Abstract
The staphylococcal virulon is controlled largely by the agr locus, a global accessory gene regulator that is autoinduced by a self-coded peptide (AIP) and is therefore a quorum sensor. The agr locus has diverged within and between species, giving rise to AIP variants that inhibit heterologous agr activation, an effect with therapeutic potential against Staphylococcus aureus: a single dose of an inhibitory AIP blocks the formation of an experimental murine abscess. As the AIP is unstable at physiological pH, owing to its essential thiolactone bond, its single-dose efficacy seems paradoxical, which has led us to analyze the in vivo kinetics of agr activation and the consequences of its blockage by a heterologous AIP. Initially, the infecting bacteria grow rapidly, achieving sufficient population density within the first 3 h to activate agr, and then enter a neutrophil-induced metabolic eclipse lasting for 2–3 d, followed by agr reactivation concomitantly with the development of the abscess. The inhibitory AIP prevents agr expression only during its short in vivo lifetime, suggesting that the agr-induced and therefore quorum-dependent synthesis of virulence factors shortly after infection is necessary for the subsequent development of the abscess lesion and bacterial survival. We confirm this finding by showing that a sterile agr+ supernatant causes a sterile abscess similar to the septic abscess caused by live bacteria. These results may provide a biological rationale for regulation of virulence factor expression by quorum sensing rather than by response to specific host signals.
Keywords: bioluminescent imaging, in vivo gene expression, bacterial pathogenesis
Although there is a large body of information on the in vitro regulation of bacterial accessory genes involved in pathogenesis (referred to here as the virulon), the in vitro environment is highly artificial and applies only imperfectly, at best, to the real-life situation. To date, however, most in vivo approaches have generated single-point transcription profiles for infecting organisms but only a handful have addressed the expression pattern of virulence determinants through time, a critical feature of any infection (1). We have begun to analyze temporal expression of staphylococcal virulence genes in vivo in animal models and to compare the results with those obtained in vitro. We have used real-time in vivo bioluminescent imaging to monitor the in vivo temporal expression of agr (accessory gene regulator), a model virulence regulator in Staphylococcus aureus (recently reviewed in ref. 2).
Agr is a complex locus that controls expression of a substantial part of the staphylococcal virulon (3, 4) consistent with its central role in pathogenesis (5–8). It consists of two divergent transcription units, driven by promoters P2 and P3 (9). The P2 operon contains four genes, agrB, agrD, agrC, and agrA, all of which are required for transcriptional activation of the agr system. AgrC is the receptor and AgrA the response regulator of a two-component signal transduction module that is autoinduced by a posttranslationally modified small peptide (AIP) (10, 11), processed by AgrB from the 46-residue agrD propeptide (12). The primary function of this four-gene unit is to activate the two major agr promoters, P2 and P3, significantly aided by a second regulatory protein, SarA (13, 14). The actual effector of agr-dependent exoprotein gene regulation, however, is the P3 transcript RNAIII, which acts primarily at the level of transcription, by an unknown mechanism (15). Being autoinduced, agr expression is population density-dependent, and agr is therefore a quorum sensor.
Evolutionary divergence within the agr locus has given rise to multiple specificity groups, of which there are four in S. aureus (10, 16) and at least 20 others in non-aureus staphylococci (17). A key feature of this diversity is that heterologous AIPs competitively inhibit agr activation by the cognate ligand (18). Indeed, a single dose of an inhibitory AIP, given along with the infecting bacteria, blocks the development of an experimental murine abscess, which, in the absence of treatment, would mature 2–3 d later (19). This effect seems paradoxical because the AIP has a short lifetime, perhaps 3 h in vivo.
We have addressed this apparent paradox by means of an imaging system that enables the monitoring of bacterial gene expression during the course of an experimental infection. This system uses a highly sensitive charge-coupled device camera to measure the bioluminescence from a luciferase-reporter carried by bacteria in vivo. With this method, we have found that infecting bacteria grow rapidly during the first 3 h, activating agr and producing toxic exoproteins. Thereafter, they enter a neutrophil-induced metabolic eclipse phase that lasts for 24–48 h after which the abscess matures, accompanied by reactivation of bacterial metabolism. Administration of an inhibitory AIP delays agr activation for only 2–4 h, but nevertheless, as noted, blocks formation of the abscess. As a sterile postexponential supernatant from an agr+ but not from an agr– strain causes a sterile lesion similar to the septic abscess, we suggest that the quorum sensing (QS) system must be activated during the first 3 h. Presumably, a 3-h delay is sufficient to block the production of toxic exoproteins and thus prevent the formation of an abscess.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions. Bacterial strains and plasmids are listed in Table 1. Bacteria were routinely grown on GL media with appropriate antibiotics and in CYGP broth (20). Cell growth was monitored by a Klett–Summerson colorimeter with a green (540 nm) filter. Inocula for in vivo studies were prepared by growing cells from Klett 5 to Klett 50 (early exponential phase). The cells were harvested, washed and resuspended in PBS, and stored at –80°C for up to 4 weeks until used. Sterile culture filtrates from late exponential-phase cells were prepared by growth of RN6734 and RN7206 in CYGP without glucose to Klett 400 followed by centrifugation and passage through a 0.2-μm filter (Nalgene).
