Abstract
Many pathogenic bacteria produce extracellular DNase, but the benefit of this enzymatic activity is not understood. For example, all strains of the human bacterial pathogen group A Streptococcus (GAS) produce at least one extracellular DNase, and most strains make several distinct enzymes. Despite six decades of study, it is not known whether production of DNase by GAS enhances virulence. To test the hypothesis that extracellular DNase is required for normal progression of GAS infection, we generated seven isogenic mutant strains in which the three chromosomal- and prophage-encoded DNases made by a contemporary serotype M1 GAS strain were inactivated. Compared to the wild-type parental strain, the isogenic triple-mutant strain was significantly less virulent in two mouse models of invasive infection. The triple-mutant strain was cleared from the skin injection site significantly faster than the wild-type strain. Preferential clearance of the mutant strain was related to the differential extracellular killing of the mutant and wild-type strains, possibly through degradation of neutrophil extracellular traps, innate immune structures composed of chromatin and granule proteins. The triple-mutant strain was also significantly compromised in its ability to cause experimental pharyngeal disease in cynomolgus macaques. Comparative analysis of the seven DNase mutant strains strongly suggested that the prophage-encoded SdaD2 enzyme is the major DNase that contributes to virulence in this clone. We conclude that extracellular DNase activity made by GAS contributes to disease progression, thereby resolving a long-standing question in bacterial pathogenesis research.
Keywords: virulence factor, Streptococcus pyogenes, neutrophil extracellular traps
Many pathogenic bacteria produce extracellular DNase, but the benefit of this enzymatic activity is not understood. It has been hypothesized that secretion of DNase provides a growth advantage by enlarging the pool of available nucleotides by DNA hydrolysis (1). Extracellular DNase activity also has been hypothesized to play a role in the dissemination and spread of infecting bacteria by liquifying pus (2–6). In addition, a recent study implied a role for extracellular DNases in the evasion of the innate immune response by degrading neutrophil extracellular traps (NETs). NETs trap and kill bacteria extracellularly, are composed of chromatin and granule proteins, and their structure is dissipated by DNase activity (7). The extracellular bactericidal activity of neutrophils directed against Shigella flexneri and Staphylococcus aureus was reduced greatly when incubated with exogenously added DNase (7).
Group A Streptococcus (GAS) is a bacterial pathogen responsible for many serious human diseases (8, 9). The wide diversity of disease manifestations caused by GAS infection is thought to be due in part to the variable number of secreted and cell-wall anchored virulence factors produced by this pathogen. These factors include capsule, M protein, streptococcal inhibitor of complement, streptococcal pyrogenic toxin superantigens, hemolysins, and several DNases (8–11). DNases were among the first secreted GAS proteins to be identified and extensively characterized (6, 12–17). However, after almost 60 years of investigation, there is no direct evidence that DNases are virulence factors (18–23). Many indirect lines of evidence implicate DNases in GAS pathogenesis. First, all GAS strains produce at least one DNase, and most strains make several distinct enzymes (13, 15–17). Second, expression of DNase is up-regulated upon interaction of GAS with human epithelial cells, polymorphonuclear leukocytes (PMNs), and oropharynx (20, 21, 24, 25). Third, patients with GAS infections seroconvert to DNases, indicating that these proteins are produced in vivo and are immunogenic (12, 14).
All GAS genomes sequenced contain several genes presumed to encode extracellular DNases (26–30; unpublished data). An important weakness of previous studies probing the pathogenesis role of secreted GAS DNases was the lack of knowledge regarding the number of genes encoding these enzymes present in the strain being used. We recently sequenced the genome of the contemporary serotype M1 GAS strain MGAS5005 (unpublished data). This strain, which is genetically representative of serotype M1 strains causing contemporary infections (unpublished data), has three genes that encode putative secreted DNases. Two of these genes are encoded by prophages (spd3 by prophage φ5005.2 and sdaD2 by prophage φ5005.3), and one is chromosomally encoded (spd). Here we studied seven isogenic mutant strains and found that extracellular DNase activity is required for normal progression of GAS infection in several animal models, in part by enhancing evasion of the innate immune system. The results resolve a longstanding issue in bacterial pathogenesis research.
Materials and Methods
Detailed protocols are provided as Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Quantitative PCR Analysis. TaqMan primer and probe sets (Table 2, which is published as supporting information on the PNAS web site) specific for proS, spd, spd3, and sdaD2 were used to perform quantitative PCR analysis. RNA was isolated by using a modified RNeasy (Qiagen, Valencia, CA) protocol. The results are presented as fold change relative to proS, a gene that is transcribed at a constant level throughout the GAS growth cycle (31).
