Abstract
Ferroportin 1 (FPN1) is transmembrane protein involved in iron homeostasis. In the duodenum, FPN1 localizes to the basolateral surface of enterocytes where it appears to export iron out of the cell and into the portal circulation. FPN1 is also abundantly expressed in reticuloendothelial macrophages of the liver, spleen, and bone marrow, suggesting that this protein serves as an iron exporter in cells that recycle iron from senescent red blood cells. To directly test the hypothesis that FPN1 functions in the export of iron after erythrophagocytosis, FPN1 was stably expressed in J774 mouse macrophages by using retroviral transduction, and release of 59Fe after phagocytosis of 59Fe-labeled rat erythrocytes was measured. J774 cells overexpressing FPN1 released 70% more 59Fe after erythrophagocytosis than control cells, consistent with a role in the recycling of iron from senescent red cells. Treatment of cells with the peptide hormone hepcidin, a systemic regulator of iron metabolism, dramatically decreased FPN1 protein levels and significantly reduced the efflux of 59Fe after erythrophagocytosis. Subsequent fractionation of the total released 59Fe into heme and nonheme compounds revealed that hepcidin treatment reduced the release of nonheme 59Fe by 50% and 25% from control and FPN1-overexpressing cells, respectively, but did not diminish efflux of 59Fe-heme. We conclude that FPN1 is directly involved in the export of iron during erythrocyte-iron recycling by macrophages.
Keywords: reticuloendothelial cell, reticuloendothelial system
Approximately two-thirds of total body iron in the adult human is present in hemoglobin in circulating red blood cells. Mature erythrocytes circulate until senescent or damaged and are then phagocytosed predominantly by reticuloendothelial macrophages of the liver, bone marrow, and spleen. After erythrophagocytosis, hemoglobin is proteolytically degraded, and the resultant heme moiety is oxidatively cleaved to release free iron, which is then either stored within the macrophage or released into the circulation. This recycling of iron from senescent red cells supplies the bone marrow with ≈20 mg of iron per day for new red cell synthesis (1).
Several lines of evidence implicate the involvement of the recently identified protein ferroportin 1 (FPN1) in iron recycling by the macrophage. FPN1, also known as IREG-1 and MTP-1, was initially identified as an iron-export protein located on the basolateral membrane of duodenal enterocytes (2-4). The abundant expression of FPN1 in reticuloendothelial macrophages of the liver, bone marrow, and splenic red pulp (5) suggests that it plays a similar iron-export role in macrophages as well. Genetic evidence for this proposed function is provided by a growing number of hemochromatosis patients identified with mutations in FPN1 (reviewed in ref. 6). The distinguishing clinical feature of these patients is iron accumulation in liver macrophages (Kupffer cells). Studies of cultured macrophages, which show marked increases in FPN1 after erythrophagocytosis (7), also strongly support a role for FPN1 in iron recycling. An export function of FPN1 has been inferred from exogenous-expression studies showing increased iron efflux from iron-loaded Xenopus oocytes injected with FPN1 cRNA (3, 4) and cellular iron depletion in HEK 293T cells transiently transfected with FPN1 cDNA (2).
Recent studies demonstrate that FPN1 expression is post-translationally regulated by hepcidin (8), a 25-aa peptide secreted by the liver in response to iron loading (9) and inflammation (10, 11). In HEK cells expressing an FPN1-GFP fusion protein, hepcidin was found to cause internalization of FPN1-GFP and its subsequent degradation in the lysosome (8). The down-regulation of FPN1 expression was associated with a decrease in the export of 59Fe after cells were loaded with 59Fe-transferrin. The negative regulatory effect of hepcidin in vitro is consistent with observations that constitutive hepcidin expression prevents iron loading in Hfe-/- mice (12), a murine model of HFE-associated hemochromatosis, and that Usf2-/- mice lacking hepcidin gene expression develop severe tissue iron overload (10).
The aim of the present study was to directly examine the role of FPN1 in iron export by the macrophage. Firm evidence to support its presumed role in iron recycling from red cells is lacking, and its transport function has not been rigorously evaluated in the context of physiological iron release. To address the role of FPN1 in the release of iron after erythrophagocytosis, the protein was stably overexpressed in murine macrophages by using retroviral transduction and suppressed by treatment with hepcidin. To model physiologic iron recycling by the macrophage, iron release was investigated after erythrophagocytosis. We report that overexpression of macrophage FPN1 enhanced the release of iron after erythrophagocytosis, and that hepcidin treatment not only reduced FPN1 protein levels but also decreased the export of red cell-derived nonheme iron from macrophages. These findings provide conclusive evidence that FPN1 functions in iron recycling from the macrophage after erythrophagocytosis.
