubMed] [Google Scholar]
- 33.Turchi, J. J., Henkels, K. M. & Zhou, Y. (2000) Nucleic Acids Res. 28, 4634-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Connelly, J. C. & Leach, D. R. F. (2004) Mol. Cell. 13, 307-316. [DOI] [PubMed] [Google Scholar]
- 35.Jin, S., Inoue, S. & Weaver, D. T. (1998) Carcinogenesis 19, 965-971. [DOI] [PubMed] [Google Scholar]
- 36.Johnson, M. A. & Jones, N. J. (1999) Mutat. Res. 435, 271-282. [DOI] [PubMed] [Google Scholar]
- 37.Perrault, R., Wang, H., Wang, M., Rosidi, B. & Iliakis, G. (2004) J. Cell Biochem. 92, 781-794. [DOI] [PubMed] [Google Scholar]
- 38.Bailey, S. M., Brenneman, M. A., Halbrook, J., Nickoloff, J. A., Ullrich, R. L. & Goodwin, E. H. (2004) DNA Repair 3, 225-233. [DOI] [PubMed] [Google Scholar]
- 39.Willmore, E., de Caux, S., Sunter, N. J., Tilby, M. J., Jackson, G. H., Austin, C. A. & Durkacz, B. W. (2004) Blood 103, 4659-4665. [DOI] [PubMed] [Google Scholar]
- 40.Block, W., Yu, Y., Merkle, D., Gifford, J., Ding, Q., Meek, K. & Lees-Miller, S. P. (2004) Nucleic Acids Res. 32, 4351-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reddy, Y. V., Ding, Q., Lees-Miller, S. P., Meek, K. & Ramsden, D. A. (2004) J. Biol. Chem. 279, 39408-39413. [DOI] [PubMed] [Google Scholar]
- 42.Dip, R. & Naegeli, H. (2004) Biochem. J. 381, 165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nickoloff, J. A. & Haber, J. E. (2001) DNA Damage Repair 3, 107-124. [Google Scholar]
- 44.Rothkamm, K., Krueger, I., Thompson, L. H. & Loebrich, M. (2003) Mol. Cell. Biol. 23, 5706-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.