Construction of Isogenic Mutant Derivative Strains. Single-, double-, and triple-isogenic mutants in the DNase genes were created from strain MGAS5005 by gene replacement with kanamycin, spectinomycin, and erythromycin resistance cassettes (32) (Table 1). A schematic outlining of the strategy used to construct the seven mutant strains is in Fig. 5, which is published as supporting information on the PNAS web site. The mutant strains were confirmed to be the correct molecular constructs by PCR, DNA sequence, Western immunoblot, and Southern blot analysis (see Fig. 7 and data not shown).
Table 1. GAS strains used in this study.
| Strain | Genotype |
|---|---|
| MGAS5005 | WT |
| NKO | spd3::KanR |
| AKO | sdaD2::SpecR |
| KKO | spd::SpecR |
| NK | spd3::KanR, spd::SpecR |
| NA | spd3::KanR, sdaD2::SpecR |
| AK | sdaD2::SpecR, spd::ErmR |
| NAK | spd3::KanR, sdaD2::SpecR, spd::ErmR |
All DNase mutant strains are derivatives of MGAS5005. KanR, kanamycin resistance cassette; SpecR, spectinomycin resistance cassette; ErmR, erythromycin resistance cassette.
Western Immunoblot Analysis of Culture Supernatant Proteins. Western blots were probed with rabbit polyclonal antibodies raised against each of the four classical GAS DNases (DNase A–D).
DNase Assays. DNase activity present in filtered culture supernatants obtained from the wild-type and mutant strains was assayed by incubating a PCR-generated fragment of GAS DNA in 1× New England Biolabs buffer 2 for 20 min at 37°C. The samples were analyzed by agarose gel electrophoresis and examined visually. Supporting Materials and Methods describes the quantitative DNase assay protocol.
Mouse Infection Assays. All mouse infection assays were performed in triplicate on separate occasions by using bacteria grown to mid-logarithmic phase (32). For the i.p. injection model, 16 female CD-1 mice were used per GAS strain (48 total mice per strain). The animals were injected with 250 μl of a 1 × 108/ml suspension of GAS in PBS, monitored every 2 h for 30 h, and three times daily thereafter. Kaplan-Meier survival curves were generated and analyzed for statistical significance by using the log-rank test. For the s.c. inoculation model, 16 female Crl:SKH10hrBR mice were given 100 μl of a1 × 108/ml suspension of GAS in PBS. The lesion volume was determined daily for up to 3 weeks by using the formula for a spherical ellipsoid (32). A mixed-model repeated-measures analysis was used to test for significant differences in lesion volumes. The repeated measures model was used with one within-subjects and one between-subjects factor. Statistical calculations were performed with proc mixed software (SAS Institute, Cary, NC).
Histopathology Studies. Mice inoculated s.c. with strain MGAS5005, strain NAK (triple DNase mutant), or PBS were killed after injection. Histopathology studies were performed on tissue obtained at 8, 24, 32, 48, and 56 h from five mice each infected with wild-type strain MGAS5005 and mutant strain NAK, and two control mice were killed at each time point. The control animals were injected with PBS only.
PMN Bactericidal Activity Assays. Human PMNs were isolated from venous blood of healthy donors in accordance with a protocol approved by the Institutional Review Board for Human Subjects at the National Institute of Allergy and Infectious Diseases. Killing of GAS strains by human PMNs was determined as described in ref. 24 with several modifications. About 107 cells from an overnight culture of strain MGAS5005 or the triple DNase mutant strain NAK were incubated with human PMNs (106) for 1 h at 37°C in a humidified CO2 incubator. In some assays, PMNs were preincubated with 10 μg/ml dihydrocytochalasin B (dHCB) to block phagocytosis, and/or 100 nM IL-8 to enhance degranulation and formation of neutrophil extracellular traps (7). Serial dilutions of each killing assay were made, the bacteria plated onto Todd-Hewitt yeast agar, and colony-forming units (cfus) counted after overnight growth. Percent killing by PMNs was determined with the equation 1 – (cfuPMN(±dHCB±IL-8)/cfuno treatment) × 100.
Non-Human Primate Model of GAS Pharyngitis. A non-human primate model of GAS pharyngitis was used with slight modifications (25). Briefly, two groups of four cynomolgus macaques were anesthetized before administration of 1 ml of an ≈2 × 107/ml suspension of MGAS5005 or NAK into the nares of each animal. All animals were treated in an identical manner to minimize possible variation in dosage. Throat swabs, nasal washes, peripheral blood samples, and scoring of clinical symptoms of pharyngitis were conducted on days 0, 7, 14, and 21. Throat swabs and clinical scoring only were performed on days 2, 4, 10, and 17. Clinical scoring was conducted by one trained veterinarian who was blinded to the test and control animal categories. To test for significance, generalized estimating equations were used to analyze the repeated binary outcomes in the data by using sas proc genmod software. SLO titers were measured by ELISA as described in ref. 25.