Materials and Methods
Retroviral Infection and Overexpression of FPN1 by J774 Cells. FPN1 was PCR amplified from NotI-digested pSport2 containing full-length murine FPN1 cDNA (gift of Dr. David Haile, University of Texas Health Sciences Center, Austin). Terminal XhoI sites were added by using synthetic oligonucleotides: 5′-CTCGAGGGGAGAACGCCTGGTGTCAT-3′ (sense) and 5′-CTCGAGTGCAGGCATTTATTGGGATTCT-3′ (antisense), and the 2-kb PCR product was directionally cloned into a murine stem cell virus-based biscistronic vector containing the GFP gene as a selectable marker (gift of Dr. Laurie Glimcher, Harvard School of Public Health, Boston). Orientation and sequence were verified by DNA sequencing (DNA Sequencing Core Facility, Beth Israel Deaconess Medical Center, Boston).
To produce retrovirus, HEK293T cells were cotransfected with a vector containing the viral packaging proteins gag and env, a vector containing pol, and the mouse stem cell retroviral vector containing GFP alone (GFP.RV) or GFP plus FPN1 (FPN1.RV). Vectors containing gag, env, and pol were from Dr. Gary Nolan (Stanford University, Palo Alto, CA). Lipofectamine (Invitrogen) reagent was used for transfection. Fortyeight hours after transfection, viral supernatants were collected, supplemented with 8 μg/ml hexadimethrine bromide (Sigma), and used to infect J774 macrophages. Forty-eight hours after infection, cells were analyzed by flow cytometry by using the FACSCalibur System (Becton Dickinson) to isolate GFP-positive cells. Cells expressing FPN1 were further subcloned by limiting dilution, and overexpression of FPN1 was identified by Western blotting as described below.
Antibody Production. Anti-FPN1 antiserum was generated in rabbit against peptide CGKQLTSPKDTEPKPLEGTH, corresponding to amino acids 247-264 of mouse FPN1. Antibodies specific to the FPN1 peptide were affinity purified by using a peptide-agarose column of SulfoLink coupling gel (Pierce). Anti-rat red blood cell antibodies were raised in a goat against rat red blood cell ghosts, and the IgG fraction was isolated from antiserum by using an ImmunoPure immobilized protein G column (Pierce).
Western Blot Analysis. J774 cells were grown as previously described (7). Cell monolayers were washed once in ice-cold PBS and lysed in ice-cold SDS lysis buffer [60 mM Tris-HCl, pH 6.8/2% SDS/5% glycerol/5% β-mercaptoethanol, and complete mini protease inhibitor mixture (Roche)]. Cell lysates were sonicated for 5 s and stored at -80°C until analysis. Protein content was determined by using the RC DC protein assay (Bio-Rad). Samples (60-120 μg protein) were mixed with SDS lysis buffer containing bromophenol blue and, without prior heating, were electrophoretically separated on a 10% SDS-polyacrylamide gel. Molecular weight standards (Precision Plus, dual color, Bio-Rad) were run in parallel. The protein was transferred to Optitran nitrocellulose membrane (Schleicher & Schuell). After incubating blots for 1 h in blocking solution [5% nonfat dry milk in Tris-buffered saline, pH 7.4, containing 0.01% Tween 20 (TBST)], blots were incubated overnight at 4°C with affinity-purified rabbit anti-FPN1 peptide antibodies (2.5 μg/ml blocking solution). Blots were washed in TBST and then incubated for 40 min with a 1:2,000 dilution of horseradish peroxidase-linked donkey anti-rabbit IgG (Amersham Pharmacia). Immunoreactivity was visualized by using enhanced chemiluminescence (SuperSignal WestPico, Pierce) and autoradiographic film. To control for loading, blots were stripped for 5 min in 0.5 M glycine (pH 2.8), 0.5 M NaCl, washed in TBS, blocked for 1 h in blocking buffer, then reprobed by using a 1:10,000 dilution of mouse monoclonal anti-α-tubulin clone B-5-1-2 (Sigma), and a 1:10,000 dilution of ZyMax goat anti-mouse IgG horseradish peroxidase conjugate (Zymed). Ferritin protein levels were determined as above, except that protein samples (30 μg) were boiled for 5 min before electrophoretic separation on a 12% SDS-polyacrylamide gel, and rabbit anti-horse spleen ferritin antiserum (Sigma), 1:1,000 dilution, was used at room temperature for 1 h. Autoradiograms were digitized by using gene-flash gel documentation system (SynGene), and signal intensities were quantified by densitometry by using genetools software (SynGene).
