Abstract
The Agrobacterium T-DNA transporter belongs to a growing class of evolutionarily conserved transporters, called type IV secretion systems (T4SSs). VirB4, 789 aa, is the largest T4SS component, providing a rich source of possible structural domains. Here, we use a variety of bioinformatics methods to predict that the C-terminal domain of VirB4 (including the Walker A and B nucleotide-binding motifs) is related by divergent evolution to the cytoplasmic domain of TrwB, the coupling protein required for conjugative transfer of plasmid R388 from Escherichia coli. This prediction is supported by detailed sequence and structure analyses showing conservation of functionally and structurally important residues between VirB4 and TrwB. The availability of a solved crystal structure for TrwB enables the construction of a comparative model for VirB4 and the prediction that, like TrwB, VirB4 forms a hexamer. These results lead to a model in which VirB4 acts as a docking site at the entrance of the T4SS channel and acts in concert with VirD4 and VirB11 to transport substrates (T-strand linked to VirD2 or proteins such as VirE2, VirE3, or VirF) through the T4SS.
Keywords: structure prediction, bacterial transport, NTPase, homology modeling
Pathogenic bacteria have evolved several strategies to subvert host cell defenses, including secretion of virulence factors directly into the interior of the host cell. This task requires the transport of macromolecules across both the bacterial envelope and the plasma membrane of the host cell, and uses specialized bacterial secretion systems. Type IV secretion systems (T4SSs) comprise a remarkably diverse class in both the variety of substrates transferred (proteins, DNA, or both) and their promiscuity with regard to host cell types. Besides transkingdom transfer from bacteria to eukaryote hosts, T4SSs are used widely by bacteria themselves during genetic exchange by conjugation (1). The importance of T4SSs in disease is revealed by genomic sequence comparison, which has expanded the list of bacterial pathogens harboring T4SS homologs to include Agrobacterium tumefaciens, Bartonella henselae, Bordetella pertussis, several Brucella species (suis, abortus, and melitensis), Campylobacter jejuni, Coxiella burnetii, Helicobacter pylori, Legionella pneumo-phila, Rickettsia prowazekii, Wolbachia spp, and several others (2–5). T4SSs are required for virulence in many of these pathogens.
Much understanding of the structure and function of T4SSs derives from A. tumefaciens, a soil-borne pathogen that genetically transforms plants, and manifests in nature as crown gall disease (reviewed in ref 6). Virulence functions, including the T4SS, are encoded by the Ti (tumor-inducing) plasmid. One specific Vir protein, VirD2, nicks the borders of the T-DNA element on the Ti plasmid, leading to the displacement of a single-stranded DNA segment, the T-strand. VirD2 remains covalently bound to the 5′ end of the T-strand during its transfer to the plant cell where it may aid in integration into the genome. The transformation process is similar to conjugative DNA transfer, in the polar transfer of DNA by means of a T4SS. This similarity is supported further by the ability of the vir-encoded T4SS to transfer exogenous mobilizable plasmids to recipient bacteria and plants (7, 8). Besides DNA, proteinaceous virulence factors, VirE2, VirE3, and VirF, are also transported to the plant cell (9, 10). Researchers capitalize on A. tumefaciens' natural capacity to genetically engineer plants by replacing T-DNA sequences with exogenous DNAs of interest.
The 11 genes of the A. tumefaciens virB operon (virB1-virB11) and the virD4 gene encode the necessary components of the T4SS (reviewed in refs. 1, 2, and 11). VirD4 is suggested to act as a coupling factor, bringing the T-strand and its associated VirD2 to the T4SS for translocation to plant cells (12, 13). T4SS components form several discrete structures. An exterior pilus is formed from VirB2 and VirB5. The structural core of the channel is assembled from VirB7 to VirB10. Recent evidence suggests VirB6 is likely to be also a major core component at the inner membrane interface (14). VirB1 has two functions; it acts as a transglycosylase to loosen the peptidoglycan layer, and it forms a processed product found on the exterior of the cell that may act to mediate plant cell contact (15). VirB3 does not have an assigned function to date.
Three proteins, VirB4, VirB11, and VirD4 have NTP-binding domains that likely energize T4SS assembly or function (2, 4, 16, 17). Crystal structures for VirB11 and VirD4 homologs, H. pylori HP0525 and Escherichia coli TrwB, respectively, (18, 19) show hexameric ring structures embedded in the inner membrane. VirB11 NTP-binding activity is correlated with T-DNA transfer (20, 21), and may coordinate assembly of the VirB11 hexamer (19). The TrwB Walker A NTP-binding motif is necessary for the conjugative transfer of plasmid R388 (22).