Results
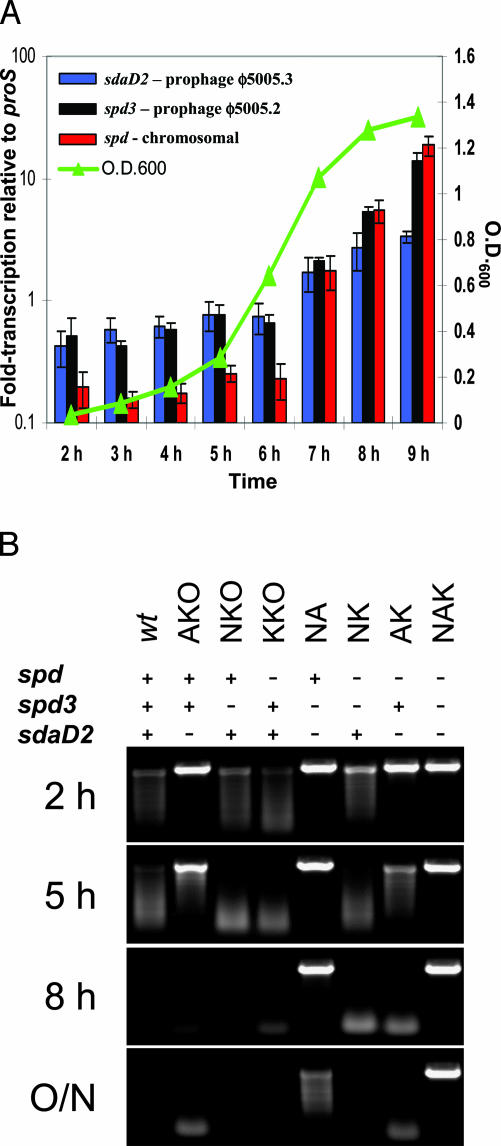
Growth-Phase-Dependent Expression of Genes Encoding Secreted DNases. The genome of strain MGAS5005 has three genes predicted to encode extracellular DNases, but it is not known whether these genes are transcribed. This issue was studied by using TaqMan quantitative PCR. We found that all three DNase genes were transcribed and the transcript level of each gene increased upon entry of the bacteria into the stationary phase of cell growth (Fig. 1A).
Fig. 1.
Characterization of DNases made by serotype M1 strain MGAS5005. (A) Transcription profiles of sdaD2, spd3, and spd. TaqMan analysis was performed on RNA purified from strain MGAS5005 grown in Todd-Hewitt yeast broth. Data are displayed as the mean (based on triplicate readings of triplicate cultures) fold change in transcription of the DNase-encoding genes relative to the control gene proS (y axis, left). The curve shows the mean OD600 of the cultures used for RNA extraction (y axis, right). All error bars are ± standard deviation. (B) DNase activity present in the culture supernatant of GAS strains used in this study. Filtered supernatants obtained from indicated strains were incubated with DNA. DNase activity was present at varying times in the supernatant of all strains except the triple DNase mutant strain NAK. The text at the top of the figure denotes presence (+) or absence (–) of intact sdaD2, spd3, and spd genes.
In Vitro Phenotypes of GAS Isogenic DNase Mutant Strains. To facilitate testing the hypothesis that extracellular DNase contributes to host–pathogen interaction, a series of seven isogenic mutant strains was constructed (Table 1). These strains represent all permutations of mutant strains possible by inactivation of the three DNase genes. The growth curves of the wild-type and seven mutant strains were identical over 9 h in Todd-Hewitt yeast media (Fig. 6, which is published as supporting information on the PNAS web site). To ensure the integrity of the DNase mutations, and also correlate the previously used DNase classification (DNase A–D) with the DNases in this study, Western immunoblots by using culture supernatant proteins and antibody to each of the four DNases (DNase A–D) were performed (Fig. 7, which is published as supporting information on the PNAS web site). The observed patterns of reactivity confirmed the integrity of the DNase mutants, and identified reactivity of anti-DNase B, C, and D antibodies with Spd, Spd3, and SdaD2, respectively (Fig. 7).
DNase activity was present in the culture supernatants of all strains, with the exception of the triple-mutant strain NAK (Fig. 1B and Fig. 8, which is published as supporting information on the PNAS web site). As expected, the culture supernatant of this strain lacked DNase activity, consistent with genetic inactivation of the three DNases genes present in strain MGAS5005. Importantly, DNase activity associated with SdaD2 was evident after only 2 h of growth (Figs. 1B and 8).