Table 1. Strains and plasmids used in this study.
| Strains/plasmids | Relevant characteristics | Source |
|---|---|---|
| Strains | ||
| RN4220 | UV-induced restriction deficient mutant | 24 |
| RN6734 | NCTC 8325-4, φ13 lysogen, agr-I | 50 |
| RN7206 | agr::tetM derivative of 6734 | 50 |
| RN9130 | RN6607 (502A), Tcr plasmid cured, agr-II | 10 |
| RN9120 | agr::tetM derivative of RN9130 | 22 |
| Plasmids | ||
| pI524 | blalR blaZ Pcr Cdr | 51 |
| pMK4-lux | pMK4 luxABCDE Cmr | 21 |
| pJW7141 | agrp3-lux | This study |
| pJW7142 | blaZp-lux | This study |
| pRN7129 | agrCA-II agrp3-blaZ Emr | 18 |
The agrp3 and blaZ promoters were cloned by PCR from chromosomal DNA isolated from RN6734 and the staphylococcal β-lactamase plasmid, pI524, respectively into the shuttle plasmid, pMK4, carrying luxABCDE from Photorhabdus luminescens, optimized for expression in Gram-positive bacteria (21). Promoter sequences were confirmed by dye terminator DNA sequencing chemistry (Skirball DNA Sequencing Core Facility). The AIP biosensor strain used for the mixed infection carries the agrp3-lux fusion in RN7206 along with the group II-specific agrCA two-component system (22). Plasmids were electroporated (23) into the S. aureus restriction-deficient strain, RN4220 (24), followed by electroporation or phage transduction into RN6734 and RN7206. Strains carrying the blaZp fusion also carried the β-lactamase control system on pI524. Plasmid retention was >95% after overnight growth in vitro in the absence of antibiotics and after recovery from 5-day-old lesions.
Murine Subcutaneous Abscess Model. Groups (n = 3 per variable examined) of hairless, euthymic SKH-1 (ISL) mice (Charles River Breeding Laboratories) were used in the murine s.c. abscess model (25). In this model, staphylococci are injected s.c. with cytodex beads, and an abscess develops at the site of injection 2–3 d later, then sloughs and drains. The organisms grow rapidly at first; the population then levels out and remains constant thereafter. Polymorphonuclear leuykocytes (PMNs) are attracted to the developing lesion by bacterial products and proinflammatory cytokines, reaching maximum numbers by ≈6 h. In one report, ≈50% of the organisms were phagocytized, the rest remaining extracellular (26).
Bacteria were thawed, diluted to appropriate cell density, mixed with sterile cytodex beads (Sigma), and injected s.c. in a volume of 0.1 ml in the flank region. For agr inhibition studies, AIP-II (10 μg) in 5% DMSO in PBS or a control dose of 5% DMSO in PBS was mixed with the bacteria immediately before injection. For PMN depletion, cyclophosphamide (200 mg/kg) was injected i.p. 4 d before infection and again 2 d before infection (26). Blood harvested by retroorbital bleed immediately before infection was analyzed by flow cytometry in a FACSCalibur instrument (Becton Dickinson), which showed that the circulating PMN population was depleted by >95% with little or no effects on other circulating white blood cell populations. Mice were fed a standard diet ad libitum and monitored daily for signs of distress. Colony-forming units per abscess were determined by plating serial dilutions of excised lesions homogenized in PBS plus 0.5% Triton X-100.
Measurement of Bioluminescence in Vivo. Mice were imaged under isoflurane inhalation anesthesia as described (21). Briefly, photons emitted from the bacteria were collected during a 1- to 5-min exposure by using the IVIS Imaging System and living image software (Xenogen, Alameda, CA). Bioluminescent images were displayed by using a pseudocolor scale (blue representing least intense and red representing the most intense signal) overlaid onto a gray-scale photographic image. For the bioluminescent images in the figures, the upper and lower limits of the overall bioluminescent signal collected by the charge-coupled device array are displayed in counts. For the graphs, the signal collected on a portion of the charge-coupled device array corresponding to the area of injected bacteria was determined in photons per sec per cm2 per steradian, referred to as relative light units (RLU), and plotted vs. time after initiation of infection.