VirB4 has an NTP-binding site comprised of residues in its Walker A and B motifs, and is thought to form part of the T4SS entry pore because of its predicted localization in the inner membrane (1–3, 5). VirB4, along with VirB7–B11, represent the most conserved T4SS components in a diverse range of pathogens (2, 3, 5). This high conservation predicts VirB4 plays a prime role in T4SS structure and/or function. Here, we perform bioinformatics analyses that predict a homology between the C-terminal domain (residues 425–789) of VirB4 and the cytoplasmic domain of TrwB, the VirD4 homolog from plasmid R388 of E. coli. These results lead to a model for the assembly and function of the T4SS where VirB4 forms a dock at the entrance of the channel to facilitate transport of T4SS DNA or protein substrates.
Materials and Methods
The VirB4 sequence used in these experiments derives from A. tumefaciens strain C58 (GenBank accession no. NP_536288, SwissProt accession no. VIB4_AGRT5, and Uniprot accession no. P17794). Bioinformatics tools described below were drawn from both publicly available resources and tools developed by the Berkeley Phylogenomics Group (which can be accessed at http://phylogenomics.berkeley.edu/resources). Protein sequences were obtained from the UniProt database (which can be accessed at www.uniprot.org). Unless otherwise noted, sequences are referred to by using their UniProt accession number. The Protein Data Bank (PDB) was used as a source of 3D crystal structures in modeling. All supplementary materials, including multiple sequence alignments (MSAs), can be accessed at http://phylogenomics.berkeley.edu/VIRB4.
Web Servers, Homology Predictions, and Modeling. The phylofacts structure prediction hidden Markov model (HMM) library (which can be accessed at http://phylogenomics.berkeley.edu/phylofacts) was used to identify structural domains. The flowerpower web server was used to identify homologs included in alignments (which can be accessed at http://phylogenomics.berkeley.edu/flowerpower). The muscle server (which can be accessed at http://phylogenomics.berkeley.edu/cgi-bin/muscle/input_muscle.py) was used to construct MSAs. Other web servers providing protein structure prediction include 3d-pssm (which can be accessed at www.sbg.bio.ic.ac.uk/~3dpssm), superfamily (which can be accessed at http://supfam.mrclmb.cam.ac.uk/SUPERFAMILY), and smart (which can be accessed at http://smart.embl-heidelberg.de). The psi-blast web server at the National Center for Biotechnology Information (which can be accessed at www.ncbi.nlm.nih.gov) and the pfam web server (which can be accessed at www.sanger.ac.uk/Software/Pfam) were used in supplementary analyses.
HMM software from the University of California, Santa Cruz sequence alignment and modeling (SAM) system suite (which can be accessed at www.cse.ucsc.edu/research/compbio/sam.html) was used to construct the HMMs used for database searches. For consistency in HMM homolog searches of databases of different sizes, E values were computed by using a database size of 1,000.
Transmembrane (TM) prediction was performed on VirB4 and a representative set of VirB4 homologs gathered by using the flowerpower server, and made nonredundant at 80% identity. To ensure that the proteins included in this analysis had the same overall domain architecture, we disallowed any sequences that were significantly longer or shorter than VirB4 and required at least 25% identity to VirB4. This set [VirB4 (P17794), Q8KIM8, Q9R2W4, Q9PHJ8, Q8PJC1, Q9A5M8, Q7WDU1, Q7 × 263, Q981S5, Q8RPM6, Q8FXK3, Q7PAP3, Q46698, Q9RLS1, Q7 × 3L6, Q8PRI7, and Q9EUF8] were submitted to the tmhmm server version 2.0 (which can be accessed at www.cbs.dtu.dk/services/TMHMM). We also submitted VirB4 to the hmmtop server (which can be accessed at www.enzim.hu/hmmtop) and to the das server (which can be accessed at www.sbc.su.se/~miklos/DAS/maindas.html).
MSA of TrwB and VirB4 Proteins. Homologs to the VirB4 C-terminal domain were identified by using flowerpower. The corresponding regions of these sequences were aligned by using muscle (to construct the VirB4 Cterm MSA), and the alignment was cropped to include only those columns matching the C-terminal domain of VirB4. Indel characters were removed to produce a set of 92 unaligned C-terminal domains for VirB4 and homologs. This process was repeated for the TrwB proteins, yielding 44 C-terminal domains. The muscle server was used to construct a joint MSA of the TrwB and VirB4 C-terminal domains (the VirB4-TrwB NTPase MSA). To construct a representative MSA shown in Fig. 4, we selected sequences from the VirB4-TrwB NTPase MSA by first removing sequences having >80% identity to others to produce a nonredundant set of proteins. From this set, we identified the top four sequences (based on percent identity) to both VirB4 (P17794) and TrwB (Q04230). This procedure produced a set of proteins with pairwise identities to VirB4 ranging from 36.7% to 39.3%. The same procedure was repeated to select four related sequences to TrwB (with pairwise identities to TrwB ranging from 28.8% to 43.9%). The belvu software (which can be accessed at www.cgb.ki.se/cgb/groups/sonnhammer/Belvu.html) was used for alignment editing and display.