Of interest, the triple-mutant strain (NAK) had a striking phenotype in overnight cultures that was characterized by clumped aggregates readily visible with the unaided eye. Light and electron microscopy examination revealed these clumps to be large aggregates of GAS chains (Fig. 9A, which is published as supporting information on the PNAS web site). The aggregates formed by the triple-mutant strain NAK were many-fold larger and occurred at a much higher frequency than clumps present in the overnight cultures of the wild-type parental strain and the other mutant strains. To test the hypothesis that the lack of extracellular DNase activity contributed to formation of these structures, exogenous DNase was added to an overnight culture of the strain NAK. The aggregates dissipated rapidly after 1 h of incubation (data not shown). Similarly, addition of the supernatant from an overnight culture of strain MGAS5005 to an overnight culture of strain NAK also resulted in the dissipation of aggregates (data not shown). These results imply that accumulation of extracellular DNA contributed to the clumping phenotype. To further test this hypothesis, TaqMan PCR analysis was used to assess the relative amount of extracellular DNA that accumulated in the cultures of mutant strain NAK and strain MGAS5005. The overnight culture of the mutant strain had 5 orders of magnitude more DNA compared with the wild-type parental strain MGAS5005 (Fig. 9B). Taken together, the data strongly suggest that lack of extracellular DNase activity was a key factor contributing to formation of the bacterial aggregates.
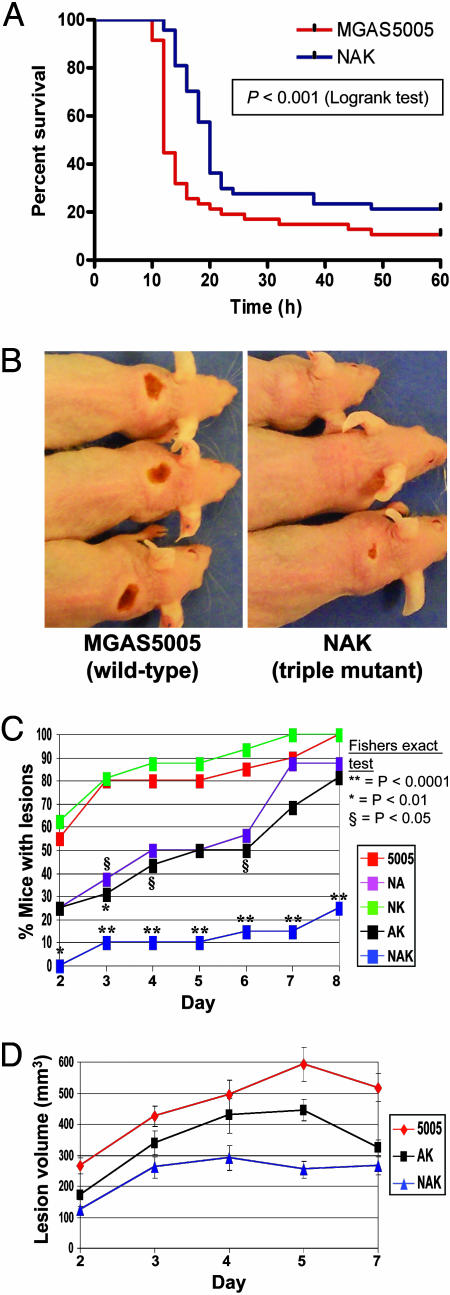
DNase Production and Mouse Infection Pathogenesis. As a first test of the hypothesis that DNase production contributes to GAS pathogenesis, the virulence of wild-type strain MGAS5005 was compared with the triple DNase mutant strain NAK by i.p. injection into outbred mice (32). Experiments done in triplicate revealed that the NAK mutant strain was significantly less virulent (Fig. 2A; P < 0.001).
Fig. 2.
Extracellular DNase activity contributes to infection pathogenesis in two mouse models of GAS disease. (A) Inactivation of DNase genes significantly decreases lethality after i.p. injection. Female CD-1 mice were injected with 2.5 × 107 cfu of strain MGAS5005 (wild-type) or strain NAK (triple DNase mutant), and survival was monitored. Forty-eight mice per GAS strain were used. (B) Lack of DNase production results in significantly decreased soft-tissue disease. Female Crl:SKH1-hrBR mice were injected s.c. with 1 × 107 CFU of GAS strain MGAS5005 (wild-type), NAK (triple DNase mutant), and NA, NK, and AK (double DNase mutants) and monitored for formation of skin lesions. Inactivation of DNase genes resulted in delayed and reduced formation of skin lesions. Shown are images of representative mice infected with strain MGAS5005 and strain NAK on day 3 postinfection. (C) SdaD2 is necessary and sufficient to produce wild-type levels of skin lesions. Data points labeled with an asterisk indicate significant differences between strain MGAS5005 (wild-type) and mutant strains by using Fisher's exact test (**, P < 0.0001; *, P < 0.01; §, P < 0.05). (D) Spd3 is insufficient to produce wild-type lesion volumes. Shown are the mean lesion volumes (± standard error of the mean) over time, displaying significant differences between MGAS5005 (wild-type) and the DNase mutant strains AK (sdaD2::SpecR and spd::ErmR, spd3+; P < 0.001) and NAK (spd3::KanR, sdaD2::SpecR, and spd::ErmR; P < 0.0001).