Determination of AIP-II Half-Life. To approximate the active lifetime of AIP, a synthetic version of AIP-II (27) was incubated in PBS or rabbit serum at 37°C for 4 h. The sample was diluted, and a dose–response assay for activation of β-lactamase induction of strain RN7206 carrying the agrCA-II agrp3-blaZ reporter fusion was performed as described (18, 28).
Results
Development and in Vivo Testing of the Luciferase Reporter System. We generated derivative of several test strains by introducing previously constructed staphylococcal plasmids containing the agr P3 promoter (agrp3) or the staphylococcal pI524-derived β-lactamase promoter (blaZp) transcriptionally fused to a modified version of the Photorabdis luciferase operon (21). In this configuration, the luciferase operon directs the synthesis of the native luciferase substrate. In vitro, in agr+ host strains, the agrp3 promoter shows classical midexponential agr induction (29); in agr– strains the agrp3 promoter is expressed very weakly throughout growth. The blaZp promoter is expressed at the same constant rate throughout growth in agr+ and agr– stains (30) (Fig. 5, which is published as supporting information on the PNAS web site). To test the system in vivo, we injected early exponential-phase bacteria plus cytodex beads s.c. into the flank region of hairless mice and followed the progress of the infection in the living animals with the IVIS imaging camera (31). Initial tests were performed with graded bacterial inocula, from 106 to 108 organisms.
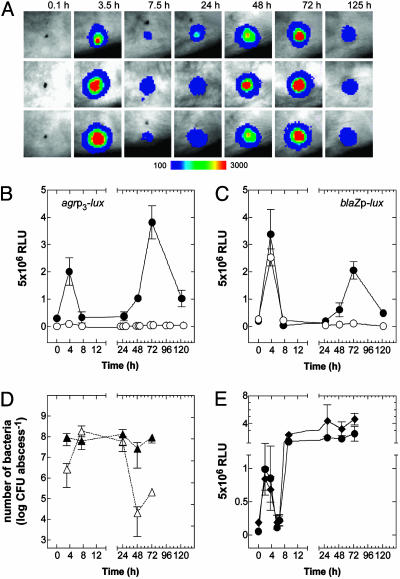
Fig. 1A shows a representative set of images of three different mice taken at various times after infection with RN6734, an agr+ strain (agr-I) containing the agrp3-lux fusion. As can be seen, there is very little signal immediately after infection. The signal then develops rapidly, reaching a peak by ≈3 h, and subsequently declines equally rapidly, remaining in an “eclipse” state for 36–48 h, after which it sharply increases once again. The second activation peak probably represents renewed growth of the bacteria in the necrotic tissue associated with the abscess. The results for three mice are shown to illustrate reproducibility. In subsequent experiments, we usually used 108 agr-I bacteria, which produced a visible s.c. lesion more reliably than lower inocula. The quantitative luciferase data obtained from these experiments are plotted in Fig. 1B, along with corresponding data for RN7206, an agr– but otherwise isogenic strain carrying the same agrp3-lux fusion and for agr+ and agr– bacteria carrying the blaZp-lux fusion (Fig. 1C). Predictably, comparisons within the first 6 h show the rapid rise and decline of the agrp3-lux signal for both agr+ and agr– strains, but with the signal amplitude of the latter amounting to ≈1/20 of the former and representing the basal activity of the agrp3 promoter. The constitutive blaZp also showed this sharp rise and decline, with the same amplitude and the same magnitude for both agr+ and agr– strains.
Fig. 1.
Expression of agr during infection. (A) Bioluminescent activity of agr+ (agr-I) bacteria carrying agrp3-lux after s.c. injection in the flank region of SKH-1 mice. Signal intensity is indicated by a pseudocolor scale. (B and C) RLU per lesion were plotted as a function of time for an infecting dose of 108 bacteria carrying either agrp3-lux (B)or blaZp-lux in agr+ (•) and agr– (○)(C) backgrounds in groups of three mice. (D) Viable counts of agr+ (▴) and agr– (▵) bacteria were enumerated at different points during infection. (E) Activity of agrp3-lux (•) and blaZp-lux (♦) in agr+ bacteria after infection of PMN-depleted mice.