Erythrophagocytosis and Hepcidin Treatment. The 59Fe-labeled rat erythrocytes were generated in vivo as previously described (13). The animal protocol was approved by the University of Florida Institutional Animal Care and Use Committee. Radiolabeled erythrocytes were stored in Alsever's solution (Sigma) at 4°C until use. To measure 59Fe release, J774 cell monolayers (≈1 × 106 cells per well of a six-well plate) were incubated at 37°C with 59Fe-labeled red cells (1 × 107) preopsonized with rabbit anti-rat red blood cell IgG (Research Diagnostics). After 1.5 h, noningested red cells were hypotonically lysed by double-processed tissue culture water (Sigma). After three washes in Dulbecco's PBS, fresh media were added, and cells were incubated at 37°C. Twenty-two hours later, supernatant media were centrifuged to pellet any floating cells, and an aliquot was used to measure 59Fe. Cells were harvested into PBS for 59Fe analysis. Radioactivity in each fraction were measured by using a Cobra II Gamma Spectrometer (Packard). The percentage of 59Fe release was calculated according to the following equation: % 59Fe release = [(cpm in supernatant)/(cpm in supernatant + cpm in cells)] × 100. After analysis, supernatants were acidified by using HCl and extracted with cyclohexanone to separate 59Fe-heme into the organic layer and nonheme 59Fe into the aqueous layer (14). To determine FPN1 protein levels after erythrophagocytosis, non-radiolabeled rat red blood cells were used. Goat anti-rat red blood cell IgG was used as opsonin instead of rabbit IgG because the latter interferes with Western analysis.
Synthetic human hepcidin was obtained from Peptides International. This hepcidin contains 25 aa with four disulfide bridges in the same positions as do other synthetic forms of hepcidin (15). To test the effect of hepcidin on FPN1 protein levels and the release of nonheme 59Fe, hepcidin (700 nM) was added immediately after the 1.5-h erythrophagocytosis period and incubated for the times indicated.
Statistical Analysis. The 59Fe release data are shown as means ± SEM. Statistical analysis was performed by one-way ANOVA and Tukey's post test using prism (version 4.02 for Windows, GraphPad, San Diego).
Results
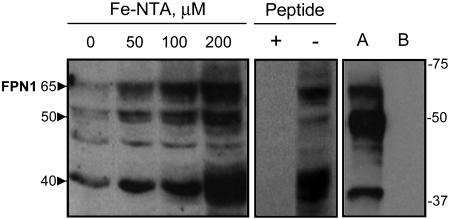
Characterization of the Immunoreactivity of an Affinity-Purified Anti-FPN1 Peptide Antibody. Rabbit polyclonal antisera were raised against a peptide corresponding to amino acids 247-264 of mouse FPN1 and affinity purified. Use of the affinity-purified antibodies in Western analysis of J774 cells treated with increasing amounts of ferric nitrilotriacetic acid revealed three immunoreactive, iron-responsive bands migrating with apparent molecular masses of ≈65, 50, and 40 kDa (Fig. 1 Left). Immunodetection using the affinity-purified antisera is blocked by the presence of a molar excess of FPN1 peptide (Fig. 1 Center). All three FPN1 bands are detected in lysates of HEK293T cells transfected with full-length mouse FPN1 cDNA (Fig. 1 Right). Because the band at 65 kDa corresponds most closely to the predicted molecular mass of FPN1 (62 kDa) and the immunoreactive bands at 50 and 40 kDa most likely represent degradation products, all subsequently presented immunoblots will focus on this band. It should be noted that immunoreactive bands of similar masses were detected using a different anti-FPN1 antibody (7).
Fig. 1.
Characterization of the immunoreactivity of affinity-purified anti-FPN1 peptide antibody. J774 cells were treated with 0-200 μM ferric-nitrilotriacetic acid for 48 h. Cell lysates (60 μg of protein per lane) were electrophoresed on a 10% SDS-polyacrylamide gel, transferred to nitrocellulose, and blotted with anti-FPN1 antibody as described in Materials and Methods. The position and masses (in kDa) of molecular weight markers are indicated on the right. The approximate molecular masses (kDa) of the main iron-responsive immunoreactive bands are indicated by arrowheads on the left. To confirm specificity of antiserum by peptide competition, lysates from J774 cells treated with 200 μM ferric-nitrilotriacetic acid for 48 h were blotted with anti-FPN1 antibody preincubated for 1 h in the presence (+) or absence (-) of a 100-fold molar excess of FPN1 peptide. In addition, lysates (15 μg per lane) were analyzed from HEK292T cells transfected with pSport2 CMV vector containing full-length mouse FPN1 cDNA (lane A) or pSport2 CMV vector alone (lane B). Ponceau staining indicated equivalent protein loading among lanes (not shown).