We next assessed whether genetic inactivation of the genes encoding secreted DNases detrimentally affected the ability to cause disease in a mouse model of soft-tissue infection (33). s.c. injection of immunocompetent hairless mice was performed with strain MGAS5005, the three double DNase mutant strains, and the triple-mutant strain NAK. Compared with the wild-type strain, the NAK mutant strain had a marked reduction in the frequency and severity of skin lesions (Fig. 2 B and C). Three different lesion profiles were identified, corresponding to strains causing high (MGAS5005, NK), moderate (NA, AK), or low (NAK) pathology. These data show that possession of an intact sdaD2 gene alone (but not spd or spd3) is sufficient to produce wild-type levels of lesion formation. The lesion volumes of mice infected with strains AK and NAK also differed significantly from the wild-type (Fig. 2D; P < 0.001 and P < 0.0001, respectively).
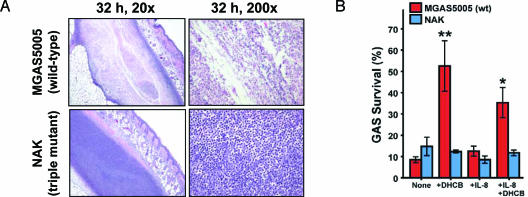
DNase Activity Contributes to Evasion of the Innate Immune Response.The difference in lesion character observed after s.c. infection with strains MGAS5005 and NAK was investigated by performing histopathologic analysis of the infection site (Fig. 3A). Mice infected with wild-type strain MGAS5005 formed abscesses as early as 8 h after injection. These abscesses had a central area of necrosis consisting of lysed PMNs and apparently viable bacteria (Fig. 3A Upper). In striking contrast, animals receiving the triple-mutant strain NAK had a solid sheet of viable PMNs at all five time points assayed (Fig. 3A). Strain MGAS5005 cells were present in high numbers in lesions throughout the experiment, whereas strain NAK bacteria were not obvious after the initial time point (8 h). The increased clearance from the injection site of strain NAK relative to strain MGAS5005 reflects the reduced frequency and severity of soft-tissue pathology observed in mice inoculated with the mutant bacteria.
Fig. 3.
Secreted DNases contribute to the evasion of host innate immunity. (A) Histopathologic analysis of the skin lesions of mice infected s.c. with strain MGAS5005 (wild-type) or NAK (triple DNase mutant). Strain MGAS5005 formed necrotic lesions containing GAS and lysed PMNs at all time points. Lesions formed by strain NAK lacked necrosis and had sheets of viable PMNs with bacteria not obvious after the initial (8 h) time point. Shown are representative images from the 32-h time point. (B) Secreted GAS DNases protect against extracellular killing by human PMNs. Extracellular killing of the wild-type GAS strain MGAS5005 by human PMNs occurred at significantly lower levels than the triple DNase mutant strain NAK (**, P < 0.001; *, P < 0.05, ANOVA with Bonferroni's posttest for multiple comparisons). Human PMNs were preincubated with 10 μg/ml of dHCB to inhibit phagocytosis and/or 100 nM IL-8 to enhance degranulation. Results are the mean ± SEM of three experiments.
To test the hypothesis that the preferential clearance of mutant strain NAK from the site of infection was related to differential extracellular killing by neutrophils, we performed in vitro GAS killing assays by using human PMNs (Fig. 3B). PMNs were pretreated with dihydrocytochalasin B to block phagocytosis and facilitate measurement of extracellular GAS killing. There was significantly less killing of strain MGAS5005 by dHCB-treated PMNs compared with triple-mutant strain NAK (P < 0.001), whereas the levels of killing by untreated PMNs were similar with the two strains. We propose that the lack of a requirement for IL-8 stimulation of extracellular killing is attributed to the stimulatory effects of GAS. These data demonstrate that one function of secreted GAS DNases is to protect against extracellular killing by PMNs, presumably through the degradation of NETs.