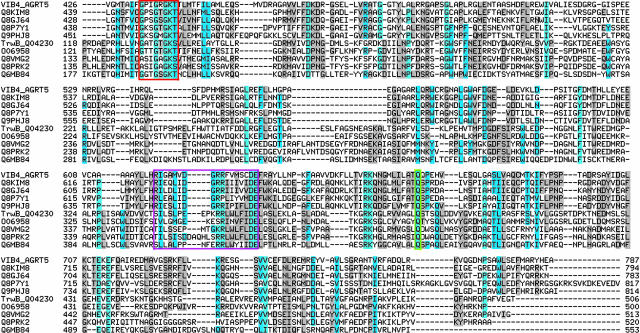
Fig. 4.
MSA of VirB4 and TrwB and selected homologs along their NTP-binding domains. Sequence homologs to VirB4 are shown directly below VirB4, followed by TrwB and homologs. Walker A residues are boxed in red, Walker B residues are boxed in magenta, and the conserved glutamine is highlighted in green.
Comparative Model Construction. Comparative (homology) model construction of both the monomer and hexamer structures was performed by using modeller software (23) provided through the Berkeley Phylogenomics Group web server (which can be accessed at http://phylogenomics.berkeley.edu/homology_model). The pairwise alignment of TrwB and VirB4 (Fig. 2), used as the basis for the homology model (Fig. 3), was produced in a multistep process. The VirB4 Cterm MSA was used to construct an HMM by using sam w0.5 software. This HMM was used to score and select TrwB homologs by using the hmmscore program, and an MSA of top-scoring TrwB homologs and VirB4 and homologs to this HMM was obtained with the align2model program. The pairwise alignment of VirB4 and TrwB was extracted from this multiple alignment. The alignment is displayed by using the boxshade web server (which can be accessed at http://bioweb.pasteur.fr/seqanal/interfaces/boxshade.html).
Fig. 2.
VirB4-TrwB pairwise alignment. The cytoplasmic domain of TrwB (residues 122–507) and the C terminus of VirB4 (residues 425–789) are aligned, with identical and similar residues shown in black and gray, respectively. Boxed areas highlight the Walker A and B motifs. The Walker A motif is conserved across VirB4 and TrwB (VirB4, G433-T440; TrwB, G130-S137). The Walker B motif is found in its entirety in VirB4 (R619-E635), whereas a truncated version is evident in TrwB (R349-E357).
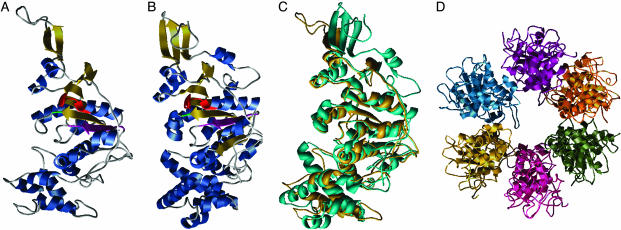
Fig. 3.
Homology model for the C terminus of VirB4. (A) VirB4 monomer model. α-helices are blue and β-sheets are gold. The Walker A motif (G433-T440) is red, the Walker B motif (R619-E635) is magenta, and the conserved Q668 is green. (B) Monomer of TrwB, using the same coloring as for VirB4. (C) Superposition of VIRB4 (turquoise) and TrwB (gold). (D) Predicted hexameric structure of VirB4.
Results
Predicted Homology Between VirB4 and TrwB. Our long-term goal is to understand the structure and function of the T4SS of Agrobacterium. To this end, it is critical to determine the 3D structure of individual T4SS components. VirB4, at 789 aa, is the largest T4SS component, and has domains that may be exposed to the cytoplasm and periplasm by spanning the inner membrane (24), providing a rich source of possible structural domains. Thus, we set out to use bioinformatics approaches coupled with the growing availability of structural information in the databases to predict the structure of VirB4.
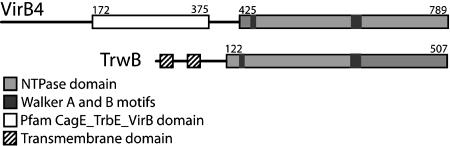
As diagrammed in Fig. 1 (and detailed in Figs. 2, 3, 4, 5 below), our analyses support a prediction that the C terminus of A. tumefaciens VirB4 (starting at D425) and the cytoplasmic domain of the E. coli TrwB-coupling protein are related by divergent evolution from a common ancestor and share a similar structural fold and function. TrwB is a hexameric integral membrane protein of 507 aa encoded by the R388 plasmid (18, 25), and is suggested to function in moving the relaxosome DNA–protein complex to the T4SS. TrwB has an N-terminal TM-spanning region (residues 1–70) and a 437-aa C-terminal domain with a solved structure [Protein Data Bank (PDB) ID code 1E9RA] classified by the Structural Classification of Proteins database to the RecA protein-like (ATPase-domain) family of P-loop-containing nucleoside triphosphate hydrolases (26). The N terminus of VirB4 contains a conserved domain (Pfam CagE_TrbE_VirB) of unknown structure from residues 172–375.