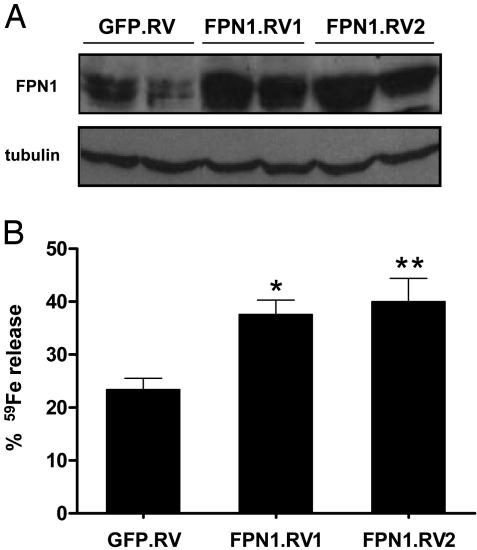
Overexpression of FPN1 in Macrophages Increases the Release of 59Fe After Phagocytosis of 59Fe-Labeled Red Cells. To overexpress FPN1, J774 macrophages were transduced with a biscistronic retroviral vector containing murine FPN1 plus GFP (FPN1.RV). Transduction of J774 cells with a retrovirus containing GFP alone (GFP.RV) served as control. As shown in Fig. 2A, two subclones (FPN1.RV1 and FPN1.RV2) were identified to overexpress FPN1 by Western analysis. Densitometric analyses of the 65-kDa bands indicate that mean FPN1 protein levels in FPN1.RV1 and FPN1.RV2 cells were 2.8 and 3.2 times the level in GFP.RV cells, respectively. The observed differences in FPN1 protein levels do not reflect differences in protein loading because each lane contains equivalent amounts of the constitutively expressed protein tubulin (Fig. 2 A).
Fig. 2.
Overexpression of FPN1 by retroviral transduction increases the release of 59Fe after phagocytosis of 59Fe-lableled red blood cells. (A) J774 cells were transduced with retroviral vectors containing GFP (GFP.RV) or GFP plus FPN1 (FPN1.RV), and GFP-expressing cells were selected by flow cytometry. Individual clones overexpressing FPN1 (FPN1.RV1 and FPN1.RV2) were subsequently identified by Western analysis. Cell lysates (120 μg of protein per lane) were blotted with anti-FPN1 antibody as described for Fig. 1. (A) Results from two different cell lysates, obtained on different days, from each of the three groups. As a control for lane loading, the blot was stripped and reprobed for tubulin. (B) J774 macrophages were incubated with 59Fe-EIgG for 1.5 h. After removal of noningested 59Fe-EIgG, cells were incubated for 22 h. Supernatant and cell fractions were collected and cpm determined by gamma counting. Percentage of iron release was calculated as [(cpm in supernatant)/(cpm in supernatant + cpm cells)] × 100. Each bar represents the mean ± SEM of five independent experiments performed in triplicate. Asterisks indicate a difference from GFP.RV: *, P < 0.05; **, P < 0.01.
The effect of FPN1 overexpression on iron recycling by J774 macrophages was investigated by measuring the percentage of 59Fe released 22 h after phagocytosis of opsonized 59Fe-labeled erythrocytes (59Fe-EIgG). Compared to GFP.RV cells, FPN1.RV1 and FPN1.RV2 cells released more 59Fe, 60% (P < 0.05) and 70% (P < 0.01), respectively (Fig. 2B). The increased release seemed to be independent of the extent of phagocytosis because FPN1.RV1 cells typically ingested 50% fewer 59Fe-EIgG than did GFP.RV cells. To exclude this difference as a potential variable, subsequent studies were performed with FPN1.RV2 cells, which phagocytosed similar amounts of 59Fe-EIgG as did GFP.RV cells.