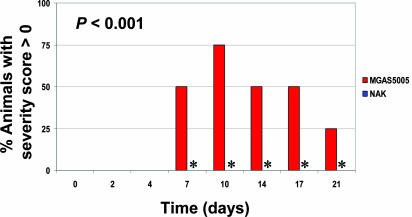
Extracellular DNases and GAS Pharyngitis. The results of the mouse infection studies and human PMN experiments supported the hypothesis that extracellular DNase contributes to disease progression. Although GAS is a common cause of invasive infections, the most common manifestation of disease is pharyngitis. Thus, it is important to understand the role of extracellular DNases in this type of infection. To address this issue, we used a recently developed non-human primate model of GAS pharyngitis (25). Importantly, only animals infected with strain MGAS5005 developed pharyngeal erythema to a level warranting a severity score (Fig. 4). All four animals infected with strain MGAS5005 had a score of 1 on at least one occasion during the experiment, a finding that was of statistical significance (P < 0.001). Although only animals infected with strain MGAS5005 developed erythema, all eight monkeys had bona fide GAS infections, as assessed by culture positivity and increased SLO antibody titers (Fig. 10, which is published as supporting information the PNAS web site). Tonsil size also trended toward higher values in animals infected with strain MGAS5005 (data not shown). However, the differences observed did not reach statistical significance, possibly as a consequence of the small number of animals in each group. Comparison of the number of cfus isolated from throat swabs of the two groups of animals identified no obvious differences.
Fig. 4.
Extracellular DNase contributes to GAS pharyngitis in a non-human primate model. Two groups of four cynomolgus macaques were infected with strain MGAS5005 or triple-mutant strain NAK by dribbling into the nares. Shown are the percent of animals within each group that had a severity score of >0, indicating pharyngeal erythema, on the indicated days. Asterisks highlight that no NAK-infected animals received severity scores, whereas MGAS5005 animals did.
Discussion
A long-standing question in bacterial pathogenesis research has been what benefit, if any, is gained by elaboration of extracellular DNase. This question has been an especially important issue in GAS research because all strains produce one or more DNases. Although the presence of extracellular DNase in the culture supernatants of GAS strains was described almost 60 years ago (2, 6), after decades of study it was not known whether GAS DNase contributes to pathogenesis. Our studies of isogenic mutant strains in three animal models of infection, and in vitro studies with human PMNs, show that DNase contributes to normal GAS disease progression. It is possible that the inability to demonstrate a detrimental effect on virulence of DNase gene inactivation in previous studies was due to mutagenesis of only one of several DNase genes present in the strains used (18, 19).
Our studies do not resolve the important question of why most GAS strains produce two or more distinct extracellular DNases. We believe there are at least four possible explanations. Production of multiple antigenically distinct DNases would preserve the ability to degrade DNA when GAS infects a host with enzyme-inhibiting antibodies to one DNase. It is also possible that production of multiple DNases with different substrate cleavage specificities or other characteristics (e.g., pH profile, cation requirement, RNase activity, etc.) would provide a survival advantage by enhancing the range of conditions across which the enzymatic activity functions. A third possibility is that possession of multiple DNase genes, including chromosomally encoded and prophage-encoded variants, enhances the likelihood that DNase activity will be made at distinct phases of the infection cycle. Differential regulation of expression would increase the probability that DNase activity is made at all times that GAS encounters innate immune functions. Inasmuch as many of the GAS DNases are encoded by prophages, a fourth possibility is that some of these enzymes contribute to the bacteriophage life cycle. For example, it is possible that degradation of GAS DNA released during the lytic cycle assists phage particles to disseminate and infect a new GAS host cell. Regardless, it will be important in future studies to investigate the contributions of each DNase to the biology of GAS by using the seven isogenic mutant strains described herein.
Genome sequencing, DNA-DNA microarray, PCR profiling, single-nucleotide polymorphism analysis, and several other genetic analysis techniques have found that strain MGAS5005 is genetically representative of a clone that is responsible for most contemporary episodes of infections caused by serotype M1 GAS strains (refs. 34 and 35; unpublished data). This clone evolved recently from a less virulent ancestor through a series of recombination events, including one involving acquisition of prophage φ5005.3-encoding SdaD2 (unpublished data). Several lines of evidence suggest that SdaD2 is the most important of the three DNases made by strain MGAS5005 in host–pathogen interactions. The sdaD2 gene is transcribed early in growth, a time that may be important for initial host–pathogen interactions, and continues at a high level throughout the growth cycle (Fig. 1). Consistent with early transcription, SdaD2 extracellular DNase activity was detected early in the GAS growth cycle (Fig. 1B). Moreover, under the conditions assayed, SdaD2 was responsible for most of the extracellular DNase activity (Figs. 1B and 8). Importantly, inactivation of the sdaD2 gene had a more pronounced detrimental effect on virulence than did mutation of spd or spd3 (Fig. 2C). Taken together, the data suggest that acquisition of sdaD2 was important in the evolution of the clone now responsible for virtually all contemporary episodes of serotype M1 infections.