Fig. 1.
Domain organization of VirB4 and TrwB. VirB4 and TrwB share a C-terminal NTP-binding domain but have different N termini. The N terminus of VirB4 matches the pfam domain CagE_TrbE_VirB, but has no known function or structure, whereas the TrwB N terminus contains two predicted TM helices.
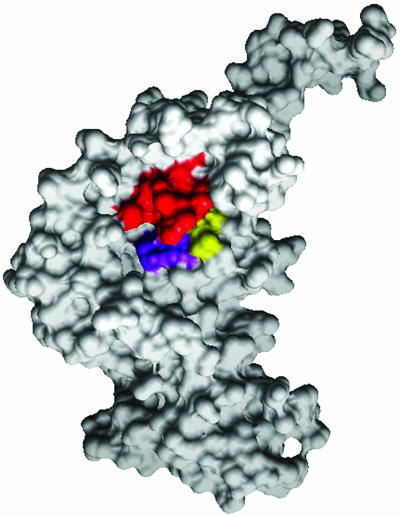
Fig. 5.
Surface image of VirB4 monomer, highlighting the NTP-binding cleft. To better display the NTP-binding cleft, we show the structure from a different perspective, corresponding to the back side of Fig. 3A. The conserved lysine (K) of the Walker A motif is red, the conserved DE of the Walker B motif is purple, and the conserved glutamine (Q) is highlighted in yellow.
Fig. 2 shows a pairwise alignment between VirB4 and TrwB. Whereas the overall sequence similarity in this alignment is low (15% identity, 21% identity plus similarity in the alignment shown), agreement at known functional motifs and at other conserved positions defining the VirB4 and TrwB families separately, and global alignment of the C-terminal 365-residue domain of VirB4 to the entire TrwB cytoplasmic domain, enable us to predict that the C terminus of VirB4 is homologous to TrwB. A comparative model for this domain of VirB4 based on the structure of TrwB is shown in Fig. 3. Our structure prediction analyses are described below.
Domain Structure Prediction. We submitted the full VirB4 sequence to a number of domain prediction web servers. Whereas blast and other simple pairwise sequence comparison methods were unable to detect any homologous folds for VirB4, structure prediction servers employing libraries of profiles or HMMs (27) produced the strongest evidence for a predicted structure of the VirB4 C terminus. Submission of VirB4 to smart (28) found a match to the TrwB cytoplasmic domain (Structural Classification of Proteins domain d1e9ra) with an expectation value (E value) of 2.0 × 10–44. The Berkeley phylofacts structure prediction server predicted the same homology with an E value of 3.69 × 10–21. The 3d-pssm (29) and superfamily (30) structure prediction servers also predicted this structure, albeit with weaker E values (5.1 × 10–4 and 0.038, respectively).
Parallel analyses in the reverse direction supported the same prediction. First, we constructed an HMM for the full-length VirB4 sequence and global homologs (the VirB4 global HMM), and scored the PDB data set of proteins whose structures have been solved. The top-scoring hit (with an E value of 0.01) was the TrwB cytoplasmic domain (PDB ID code 1E9RA). Examination of the alignment of the TrwB cytoplasmic domain to the HMM showed that TrwB matches the C terminus of VirB4, with agreement at the Walker A and B motifs. Next, an HMM constructed from the VirB4 Cterm MSA was used to detect and align additional sequences from UniProt, by using an E value cutoff of 10–05. The new alignment was used to construct a revised HMM, followed by scoring PDB once again. This time, the TrwB structure again received the top score but with a stronger E value (1.5 × 10–06). Finally, to avoid spurious matches between VirB4 and TrwB based on only local similarity at the highly conserved Walker A motif, we removed the initial columns of the VirB4 Cterm MSA up to and just past the Walker A motif to construct an HMM lacking this conserved motif. This HMM also detected the TrwB structure and homologs (albeit with slightly less significant scores). This three-stage analysis clearly identifies the local region of homology between VirB4 and TrwB (step 1), confirms the significance of the match through intermediate sequences (step 2), and reduces the likelihood of a spurious match based simply on agreement at the conserved Walker A motif (step 3).
By contrast, psi-blast searches at the National Center for Biotechnology Information database produced mixed results. A psi-blast search against the nonredundant protein database by using VirB4 as a query yielded no proteins of solved structure in the first five iterations of psi-blast. However, submitting TrwB as the query identified several VirB4 homologs with significant E values in the second iteration (e.g., O24862 VirB4 homolog of H. pylori 26695 with an E value of 2 × 10–8) and identified A. tumefaciens VirB4 in the sixth iteration with an E value of 9 × 10–46.
Finally, a similar analysis of two A. tumefaciens VirB4 homologs, H. pylori tfts3 ORFG (Q8RMY9) and Ruegeria sp. PR1b RC161 (Q8KW29), supported the same TrwB homology.