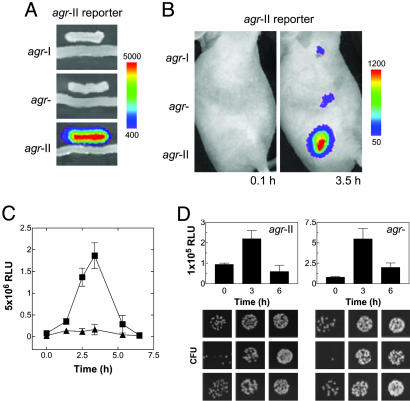
Agr Transactivation. Given the observed parallel between the agrp3-lux and blaZp-lux signals during the first 3 h, it was possible that the agrp3-lux signal represented only growth of the bacteria and not specific agr activation. To test for specific agr activation we used a mixed infection strategy in which one of the infecting strains would generate a luciferase signal in response to AIP produced by the other (Fig. 2A). The responding strain, in effect, a biosensor, carried agrp3-lux plus agrAC-II and the other was a WT agr-II strain. As shown in Fig. 2B, a strong luciferase signal was produced in the mixed infection with effectively the same kinetics as seen in mice singly infected with the WT strain carrying agrp3-lux (Fig. 1 A). Also shown are mixed infections with the biosensor plus either of two control strains, one producing AIP-I, the other producing no AIP. With either of the latter, only a weak signal representing the basal activity of the agrp3-lux was observed. Note that this basal activity is not inhibited by an antagonistic AIP (18), so that similar signals are seen with the agr-null and agr-I coinfecting control strains. Because the bacteria were washed and there was no immediate increase in the signal, it can be concluded that AIP-II was produced in vivo rather than being injected along with the bacteria. Thus, this response confirms the production and transactivity of this peptide by the infecting bacteria.
Fig. 2.
Autoinduction of agr in vivo.(A) The group-specific AIP-II biosensor strain responds in trans only to an agr-II strain producing its cognate autoinducer, AIP-II, on solid media. (B) A total of 108 agr-I, agr– (agr-II::tetM), or agr-II were mixed with equal numbers of the AIP-II biosensor, and a s.c. infection was initiated (n = 3) followed by in vivo imaging. The results with a single mouse represent the series. (C) Bioluminescence generated from 5 × 106 agr-II carrying agrp3-lux (▪) or blaZp-lux (▴) as described in Fig. 1. (D) (Upper) blaZp-lux activities during infection with agr-II and its agr-null derivative. (Lower) The corresponding bacterial colonies obtained by plating identical serial dilutions of the homogenized tissue excised from the site of infection at 0, 3, and 6 h. Each panel shows the bacterial colonies from a single lesion.
Effect of Inoculum on agr Activation Kinetics. We next addressed the failure of agrp3-lux to show the expected activation kinetics in vivo, despite the above demonstration of agr activation in trans. One possibility was that the high infecting dose was responsible. To test this notion, we used a more virulent strain, RN9130 (agr-II), that could reliably produce an abscess at a lower infecting dose than RN6734. After injection of 5 × 106 bacteria, containing either agrp3-lux or blaZp-lux, we observed the expected burst of agrp3 activity, ≈10-fold greater than the increase in blaZp activity (Fig. 2C). The latter was nearly parallel to the clear increase in bacterial numbers (Fig. 2D), confirming the utility of blaZp-lux as a monitor of bacterial growth during this period. The difference in amplitude between agrp3 and blaZp was not observed with an agr-null derivative of RN9130 (RN9120). Aside from its clear activation of agrp3, RN9130 at 5 × 106 showed temporal patterns of agrp3 and blaZp activity almost identical to those of RN6734 at 108.
The Decline Phase. Growth of agr+ and agr– bacteria appears to level off after 3 h, which may represent entry of the bacteria into stationary phase, as is normally observed in vitro. This phase is followed by a sharp decline in luciferase activity for both agrp3-lux and blaZp-lux. Given that luciferase activity (catalyzed by LuxAB) depends on the endogenous synthesis of the luciferase substrate (by LuxCDE) plus the required cofactors FMNH2 and O2, it is a sensitive indicator of the metabolic state of the organism (31) as well as a very sensitive reporter of transcription (and translation) activity and thus it seems likely that lack of substrate is responsible for the decline. We have observed a similar reduction in luciferase activity after bacterial growth has ceased on solid media in vitro. Bioluminescence is immediately restored by exogenous substrate or soon after the infusion of fresh media (unpublished results), suggesting a metabolic effect on substrate availability. Alternatively, immediate host responses to infection may be inducing a stress response that down-regulates expression of agrp3 and blaZp; the action of complement, acute-phase proteins in inflammatory fluids, or nitric oxide released by local host cells may represent candidate factors that exert this effect.