We reported that spd was the mostly highly transcribed of 18 proven or putative virulence genes studied during human acute GAS pharyngitis, a result supporting the idea that DNase is important in pharyngeal infection (25). The results of our non-human primate experiment confirmed that extracellular DNase is required for normal GAS pharyngitis progression. Several possibilities may account for the significant difference in severity scores between MGAS5005- and NAK-infected animals. One possibility is that digestion of DNA in mucus enhances access of strain MGAS5005 and its secreted inflammatory factors to the host cells in the posterior pharynx. It is also possible that DNase is directly responsible for the increase in the severity score observed in the animals infected with the wild-type strain. Although thus far there is no evidence that GAS DNase is directly cytotoxic or proinflammatory, we note that several DNases made by bacterial pathogens are cytolethal (36, 37). Regardless of the exact mechanism, our results provide the impetus for future investigations into the processes whereby extracellular DNase contributes to upper respiratory tract disease.
A key discovery arising from our studies was that extracellular DNase protects GAS from extracellular killing by PMNs. We believe the protective activity of DNases is through degradation of NETs, although we have not proven this conclusion directly. Whatever the process, DNase production represents a heretofore uncharacterized mechanism used by pathogenic bacteria to evade innate immune function. Purulent exudates form at the site of bacterial infections and are characterized by high concentrations of PMNs and DNA, much of it released from dead and dying host cells. Studies conducted >50 years ago showed that GAS DNase depolymerized DNA present in exudates, both in vitro and in humans, resulting in greatly decreased viscosity of the exudates (3). We hypothesize that this reduction in viscosity enhances GAS dissemination from the site of infection not only by degrading nucleic acids released from cells destroyed by other host and pathogen factors (6), but also by reducing aggregation of bacteria (Fig. 9A). It is very possible that GAS extracellular DNases have additional functions yet to be discovered. However, our discovery that DNase assists escape from PMN extracellular killing documents a previously uncharacterized strategy used by a human bacterial pathogen to thwart innate immunity, proliferate, and cause disease.
Supplementary Material
Acknowledgments
This article is dedicated to the memory of Dr. Maclyn McCarty, a pioneer in streptococcal DNase research. We thank Patricia Ferrieri (University of Minnesota) for providing antisera to the four GAS DNases (DNase A–D) and D. Dorwood, S. M. Ricklefs, and R. Ireland for important contributions to the success of this study.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: cfu, colony-forming unit; dHCB, dihydrocytochalasin B; GAS, group A Streptococcus; NET, neutrophil extracellular trap; PMN, polymorphonuclear leukocyte.
References
- 1.Fox, J. B. & Holtman, D. F. (1968) J. Bacteriol. 95, 1548–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tillett, W. S., Sherry, S. & Christensen, L. R. (1948) Proc. Soc. Exp. Biol. Med. 68, 184–188. [DOI] [PubMed] [Google Scholar]
- 3.Sherry, S., Johnson, A. & Tillett, W. S. (1949) J. Clin. Invest. 28, 1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherry, S. & Goeller, J. P. (1950) J. Clin. Invest. 29, 1588–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson, A. J. (1950) J. Clin. Invest. 29, 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarty, M. (1948) J. Exp. Med. 88, 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinkmann, V., Reichard, U., Goosmann, C., Fauler, B., Uhlemann, Y., Weiss, D. S., Weinrauch, Y. & Zychlinsky, A. (2004) Science 303, 1532–1535. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham, M. W. (2000) Clin. Microbiol. Rev. 13, 470–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musser, J. M. & Krause, R. M. (1998) in Emerging Infections, ed. Krause, R. M. (Academic, New York), pp. 185–218.
- 10.Fischetti, V. A. (1989) Clin. Microbiol. Rev. 2, 285–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoe, N. P., Nakashima, K., Lukomski, S., Grigsby, D., Liu, M., Kordari, P., Dou, S. J., Pan, X., Vuopio-Varkila, J., Salmelinna, S., et al. (1999) Nat. Med. 5, 924–929. [DOI] [PubMed] [Google Scholar]
- 12.McCarty, M. (1949) J. Exp. Med. 90, 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wannamaker, L. W. (1958) J. Exp. Med. 107, 797–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wannamaker, L. W. (1964) in The Streptococcus, Rheumatic Fever, and Glomer-ulonephritis, ed. Uhr, J. W. (Williams & Wilkins, Baltimore), pp. 140–165.