Thus, the predicted homology between the C terminus of VirB4 and the cytoplasmic domain of TrwB is supported by several independent lines of evidence. Consensus approaches such as this have been shown in previous Critical Assessment of protein Structure Prediction experiments to be very reliable predictors of homology, even in cases where sequence identity is vanishingly low (31).
Attempts to predict the 3D structure of the N terminus of VirB4 were unsuccessful. We await more entries of solved structures into the PDB database to probe for structural homologs to this region in the future.
TM Prediction. VirB4 was previously predicted to contain several TM domains; two in the N terminus, and two in the C terminus (24). Our predicted globular structure for the C-terminal domain of VirB4 disallows the presence of TM domains in this region. In addition, our analyses of VirB4 and a representative set of global homologs by using several TM prediction servers do not support the presence of TM domains. Because TM prediction is known to have high false-positive and -negative rates (32), experimental investigation is required to confirm or refute the presence of TM domains in VirB4. In fact, a biological study that assayed for alkaline phosphatase activity in fusions to VirB proteins failed to predict TM domains for VirB4 (33).
Comparative Model Construction and Structure Analysis. The accuracy of a comparative model is directly related to the accuracy of the pairwise alignment between the target (unknown structure) and template protein whose structure is known, and correlates highly with the pairwise sequence identity. Structural analyses of homologous proteins having <20% pairwise identity show that only a fraction of their residues can be structurally superposed. This finding obviously imposes an upper limit on the accuracy of a comparative model based on a very remotely related template (34). To improve model accuracy, we constructed and examined a number of alignments. Our final model, based on agreement at the Walker A and B motifs and other conserved residues, was produced by using the alignment shown in Fig. 2. Fig. 3 shows the predicted VirB4 monomer (Fig. 3A), the TrwB structure (Fig. 3B), the superposition of the predicted VirB4 and TrwB monomer structures (Fig. 3C), and a model for the VirB4 hexamer (Fig. 3D). These models display the proximity of the Walker A and B motifs in the individual and superposed structures, which is consistent with their functional cooperation. The presence of Walker A and B motifs in the VirB4 protein has been noted by others (16). Here, we extend this observation from agreement at local sequence motifs to predict an evolutionary relationship and similarity at the 3D-structure level between the C terminus of VirB4 and the TrwB cytoplasmic NTP-binding domain.
Fig. 4 presents a representative MSA for VirB4, TrwB, and their homologs, along their region of predicted homology (the VirB4 C terminus and the TrwB cytoplasmic domain). Selected sequence motifs conserved across the set are highlighted in this MSA for comparison with their location on the homology models presented in Fig. 3. The Walker B motif was originally defined as [R/K]X3GX3Lh4D, where h is any hydrophobic residue (35). Subsequent analysis of a 19-member VirB4 family of homologs redefined the Walker B motif as h4DE[A/F]W (16). Our sequence analysis of an evolutionarily wide spectrum of TrwB and VirB4 homologs shows only the DE residues of the Walker B motif to be highly conserved; all other positions (including the terminal tryptophan, which is found in most VirB4 homologs, but not in TrwB and homologs) show variability. Structure analysis reveals that only the conserved DE residues are structurally proximal to the Walker A motif (particularly the terminal conserved GK[S/T]), whereas the remainder of the Walker B motif forms a β-strand that presumably allows for more substitutions (conserving structure). In addition, the model in Fig. 3 and the MSA in Fig. 4 display a conserved glutamine found in virtually all homologs to both TrwB and VirB4. This residue (Q668 in VirB4 and Q386 in TrwB) is structurally proximal to both the conserved lysine (K439 in VirB4 and K136 in TrwB) in the Walker A motif and the conserved DE (residues 634 and 635 in VirB4, and residues 356 and 357 in TrwB) of the Walker B motif. The combination of family-wide conservation and structural proximity to the functionally critical Walker A and B motifs leads us to predict this residue participates in NTP-binding function. The proximity of the glutamine to the Walker A and B motifs is even more evident in the space-filling model of the C terminus of VirB4 shown in Fig. 5.
The VirB4 hexameric structure prediction is supported not only by structural homology between the TrwB and VirB4 monomers but also by examination of patterns of conserved residues across the VirB4 and TrwB MSAs. Most notably, both structures have core NTP-binding sites, a property shared with numerous hexameric protein structures with NTP-binding function (36). Furthermore, joint analysis of the TrwB hexamer structure and conservation patterns in the MSA of VirB4 and TrwB homologs reveals the presence of similarly placed conserved residues on the interface between monomers (e.g., the RK motif at positions 657–658 in VirB4 and positions 376–376 in TrwB); such a hydrophilic interface between monomers of TrwB has been noted (18).
Discussion
A variety of structure prediction methods and comparative sequence analysis of VirB4 and TrwB homologs support a prediction that the C terminus of the A. tumefaciens VirB4 monomer is related by divergent evolution to the cytoplasmic domain of TrwB, and, as with TrwB, the VirB4 protein forms a hexamer. This result is consistent with earlier data, demonstrating that B4 forms dimers or multimers (37).