The Eclipse Phase: Effect of Neutrophils. We next considered the eclipse period. Because viable counts of agr+ bacteria taken from the lesions during this time showed no decrease (Fig. 1C), consistent with previous studies using this model (26, 32), the eclipse cannot be accounted for by loss of bacterial viability. We suggest, therefore, that the eclipse represents metabolic shutdown, perhaps owing to phagocytosis by PMNs attracted to the site of infection. Measurements of luciferase activity at intervening time points (9.5 and 19.5 h) showed the same very low activities as depicted in Fig. 1. It is conceivable, but unlikely, that promoter activation occurs between these time points. It has been reported that the numbers of PMNs recruited to the site of infection in this model are maximal at 6 h (26), corresponding to the onset of the eclipse period. To test for a possible role of PMNs, we infected mice made neutropenic by treatment with cyclophosphamide. Fig. 1E shows the kinetics of luciferase production by RN6734 carrying agrp3-lux or blaZp-lux in neutropenic mice. Here, there is the same initial rise and decline of activity of both agrp3 and blaZp, as seen with normal mice, but there is no eclipse; both signals rapidly rise again to a high plateau and remain there for the next 2 d. This reactivation may be caused by the availability of nutrients provided by the influx of inflammatory fluids in response to the infection. Inexplicably, a characteristic lesion is formed by agr+ bacteria in PMN-depleted mice.
Thus the PMNs appear to be responsible for the eclipse, but not for the sharp decline in luciferase activity after the initial rise, because this effect evidently precedes the arrival of PMNs at the infection site. If, indeed, the eclipse phase is caused by metabolic shutdown by the PMNs, then one would ask how this happens. Previous studies with this model suggest that some 50% of the bacteria have been ingested by PMNs at this point during the progress of the infection (26), virulent S. aureus survive intracellularly (33), and PMNs obtained from an abscess are unable to kill S. aureus (34). In contrast, agr– staphylococci are rapidly eliminated in vivo, as reported in ref. 35 and confirmed here (Fig. 1D), and they failed to form an abscess or reactivate agrp3 or blaZp at 48–72 h (Fig. 1 B and C). Temporary residence within phagocytes is, incidentally, thought to be partly responsible for the refractoriness to antibiotics of staphylococci in an abscess (36, 37). Metabolic shutdown would presumably contribute importantly to this refractoriness.
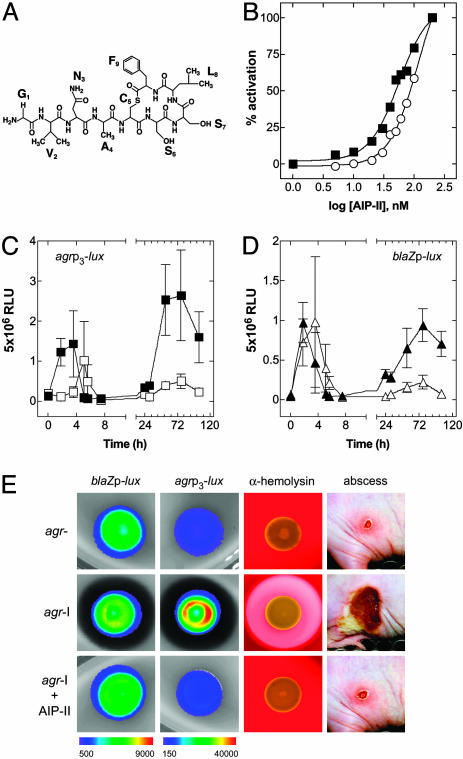
The Single-Dose Paradox. These results have set the stage for an understanding of the long-term effects of a single dose of an antagonistic AIP. A single dose of an antagonist AIP-II (Fig. 3A) coinjected with the agr-I bacteria blocks the subsequent formation of an abscess (19); under these conditions agr activation is inhibited for some 3 h but not eliminated (Fig. 3C), whereas blaZp activation is not significantly affected (Fig. 3D). This result suggests that the effective biological lifetime of the peptide in vivo is ≈3 h. As one would expect, this lifetime is considerably shorter than the in vitro lifetime (≈4 h in normal rabbit serum; Fig. 3B) because the AIP is inherently unstable at physiological pH, would almost certainly be cleared by the kidneys, and might be degraded by thioesterases and peptidases or inactivated by phagocyte-derived NADPH oxidases (38). A remarkable conclusion from this experiment is that the transient (≈3 h) inhibition of agr activation by the inhibitory AIP was sufficient to attenuate abscess formation and prevent the agr and blaZ reactivation, all of which occur 36–48 h later. In other words, agr-dependent events occurring within the first 3 h, in this model, are necessary (and probably sufficient) for the subsequent development of an abscess. Notably, the inhibitory AIP acts similarly on solid media in vitro, blocking the luciferase activity of the agrp3-lux construct and agr-induced α-hemolysin production without affecting blaZp-lux expression (Fig. 3E).
Fig. 3.