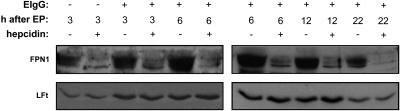
Hepcidin Decreases FPN1 Protein Expression in Macrophages. Treatment of HEK293 cells exogenously expressing FPN1 with hepcidin (500-700 nM) has recently been shown to cause the rapid degradation of FPN1 and an increase in cellular iron retention (8). In the present study, treatment of FPN1.RV2 J774 cells with 700 nM hepcidin for 3 h promoted a dramatic reduction in FPN1 protein levels (Fig. 3, lanes 1 and 2). To test the effect of hepcidin on FPN1 protein levels after erythrophagocytosis, macrophages were incubated for 1.5 h in the presence of EIgG. After phagocytosis and removal of noningested EIgG, hepcidin (700 nM) or sterile water (as control) was added to the culture medium, and cell lysates were obtained after 3, 6, 12, and 22 h. Similar to our previous results with J774 cells (7), ingestion of EIgG caused FPN1 protein levels to increase after 6 h and return to basal levels by 22 h (Fig. 3). However, at each time point after erythrophagocytosis, hepcidin treatment effectively reduced FPN1 protein levels. The effect of hepcidin was rapid; marked reductions in FPN1 protein levels were observed after treatment of macrophages for only 1 h (data not shown). Identical results were observed using synthetic human hepcidin obtained from the laboratory of Dr. Tomas Ganz (University of California, Los Angeles) (data not shown). For comparison, the effect of hepcidin on levels of the iron-storage protein, ferritin, was also assessed in FPN1.RV2 J774 cells after erythrophagocytosis. Although red cell ingestion increased the expression of ferritin, treatment with hepcidin had no effect on ferritin protein levels (Fig. 3).
Fig. 3.
Hepcidin treatment markedly decreases FPN1 protein levels but does not affect light-chain ferritin (LFt) levels after erythrophagocytosis (EP). FPN1.RV2 J774 cells were incubated with or without EIgG for 1.5 h. After removal of noningested EIgG, macrophages were treated with or without 700 nM hepcidin and incubated for 3-6 h (Left) or 6-22 h (Right). Cell lysates (60 μg and 30 μg per lane) were blotted with anti-FPN1 antibody and anti-ferritin antiserum, respectively.
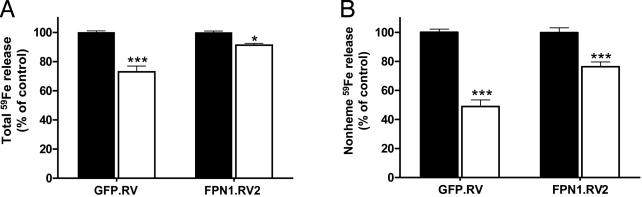
Hepcidin Decreases the Release of 59Fe from Macrophages After Phagocytosis of 59Fe-Labeled Red Cells. In light of the marked decrease in FPN1 protein levels on hepcidin treatment (Fig. 3), we next examined the effect of hepcidin on the release of 59Fe after phagocytosis of 59Fe-labeled erythrocytes. Hepcidin-treated GFP.RV cells released 27% less total 59Fe than did untreated GFP.RV cells (P < 0.001, Fig. 4A). Similarly, total 59Fe release was reduced by 9% in FPN1.RV2 cells relative to untreated controls (P < 0.05, Fig. 4A). To determine whether the decrements in total 59Fe release represented reduced release of 59Fe-heme or nonheme 59Fe, the amount of 59Fe in each fraction was determined after cyclohexanone extraction. Hepcidin treatment significantly reduced only the nonheme 59Fe component by 51% in GFP.RV cells (P < 0.001) and by 24% in FPN1.RV2 cells (P < 0.001) (Fig. 4B). In contrast, the percentage of 59Fe-heme released after hepcidin treatment was found to be increased by 22% in GFP.RV cells (P < 0.05) but did not differ in FPN1.RV2 cells (data not shown).
Fig. 4.
Hepcidin treatment decreases the release of nonheme 59Fe after phagocytosis of 59Fe-labeled red blood cells. (A) GFP.RV and FPN1.RV2 J774 cells were incubated with EIgG for 1.5 h. After removal of noningested EIgG, macrophages were treated with 700 nM hepcidin (open bars) or an equal amount of sterile water as control (solid bars), and the release of 59Fe after 22 h was determined as described in the legend of Fig. 2. (B) The percentage of total 59Fe released as nonheme 59Fe was determined after extracting the supernatant media with cyclohexanone to remove 59Fe-heme. To account for day-to-day variability in red cell preparations and erythrophagocytic activity, results are shown as relative differences of 59Fe release as compared to the untreated control (100%). Data are expressed as means ± SEM for three independent experiments, performed in triplicate. Asterisks indicate a difference from respective untreated controls: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
In previous studies of macrophage iron recycling, we observed that FPN1 protein levels in J774 cells increased markedly after erythrophagocytosis, supporting its proposed role in the release of iron (7). Here, we report that changes in the steady-state levels of FPN1 in J774 macrophages modulate the release of 59Fe after phagocytosis of 59Fe-labeled red blood cells. These new data are significant because they demonstrate a direct functional role for FPN1 in iron recycling by the macrophage.