Below, we comment on the function of VirB4 in the context of the Agrobacterium-specific T4SS, by discussing the Agrobacterium-specific TrwB homolog, VirD4. VirD4 (P18594) and TrwB are related by divergent evolution along their NTP-binding domains, where they share a common fold and molecular function (data not shown, see also ref. 25). Whereas pairwise sequence comparison using blast shows no significant similarity, HMM searches support the predicted homology (e.g., an E value of 4.96 × 10–38 for VirD4 similarity to the phylofacts TrwB HMM). The low sequence similarity between VirD4 and TrwB (15% identity) is not surprising because they interact with their cognate relaxosomes and T4SSs, which may be evolutionarily related but divergent.
That VirB4 and VirD4 have similar structures predicts that both proteins interact with other T4SS components in a spatially related manner. How might this occur? There are several alternative models, starting from the assumption that both VirB4 and VirD4 can interact with the VirB11 hexamer at the entrance of the T4SS. This assumption is supported by two lines of evidence. First, VirB11 and VirB4 interact in two-hybrid analyses (38). Second, whereas T-complex (T-strand linked to VirD2) coupled to VirD4 interacts with VirB11, effective transport through the T4SS requires VirB4 (39). One model might propose that VirB4 and VirD4 compete for binding to VirB11. However, this model would lead to the exclusion of VirB4 (or VirD4) from some T4SS channels. It is more likely that VirB4 is essential to the assembly and/or function of each channel because of the strong conservation of VirB4 in T4SSs (2–5). One possibility is that VirB4 may be used as a scaffold to build the shape of the entrance in concert with VirB11 and other T4SS components. Once this shape is constructed, VirB4 may be displaced by VirD4. Although this model cannot be discounted at this time, we favor a model where VirB4 is used both structurally and functionally during transport of substrate through the channel.
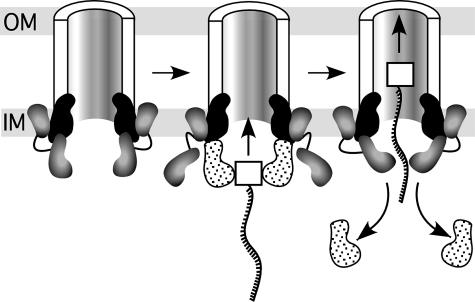
In the model in Fig. 6, the N terminus of VirB4 anchors VirB4 to the inner membrane, and the C-terminal hexamer of VirB4 is stacked with the VirB11 hexamer. The N terminus likely forms an independent structural domain, and may be separated from the C terminus by an unstructured hinge that potentially allows each domain to act independently. We then propose that VirB4 undergoes a conformational change in the presence of VirD4, so that VirD4 (carrying the VirD2–T-strand complex) now stacks with VirB11. The similar structures of the C terminus of VirB4 and the cytoplasmic domain of VirD4 would allow subunit exchange. In fact, VirD4 has been suggested to exist in equilibrium between monomers and hexamers (40), and this state may facilitate subunit exchange with VirB4. By repeated subunit exchange, VirD4 would be displaced, and the T-complex would move up to the VirB11 hexamer, facilitated by VirB4. In this model, either VirD4 also brings protein substrates (such as VirE2, VirE3, or VirF) to the T4SS for transport as above, or VirD4 interaction with VirB4 activates the channel to then allow proteins substrate transport via VirB4.
Fig. 6.
Model for the function of VirB4 during the type IV secretion process. VirB4 is shaded in gray as two domains linked by a hinge (solid line). VirB11 and VirD4 are black and stippled, respectively. For simplicity, only two subunits of VirB4, VirD4, and VirB11 are shown, and the rest of the T4SS is drawn as a half cylinder. VirD2 is shown as a white square attached to the T-strand (wave ladder-like line). Arrows indicate subunit exchange between VirB4 and VirD4. See text for details. OM, outer membrane; IM, inner membrane.
The model in Fig. 6 suggests that hexamers of both VirB11 and VirB4 are stacked to form the entrance to the T4SS; their NTP-binding activities may then act in concert to facilitate substrate transport. There is precedent for such two-tiered hexameric architecture in the p97 AAA ATPase (41). Furthermore, given the similarity of the structures of VirB4 C terminus and VirD4, a second two-tiered structure may form when monomers of the C terminus of VirB4 are displaced by the VirD4 hexamer carrying the T-complex. Such exchange may be facilitated by VirD4 association with the inner membrane through its predicted N-terminal TM (33), and by association with VirB10 (42, 43). That antibodies to VirB4, VirB11, or VirD4 each precipitate complexes of all three proteins supports an intimate interaction between all three hexamers at the entrance to the T4SS channel (39).