In vivo and in vitro consequences of agr inhibition by a synthetic QS peptide antagonist. (A) Structure of the agr-II autoinducer AIP-II, an agr-I signaling antagonist. (B) Determination of AIP-II half-life. The y axis for the graph indicates the percentage of maximal activation of agrp3-blaZ, and the x axis represents the concentration of AIP-II. The EC50 in PBS was 56.1 nM, and in serum it was 134 nM, indicating that the AIP-II half-life in vivo is <4h.(C and D) The effect of AIP-II on 108 agr-I bacteria carrying agrp3-lux (▪, untreated; □, AIP-II treated) (C) and blaZp-lux (▴, untreated; ▵, AIP-II treated) (D). (E) The effect of AIP-II on bioluminescence of agr-I bacteria carrying blaZp-lux or agrp3-lux when grown on solid media (24 h), on α-hemolysin production when grown on 5% sheep blood agar (24 h), and on lesion size in the murine s.c. model (at 96 h) in comparison with agr–.
Components of the agr-Triggered Virulon Cause a Sterile Abscess. The activation of agr-I, when blocked by AIP-II, is not only delayed, but is also weaker than in the absence of any inhibitory peptide. More importantly, the delayed activation occurs at about the same time that the eclipse period starts. It will be recalled that at least some of the agr up-regulated virulence factors are produced as long as 2 h after agrp3 activation in vitro (29), and if the eclipse involves these, then they would probably not be produced after the delayed activation of the agr system. That is, the antagonistic AIP delays production of the abscess-inducing factors for just long enough to enable the PMNs to take over.
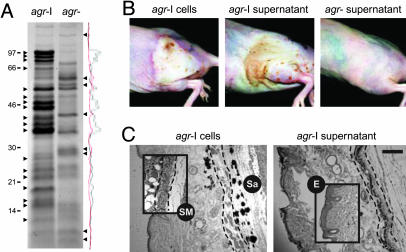
To test for a role of agr-induced factors (exoproteins) we injected sterile supernatant from postexponential cultures of agr+ (agr-I) and agr– strains instead of viable bacteria. As seen in Fig. 4A, there is a dramatic difference in exoproteins elaborated by these strains. Remarkably, the agr+ supernatant produced a sterile lesion superficially very similar, although less well demarcated, and with the same kinetics as viable agr+ organisms (Fig. 4B). We note that this effect was observed many years ago, using supernatants from uncharacterized strains (39). Neither the agr– nor a boiled agr+ supernatant (data not shown) produced a detectable lesion. To compare the septic and sterile lesions microscopically, we killed several mice and prepared histological sections. As shown in Fig. 4C, the septic abscess consists of large numbers of PMNs, Gram-positive cocci, and local tissue necrosis, whereas the sterile lesion lacks the easily identified cocci. Our interpretation is that agr-induced exoproteins are responsible for the abscess and are not made in sufficient quantities when agr expression is blocked, even temporarily, by an AIP antagonist or is eliminated by disruption of the agr locus.
Fig. 4.
Histological comparison of lesions caused by agr+ (agr-I) cells and sterile supernatant. (A) Exoproteins from agr+ and agr– supernatants were precipitated with 10% trichloroacetic acid and separated by 10% Tricine SDS/PAGE. After staining with Coomassie brilliant blue and densitometry analysis, several bands were identified that were differentially expressed between the two strains. Sterile culture filtrate was diluted up to 1:4 in PBS, and 0.1 ml was injected without cytodex beads. (B) Gross lesions at 48 h caused by agr+ cells, agr+ supernatant, and agr– supernatant. (C) Histological sections of the lesions caused by agr+ cells and agr+ supernatant were stained with hematoxylin and eosin. A Gram stain indicates the dark-stained regions of the lesion caused by agr+ cells that represent clumped staphylococci (Sa). (Insets) For comparison the integrity of the s.c. musculature (SM) (demarcated with dashed lines) and the epidermis (E) in a lesion caused by agr– cells is illustrated. (Scale bar: 200 μM.)
Discussion
The results presented here delineate the natural history of staphylococcal abscess formation in terms of the temporal pattern of agr expression. Entry of a relatively small number of staphylococci into the s.c. tissue of the mouse leads to rapid growth and activation of the agr QS system (≈3 h) and is followed by an equally rapid decline (≈7 h). The decline in activity could represent entry of the bacteria into the stationary phase or metabolic inhibition by host factors and is independent of PMNs because it occurs in neutropenic mice. A PMN-dependent metabolic eclipse period ensues, lasting for ≈48 h, during which agr– bacteria are killed and agr+ bacteria form the lesion. This phase is followed, with agr+ but not with agr– organisms, by the reactivation of both promoters and the maturation of the abscess (≈48–72 h). Administration of an inhibitory AIP along with the bacteria transiently inhibits agr (but not blaZ) activation and blocks subsequent formation of the lesion. The inhibitory effects of the antagonistic AIP last just long enough for the PMN-induced eclipse to be initiated. Because S. aureus supernatant is cytotoxic to PMNs in vitro (40), we suggest that the early production of exoproteins may establish an inhospitable environment for the incoming PMNs and cause the influx of tissue fluids. This feature could enhance survival of extracellular and ingested bacteria and provide a new source of nutrients, facilitating later reactivation.