To overexpress FPN1 in the J774 macrophage cell line, we used retroviral transduction because it is not possible with existing technologies to transfect macrophages with high efficiency. Moreover, constitutive expression of very high levels of FPN1, such as those achieved after transfection, has been reported to be toxic to cells in culture (16). Retroviral transduction offers two additional advantages over transient transfection. First, because genes delivered into the cell by retroviral transduction are integrated into the host genome, this approach allows for the generation of stable cell lines overexpressing FPN1. Second, retrovirally transduced genes are often expressed at more modest levels and may thus provide a better representation of functional changes reflecting activity of native protein levels. The stable J774 clone FPN1.RV2 established by our study displays basal FPN1 protein levels that are 3.2 times those measured in the GFP.RV control cells. The increased FPN1 expression in this clonal line was associated with a significant 70% increase in the export of 59Fe after erythrophagocytosis, but overexpression of FPN1 at this level did not seem to be toxic or to impair other cell functions.
Hepcidin, a negative regulator of iron export, was used to examine the effects of FPN1 down-regulation. Similar to the report by Nemeth et al. (8), our study reveals that treatment with hepcidin rapidly induces a dramatic reduction in FPN1 protein levels and further demonstrates that the macrophage is a target for this hormone's action. Moreover, the peptide effectively down-regulated FPN1 when endogenous levels of the protein are markedly up-regulated after erythrophagocytosis. The suppression of FPN1 expression may result from increased degradation or impaired synthesis. Importantly, the loss of FPN1 was associated with a decrease in 59Fe release after ingestion of 59Fe-labeled red cells. FPN1 appears to specifically mediate the export of nonheme 59Fe because hepcidin treatment decreased the export of nonheme 59Fe but not 59Fe-heme. This observation is consistent with studies of iron-loaded Xenopus oocytes injected with FPN1 cRNA, which demonstrate FPN1-induced transport of nonheme ferrous iron (3, 4). Nemeth et al. (8) have also shown the overexpression of FPN1-GFP was associated with increased release of 59Fe taken up by transferrin-mediated endocytosis, presumably in a nonheme form. The model that FPN1 mediates nonheme iron transport in the macrophage is further supported by recent studies that used small interfering RNAs to specifically target FPN1 (17). Suppression of FPN1 expression in human macrophages resulted in a significant accumulation of nonheme iron, as measured by an increase in Perls staining and enhanced synthesis of H-ferritin.
Because 80% of the iron in the circulation represents iron released from reticuloendothelial macrophages after red cell catabolism (18), fluctuations in macrophage iron efflux markedly affect serum iron concentrations. Inflammation, for example, causes a blockade of reticuloendothelial iron release and reduces serum iron levels (19, 20). Using mouse models of acute and chronic inflammation, Yang et al. (5) found that the reduced serum iron levels were associated with decreased FPN1 expression in reticuloendothelial macrophages of the spleen, liver, and bone marrow, suggesting a causative role for FPN1. The hypoferremia of inflammation does seem to require the participation of hepcidin because the drop in serum iron levels secondary to inflammation induced by turpentine injection was shown to be completely blunted in hepcidin-deficient mice (21). In the present study, we establish a direct relationship between hepcidin, FPN1 expression, and iron release from the macrophage after erythrophagocytosis, and thus further strengthen the association between macrophage FPN1 expression and serum iron levels. It should be noted, however, that hepcidin treatment did not completely suppress the release of iron from J774 macrophages. Perhaps higher levels of hepcidin would have been more effective. Alternatively, the continued but diminished release of iron from the macrophage in the presence of hepcidin may reflect the physiological situation and may partially account for the characteristically mild and nonprogressive nature of the anemia of inflammation (22). Although a reduction in macrophage iron release can be produced by suppression of iron export, it may also result from increased diversion of the metal from the prerelease iron pool into cellular ferritin stores, which increase during inflammation (23, 24). Assuming that iron is released from ferritin upon protein degradation, our observation that hepcidin treatment did not change steady-state ferritin levels indicates that reductions in iron export can occur without changes in this storage form of iron in the macrophage. However, we cannot exclude the possibility that changes in rates of ferritin synthesis may have affected iron export.