Models to date have placed VirD4 hexamers or VirB4 oligomers external to the T4SS, bracketing the sides of the centrally located VirB11 entrance position. This positioning of VirB4 or VirD4 suggested that substrate is transported to the channel proper by shoot-and-pump (13) or ping-pong (39) mechanisms, requiring substrate to shuttle inward from a more lateral position toward an internal VirB11 and other T4SS entry components. The present model is simpler, with VirB4 (and VirD4) directly docked in the central position with VirB11.
A recent elegant series of experiments by using transfer DNA immunoprecipitation (TrIP) suggest an ordered stepwise pathway for the transport of the T-strand DNA substrate through the T4SS (39, 44). First, the T-complex interacts with the VirD4 coupling protein. This T-complex then interacts with VirB11, followed by VirB6 and VirB8. Although VirB6 and VirB8 interactions could not be ordered, both require VirB4. The complex then interacts with VirB9 and VirB2, in a process requiring VirB3, VirB5, and VirB10. This pathway fits with our proposed model because it places VirB4 function at one of the early steps of the transfer pathway, likely acting in concert with VirB11. Furthermore, the TrIP assay suggests the T-strand contacts VirD4, VirB11, VirB6, VirB8, VirB9, and VirB2, but not VirB4. The model in Fig. 6 accommodates these results by subunit exchange between the VirB4 C terminus and VirD4, leading to T-complex transport to the VirB11 hexamer, facilitated by a conformational change in VirB4. Finally, the TrIP assay reveals that VirD4–T-complex can interact with VirB11 in a strain producing only VirD4, and VirB7–11. This interaction could not proceed to VirB6 and VirB8 in the absence of VirB4, suggesting VirB4 may also facilitate productive binding of VirD4 to the channel, and subsequent T-strand transport (39).
That all three T4SS NTP binding proteins, VirB4, VirB11, VirD4, are predicted to form hexameric structures agrees with the solved structures of a variety of hexameric ATPases (36). It is intriguing that all three hexamers likely interact with each other in T4SSs. To accommodate these interactions and to facilitate substrate transfer, we predict that the T4SS undergoes major structural rearrangements. Many ATPases involved in a diverse array of cellular processes (including DNA replication and recombination, proteolysis, membrane fusion, and molecular motors) undergo alterations in structure during function. VirB4 interacts with itself and at least four other T4SS components, VirB1, VirB8, VirB10, and VirB11 (ref. 38 and O. Draper and P.Z., unpublished work). In the context of the T4SS channel, these additional interactions likely facilitate the function of VirB4 to shape the channel entrance and facilitate the mechanical force required during the secretion process. As suggested above, VirB4 may act as both a docking and pumping agent. Future studies will test the hypotheses presented here.
Acknowledgments
We thank John Zupan (University of California, Berkeley) for intellectual and graphical input, and Dan Kirshner (University of California, Berkeley) and Eswar Narayanan (University of California, San Francisco) for assistance in homology modeling tasks. This work was supported by National Science Foundation Presidential Early Career Award for Scientists and Engineers 0238311 (to N.K. and K.S.), R.M. was supported by a National Science Foundation Predoctoral Fellowship (to R.M.) and National Science Foundation Grant 0343566 (to P.Z.).
Abbreviations: HMM, hidden Markov model; T4SS, type IV secretion system; MSA, multiple sequence alignment; TM, transmembrane; PDB, Protein Data Bank.
References
- 1.Christie, P. J. (2004) Biochim. Biophys. Acta 1694, 219–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron, C., O'Callaghan, D. & Lanka, E. (2002) Mol. Microbiol. 43, 1359–1365. [DOI] [PubMed] [Google Scholar]
- 3.Cao, T. B. & Saier, M. H., Jr. (2001) Microbiology 147, 3201–3214. [DOI] [PubMed] [Google Scholar]
- 4.Christie, P. J. & Vogel, J. P. (2000) Trends Microbiol. 8, 354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sexton, J. A. & Vogel, J. P. (2002) Traffic 3, 178–185. [DOI] [PubMed] [Google Scholar]
- 6.Escobar, M. A. & Dandekar, A. M. (2003) Trends Plant Sci. 8, 380–386. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan-Wollaston, V., Passiatore, J. & Cannon, F. (1987) Nature 328, 172–175. [Google Scholar]
- 8.Fullner, K. J. (1998) J. Bacteriol. 180, 430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vergunst, A. C., Jansen, L. E., Fransz, P. F., de Jong, J. H. & Hooykaas, P. J. (2000) Chromosoma 109, 287–297. [DOI] [PubMed] [Google Scholar]