A fundamental conclusion from these studies is that a rapid burst of agr activity early in infection, perhaps before the innate immune system has had time to assemble a response, is critical for later survival of the bacteria in host tissues. These results may provide a biological rationale for QS activation of the staphylococcal virulon, namely a requirement for large quantities of the toxic exoproteins to neutralize the killing function of the PMNs and enable the development of the characteristic staphylococcal lesion. If the virulon were activated immediately after entry, for example, by the elevated temperature of the mammalian body, then the innate immune response might be mobilized before there was a sufficient quantity of exoproteins to counter its effects, and the organisms would be eliminated. Both agr+ and agr– bacterial multiply rapidly before PMNs are mobilized (see Fig. 2D); agr+ organisms then undergo autoinduction of the virulon, resisting killing by the influx of PMNs, whereas the agr– organisms eventually are eliminated. In support of this concept, which has also been articulated by Hentzer and Givskov (41), we have observed that administration of an agr+ supernatant along with agr– organisms protects the bacteria, which later persist in the septic abscess (unpublished results). The onset and rapid kinetics of agr autoinduction during infection also suggests that the development and administration of potential QS inhibitors for the treatment of certain types of staphylococcal infections and, perhaps, those caused by other bacteria may hold only prophylactic value.
Recent reports have suggested that agr is not expressed in vivo and is not relevant for pathogenesis. The results described above are clearly inconsistent with these reports but these inconsistencies may be explicable. Yarwood et al. (42) report that stationary-phase cells of S. aureus strain MN-NJ introduced into a sterile s.c. chamber in rabbits rapidly turn off agr RNAIII expression. We have long known that early stationary-phase cells, which are maximally induced for RNAIII, rapidly turn off RNAIII synthesis after dilution into fresh medium and similarly switch off RNAIII production after injection into mice (unpublished results). Goerke et al. (43, 44) report that staphylococci extracted from subacute or chronic lesions contain very low levels of RNAIII. These organisms were isolated from sputa of chronically S. aureus-infected cystic fibrosis patients (43) or 2–6 d after experimental infection (44), in other words, long after initiation of the infection. We suggest that agr expression is critically required for events early in infection. Because Goerke et al. did not collect any samples until 2 d after the initiation of their foreign body infection, they would not have observed early activation. Also, because agr activity critically depends on the metabolic activity of the organism it probably would not have been seen in organisms from the chronic cystic fibrosis lung because these would probably not be metabolically active. Therefore, the results of Goerke et al. are consistent with the eclipse concept and are not relevant to the well established essentiality of agr for staphylococcal pathogenicity. We also note that a similar shift from high to low metabolic state has been reported for experimental foreign body Staphylococcus epidermidis infections (45).
An interesting contrast with the observations reported here exists with S. aureus strains producing the agr-regulated superantigen, toxic shock syndrome toxin-1 (TSST-1). Local infections caused by these organisms are remarkably apurulent (46, 47), owing to the repression of exoprotein genes by the agr-induced superantigen (48). Thus the regulatory behavior of TSST-1 serves to illustrate an agr-dependent pathogenic strategy that is radically different from that used for abscess formation, which involves production of large quantities of exoproteins. Moreover, TSST-1 is encoded by a highly mobile pathogenicity island (49). Thus the agr QS system represents a fundamental regulatory paradigm that can encompass different adaptive strategies and accommodate horizontally acquired virulence determinants.
Many fundamental questions raised by this work remain to be resolved: (i) What is the cause of the resurgence in promoter activity in the absence of PMNs, after its dramatic decline? (ii) Does the eclipse represent a metabolic shutdown? (iii) Is it a consequence of phagocytosis, extracellular PMN-related factors, or both? (iv) Is the reactivation of promoter activity after the eclipse caused by growth of bacteria in necrotic tissue, to escape from phagocytes, or other factors? Answers to these questions are expected to significantly increase our understanding of the role of staphylococcal QS and PMNs in the pathogenesis of a typical staphylococcal lesion.