After phagocytosis of 59Fe-labeled erythrocytes by J774 macrophages, ≈25-30% of the total 59Fe was released as 59Fe-heme. The release of iron either as hemoglobin or heme is a consistent finding in studies of erythrophagocytosing macrophages (25-28). The mechanism of 59Fe-heme release from the macrophage is unknown; it may result from passive diffusion across the lipid bilayer (29) or it may involve a specific transporter. Support for the latter alternative is provided by the recent identification of a heme-export protein that is widely expressed among various cell types (30). Given that free heme is highly redox-active and seems to be the preferred growth substrate of certain pathogenic bacteria (31), regulated transport of heme would seem most beneficial to the host. However, although our studies directly implicate a role for hepcidin in the regulation of nonheme iron export because of changes in FPN1 levels, the peptide did not decrease levels of released 59Fe-heme. Thus, it is possible that other mechanisms influence this pool of heme iron generated by red cell catabolism.
Although our study rigorously identifies the function of FPN1 in macrophage iron recycling and its regulation by hepcidin, it also raises a number of questions. Iron status clearly plays a role in the cellular regulation of FPN1 expression, apparently at both the transcriptional and posttranscriptional levels (2, 7, 13, 32, 33). In light of systemic signaling via hepcidin, how does the macrophage integrate both organismal and cellular information relaying cues about availability and overload to export the appropriate amount of iron? Because hepatocytes are a major storage depot for iron and also express high levels of FPN1, how does regulation of FPN1 function in the macrophage by hepcidin consequently influence liver iron storage? These questions must be explored in further analyses of FPN1 function and regulation.
Acknowledgments
We thank Drs. Tomas Ganz and Elizabeta Nemeth (University of California, Los Angeles) for kindly providing a sample of synthetic hepcidin. This research was supported by the Florida Agricultural Experiment Station and National Institutes of Health Grants DK065064 (to M.D.K.) and DK56160 (to M.W.-R.) and was approved for publication as Journal Series R-10655.
Abbreviations: FPN1, ferroportin 1; EIgG, opsonized erythrocytes.
References
- 1.Knutson, M. & Wessling-Resnick, M. (2003) Crit. Rev. Biochem. Mol. Biol. 38, 61-88. [DOI] [PubMed] [Google Scholar]
- 2.Abboud, S. & Haile, D. J. (2000) J. Biol. Chem. 275, 19906-19912. [DOI] [PubMed] [Google Scholar]
- 3.Donovan, A., Brownlie, A., Zhou, Y., Shepard, J., Pratt, S. J., Moynihan, J., Paw, B. H., Drejer, A., Barut, B., Zapata, A., et al. (2000) Nature 403, 776-781. [DOI] [PubMed] [Google Scholar]
- 4.McKie, A. T., Marciani, P., Rolfs, A., Brennan, K., Wehr, K., Barrow, D., Miret, S., Bomford, A., Peters, T. J., Farzaneh, F., et al. (2000) Mol. Cell 5, 299-309. [DOI] [PubMed] [Google Scholar]
- 5.Yang, F., Liu, X. B., Quinones, M., Melby, P. C., Ghio, A. & Haile, D. J. (2002) J. Biol. Chem. 277, 39786-39791. [DOI] [PubMed] [Google Scholar]
- 6.Pietrangelo, A. (2004) Blood Cells Mol. Dis. 32, 131-138. [DOI] [PubMed] [Google Scholar]
- 7.Knutson, M. D., Vafa, M. R., Haile, D. J. & Wessling-Resnick, M. (2003) Blood 102, 4191-4197. [DOI] [PubMed] [Google Scholar]
- 8.Nemeth, E., Tuttle, M. S., Powelson, J., Vaughn, M. B., Donovan, A., Ward, D. M., Ganz, T. & Kaplan, J. (2004) Science 306, 2090-2093. [DOI] [PubMed] [Google Scholar]