- 10.Schrammeijer, B., den Dulk-Ras, A., Vergunst, A. C., Jurado Jacome, E. & Hooykaas, P. J. (2003) Nucleic Acids Res. 31, 860–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zupan, J., Muth, T. R., Draper, O. & Zambryski, P. (2000) Plant J. 23, 11–28. [DOI] [PubMed] [Google Scholar]
- 12.Kumar, R. B. & Das, A. (2002) Mol. Microbiol. 43, 1523–1532. [DOI] [PubMed] [Google Scholar]
- 13.Llosa, M., Gomis-Ruth, F. X., Coll, M. & de la Cruz, F. (2002) Mol. Microbiol. 45, 1–8. [DOI] [PubMed] [Google Scholar]
- 14.Jakubowski, S. J., Krishnamoorthy, V., Cascales, E. & Christie, P. J. (2004) J. Mol. Biol. 341, 961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llosa, M., Zupan, J., Baron, C. & Zambryski, P. (2000) J. Bacteriol. 182, 3437–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabel, C., Grahn, A. M., Lurz, R. & Lanka, E. (2003) J. Bacteriol. 185, 1045–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rashkova, S., Zhou, X. R., Chen, J. & Christie, P. J. (2000) J. Bacteriol. 182, 4137–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomis-Ruth, F. X., Moncalian, G., Perez-Luque, R., Gonzalez, A., Cabezon, E., de la Cruz, F. & Coll, M. (2001) Nature 409, 637–641. [DOI] [PubMed] [Google Scholar]
- 19.Yeo, H. J., Savvides, S. N., Herr, A. B., Lanka, E. & Waksman, G. (2000) Mol. Cell 6, 1461–1472. [DOI] [PubMed] [Google Scholar]
- 20.Krause, S., Pansegrau, W., Lurz, R., de la Cruz, F. & Lanka, E. (2000) J. Bacteriol. 182, 2761–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sagulenko, E., Sagulenko, V., Chen, J. & Christie, P. J. (2001) J. Bacteriol. 183, 5813–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moncalian, G., Cabezon, E., Alkorta, I., Valle, M., Moro, F., Valpuesta, J. M., Goni, F. M. & de La Cruz, F. (1999) J. Biol. Chem. 274, 36117–36124. [DOI] [PubMed] [Google Scholar]
- 23.Sali, A. & Blundell, T. L. (1993) J. Mol. Biol. 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 24.Dang, T. A. & Christie, P. J. (1997) J. Bacteriol. 179, 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomis-Ruth, F. X., Moncalian, G., de la Cruz, F. & Coll, M. (2002) J. Biol. Chem. 277, 7556–7566. [DOI] [PubMed] [Google Scholar]
- 26.Murzin, A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995) J. Mol. Biol. 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 27.Krogh, A., Brown, M., Mian, I. S., Sjölander, K. & Haussler, D. (1994) J. Mol. Biol. 235, 1501–1531. [DOI] [PubMed] [Google Scholar]
- 28.Letunic, I., Copley, R. R., Schmidt, S., Ciccarelli, F. D., Doerks, T., Schultz, J., Ponting, C. P. & Bork, P. (2004) Nucleic Acids Res. 32, D142–A144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. (2000) J. Mol. Biol. 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 30.Gough, J., Karplus, K., Hughey, R. & Chothia, C. (2001) J. Mol. Biol. 313, 903–919. [DOI] [PubMed] [Google Scholar]
- 31.Fischer, D. & Rychlewski, L. (2003) Protein Eng. 16, 157–160. [DOI] [PubMed] [Google Scholar]
- 32.Chen, C. P., Kernytsky, A. & Rost, B. (2002) Protein Sci. 11, 2774–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das, A. & Xie, Y. H. (1998) Mol. Microbiol. 27, 405–414. [DOI] [PubMed] [Google Scholar]
- 34.Baker, D. & Sali, A. (2001) Science 294, 93–96. [DOI] [PubMed] [Google Scholar]
- 35.Walker, J. E., Saraste, M., Runswick, M. J. & Gay, N. J. (1982) EMBO J. 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vale, R. D. (2000) J. Cell Biol. 150, F13–F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dang, T. A., Zhou, X. R., Graf, B. & Christie, P. J. (1999) Mol. Microbiol. 32, 1239–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward, D. V., Draper, O., Zupan, J. R. & Zambryski, P. C. (2002) Proc. Natl. Acad. Sci. USA 99, 11493–11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atmakuri, K., Cascales, E. & Christie, P. J. (2004) Mol. Microbiol. 54, 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hormaeche, I., Alkorta, I., Moro, F., Valpuesta, J. M., Goni, F. M. & de la Cruz, F. (2002) J. Biol. Chem. 277, 46456–45462. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, X., Shaw, A., Bates, P. A., Newman, R. H., Gowen, B., Orlova, E., Gorman, M. A., Kondo, H., Dokurno, P., Lally, J., et al. (2000) Mol. Cell 6, 1473–1484. [DOI] [PubMed] [Google Scholar]
- 42.Gilmour, M. W., Gunton, J. E., Lawley, T. D. & Taylor, D. E. (2003) Mol. Microbiol. 49, 105–116. [DOI] [PubMed] [Google Scholar]
- 43.Llosa, M., Zunzunegui, S. & de la Cruz, F. (2003) Proc. Natl. Acad. Sci. USA 100, 10465–10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cascales, E. & Christie, P. J. (2004) Science 304, 1170–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]