- 15.Tiesler, E. & Beck, U. (1976) Zentralbl. Bakteriol. Mikrobiol. Hyg. Abt. 1 Orig. A 234, 462–472. [PubMed] [Google Scholar]
- 16.Miyaka, Y., Yamada, T., Shitara, M. & Fukazawa, Y. (1985) Microbiol. Immunol. 29, 195–204. [DOI] [PubMed] [Google Scholar]
- 17.Ferreira, B. T., Benchetrit, L. C., De Castro, A. C., Batista, T. G. & Barrucand, L. (1992) Zentralbl. Bakteriol. 277, 493–503. [DOI] [PubMed] [Google Scholar]
- 18.Podbielski, A., Zarges, I., Flosdorff, A. & Weber-Heynemann, J. (1996) Infect. Immun. 64, 5349–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sriskandan, S., Unnikrishnan, M., Krausz, T. & Cohen, J. (2000) Microbiology 146, 2785–2792. [DOI] [PubMed] [Google Scholar]
- 20.Broudy, T. B., Pancholi, V. & Fischetti, V. A. (2002) Infect. Immun. 70, 2805–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banks, D. J., Lei, B. & Musser, J. M. (2003) Infect. Immun. 71, 7079–7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasegawa, T., Torii, K., Hashikawa, S., Iinuma, Y. & Ohta, M. (2002) Arch. Microbiol. 177, 451–456. [DOI] [PubMed] [Google Scholar]
- 23.Iwasaki, M., Igarashi, H. & Yutsudo, T. (1997) Microbiology 143, 2449–2455. [DOI] [PubMed] [Google Scholar]
- 24.Voyich, J. M., Sturdevant, D. E., Braughton, K. R., Kobayashi, S. D., Lei, B., Virtaneva, K., Dorward, D. L., Musser, J. M. & DeLeo, F. R. (2003) Proc. Natl. Acad. Sci. USA 100, 1996–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Virtaneva, K., Graham, M. R., Porcella, S. F., Hoe, N. P., Su, H., Graviss, E. A., Gardner, T. J., Allison, J. E., Lemon, W. J., Bailey, J. R., et al. (2003) Infect. Immun. 71, 2199–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferretti, J. J., McShan, W. M., Ajdic, D., Savic, D. J., Savic, G., Lyon, K., Primeaux, C., Sezate, S., Suvorov, A. N., Kenton, S. et al. (2001) Proc. Natl. Acad. Sci. USA 98, 4658–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smoot, J. C., Barbian, K. D., Van Gompel, J. J., Smoot, L. M., Chaussee, M. S., Sylva, G. L., Sturdevant, D. E., Ricklefs, S. M., Porcella, S. F., Parkins, L. D., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 4668–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beres, S. B., Sylva, G. L., Barbian, K. D., Lei, B., Hoff, J. S., Mammarella, N. D., Liu, M.-Y., Smoot, J. C., Porcella, S. F., Parkins, L. D., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 10078–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakagawa, I., Kurokawa, K., Yamashita, A., Nakata, M., Tomiyasu, Y., Okahashi, N., Kawabata, S., Yamazaki, K., Shiba, T., Yasunga, T., et al. (2003) Genome Res. 13, 1042–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banks, D. J., Porcella, S. F., Barbian, K. D., Beres, S. B., Philips, L. E., Voyich, J. M., DeLeo, F. R., Martin, J. M., Somerville, G. A. & Musser, J. M. (2004) J. Infect. Dis. 190, 727–738. [DOI] [PubMed] [Google Scholar]
- 31.Graham, M. R., Smoot, L. M., Lux Migliaccio, C. A., Virtaneva, K., Sturdevant, D. E., Porcella, S. F., Federle, M. J., Adams, G. J., Scott, J. R. & Musser, J. M. (2002) Proc. Natl. Acad. Sci. USA 99, 13855–13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukomski, S., Nakashima, K., Abdi, I., Cipriano, V. J., Ireland, R. M., Reid, S. D., Adams, G. G. & Musser, J. M. (2000) Infect. Immun. 68, 6542–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukomski, S., Montgomery, C. A., Rurangirwa, J., Geske, R. S., Barrish, J. P., Adams, G. J. & Musser, J. M. (1999) Infect. Immun. 67, 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Musser, J. M., Hauser, A. R., Kim, M. H., Schlievert, P. M., Nelson, K. & Selander, R. K. (1991) Proc. Natl. Acad. Sci. USA 88, 2668–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musser, J. M., Kapur, V., Szeto, J., Pan, X., Swanson, D. & Martin, D. (1995) Infect. Immun. 63, 994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lara-Tejero, M. & Galan, J. E. (2000) Science 290, 354–357. [DOI] [PubMed] [Google Scholar]
- 37.Lara-Tejero, M. & Galan, J. E. (2002) Trends Microbiol. 10, 147–152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.