- 9.Pigeon, C., Ilyin, G., Courselaud, B., Leroyer, P., Turlin, B., Brissot, P. & Loreal, O. (2001) J. Biol. Chem. 276, 7811-7819. [DOI] [PubMed] [Google Scholar]
- 10.Nicolas, G., Bennoun, M., Devaux, I., Beaumont, c., Grandchamp, B., Kahn, A. & Vaulont, S. (2001) Proc. Natl. Acad. Sci. USA 98, 8780-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemeth, E., Valore, E. V., Territo, M., Schiller, G., Lichtenstein, A. & Ganz, T. (2003) Blood 101, 2461-2463. [DOI] [PubMed] [Google Scholar]
- 12.Nicolas, G., Viatte, L., Lou, D. Q., Bennoun, M., Beaumont, C., Kahn, A., Andrews, N. C. & Vaulont, S. (2003) Nat. Genet. 34, 97-101. [DOI] [PubMed] [Google Scholar]
- 13.Chung, J., Haile, D. J. & Wessling-Resnick, M. (2004) Proc. Natl. Acad. Sci. USA 101, 2700-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krantz, S. B. & Fried, W. (1968) J. Lab. Clin. Med. 72, 157-164. [PubMed] [Google Scholar]
- 15.Hunter, H. N., Fulton, D. B., Ganz, T. & Vogel, H. J. (2002) J. Biol. Chem. 277, 37597-37603. [DOI] [PubMed] [Google Scholar]
- 16.McKie, A. T. & Barlow, D. J. (2004) Pflügers Arch. 447, 801-806. [DOI] [PubMed] [Google Scholar]
- 17.Galli, A., Bergamaschi, G., Recalde, H., Biasiotto, G., Santambrogio, P., Boggi, S., Levi, S., Arosio, P. & Cazzola, M. (2004) Br. J. Haematol. 127, 598-603. [DOI] [PubMed] [Google Scholar]
- 18.Finch, C. A. & Huebers, H. (1982) N. Engl. J. Med. 306, 1520-1528. [DOI] [PubMed] [Google Scholar]
- 19.Lipschitz, D. A., Simon, M. O., Lynch, S. R., Dugard, J., Bothwell, T. H. & Charlton, R. W. (1971) Br. J. Haematol. 21, 289-303. [DOI] [PubMed] [Google Scholar]
- 20.Fillet, G., Beguin, Y. & Baldelli, L. (1989) Blood 74, 844-851. [PubMed] [Google Scholar]
- 21.Nicolas, G., Chauvet, C., Viatte, L., Danan, J. L., Bigard, x., Devaux, I., Beaumont, C., Kahn, A. & Vaulont, S. (2002) J. Clin. Invest. 110, 1037-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, G. R. (1983) Semin. Hematol. 20, 61-80. [PubMed] [Google Scholar]
- 23.Konijn, A. M. & Hershko, C. (1977) Br. J. Haematol. 37, 7-16. [PubMed] [Google Scholar]
- 24.Birgegard, G. & Caro, J. (1984) Scand. J. Haematol. 33, 43-48. [DOI] [PubMed] [Google Scholar]
- 25.Kleber, E. E., Lynch, S. R., Skikne, B., Torrance, J. D., Bothwell, T. H. & Charlton, R. W. (1978) Br. J. Haematol. 39, 41-54. [DOI] [PubMed] [Google Scholar]
- 26.Custer, G., Balcerzak, S. & Rinehart, J. (1982) Am. J. Hematol. 13, 23-36. [DOI] [PubMed] [Google Scholar]
- 27.Costa, L. M., Moura, E. M., Moura, J. J. & de Sousa, M. (1998) Biochem. Biophys. Res. Commun. 247, 159-165. [DOI] [PubMed] [Google Scholar]
- 28.Kondo, H., Saito, K., Grasso, J. P. & Aisen, P. (1988) Hepatology 8, 32-38. [DOI] [PubMed] [Google Scholar]
- 29.Light, W. R., III & Olson, J. S. (1990) J. Biol. Chem. 265, 15623-15631. [PubMed] [Google Scholar]
- 30.Quigley, J. G., Yang, Z., Worthington, M. T., Phillips, J. D., Sabo, K. M., Sabath, D. E., Berg, C. L., Sassa, S., Wood, B. L. & Abkowitz, J. L. (2004) Cell 118, 757-766. [DOI] [PubMed] [Google Scholar]
- 31.Skaar, E. P., Humayun, M., Bae, T., DeBord, K. L. & Schneewind, O. (2004) Science 305, 1626-1628. [DOI] [PubMed] [Google Scholar]
- 32.Lymboussaki, A., Pignatti, E., Montosi, G., Garuti, C., Haile, d. J. & Pietrangelo, A. (2003) J. Hepatol. 39, 710-715. [DOI] [PubMed] [Google Scholar]
- 33.Liu, X. B., Hill, P. & Haile, D. J. (2002) Blood Cells Mol. Dis. 29, 315-326. [DOI] [PubMed] [Google Scholar]