Supplementary Material
Acknowledgments
We thank T. M. Muir (The Rockefeller University, New York) for providing a sample of synthetic AIP-II, A. Nieves and C. Williams for media and reagent preparation, A. M. Maraver and J. Lafaille for assistance with flow cytometry, B. Arias for producing histological sections, F. Diaz-Griffero for help with microscopy, and Y. Weinrauch and G. J. Lyon for helpful discussions. This work was supported by National Institutes of Health Grant RO142736 (to R.P.N.) and National Research Service Award Postdoctoral Fellowship F32AI055242 (to J.S.W.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: agr, accessory gene regulator; AIP, autoinducing peptide; QS, quorum sensing; PMN, polymorphonuclear leuykocyte; RLU, relative light units.
References
- 1.Shelburne, S. A. & Musser, J. M. (2004) Curr. Opin. Microbiol. 7, 283–289. [DOI] [PubMed] [Google Scholar]
- 2.Novick, R. P. (2003) Mol. Microbiol. 48, 1429–1449. [DOI] [PubMed] [Google Scholar]
- 3.Dunman, P. M., Murphy, E., Haney, S., Palacios, D., Tucker-Kellogg, G., Wu, S., Brown, E. L., Zagursky, R. J., Shlaes, D. & Projan, S. J. (2001) J. Bacteriol. 183, 7341–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Recsei, P., Kreiswirth, B., O'Reilly, M., Schlievert, P., Gruss, A. & Novick, R. P. (1986) Mol. Gen. Genet. 202, 58–61. [DOI] [PubMed] [Google Scholar]
- 5.Abdelnour, A., Arvidson, S., Bremell, T., Ryden, C. & Tarkowski, A. (1993) Infect. Immun. 61, 3879–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booth, M. C., Atkuri, R. V., Nanda, S. K., Iandolo, J. J. & Gilmore, M. S. (1995) Invest. Ophthalmol. Visual Sci. 36, 1828–1836. [PubMed] [Google Scholar]
- 7.Cheung, A. L., Eberhardt, K. J., Chung, E., Yeaman, M. R., Sullam, P. M., Ramos, M. & Bayer, A. S. (1994) J. Clin. Invest. 94, 1815–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillaspy, A. F., Hickmon, S. G., Skinner, R. A., Thomas, J. R., Nelson, C. L. & Smeltzer, M. S. (1995) Infect. Immun. 63, 3373–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novick, R. P., Projan, S. J., Kornblum, J., Ross, H. F., Ji, G., Kreiswirth, B., Vandenesch, F. & Moghazeh, S. (1995) Mol. Gen. Genet. 248, 446–458. [DOI] [PubMed] [Google Scholar]
- 10.Ji, G., Beavis, R. & Novick, R. P. (1997) Science 276, 2027–2030. [DOI] [PubMed] [Google Scholar]
- 11.Ji, G., Beavis, R. C. & Novick, R. P. (1995) Proc. Natl. Acad. Sci. USA 92, 12055–12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, L., Gray, L., Novick, R. P. & Ji, G. (2002) J. Biol. Chem. 277, 34736–34742. [DOI] [PubMed] [Google Scholar]
- 13.Chien, Y. & Cheung, A. L. (1998) J. Biol. Chem. 273, 2645–2652. [DOI] [PubMed] [Google Scholar]
- 14.Chien, Y., Manna, A. C. & Cheung, A. L. (1998) Mol. Microbiol. 30, 991–1001. [DOI] [PubMed] [Google Scholar]
- 15.Novick, R. P., Ross, H. F., Projan, S. J., Kornblum, J., Kreiswirth, B. & Moghazeh, S. (1993) EMBO J. 12, 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarraud, S., Lyon, G. J., Figueiredo, A. M., Gerard, L., Vandenesch, F., Etienne, J., Muir, T. W. & Novick, R. P. (2000) J. Bacteriol. 182, 6517–6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dufour, P., Jarraud, S., Vandenesch, F., Greenland, T., Novick, R. P., Bes, M., Etienne, J. & Lina, G. (2002) J. Bacteriol. 184, 1180–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyon, G. J., Wright, J. S., Christopoulos, A., Novick, R. P. & Muir, T. W. (2002) J. Biol. Chem. 277, 6247–6253. [DOI] [PubMed] [Google Scholar]
- 19.Mayville, P., Ji, G., Beavis, R., Yang, H., Goger, M., Novick, R. P. & Muir, T. W. (1999) Proc. Natl. Acad. Sci. USA 96, 1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novick, R. (1991) Methods Enzymol. 204, 587–683. [DOI] [PubMed] [Google Scholar]
- 21.Francis, K. P., Joh, D., Bellinger-Kawahara, C., Hawkinson, M. J., Purchio, T. F. & Contag, P. R. (2000) Infect. Immun. 68, 3594–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyon, G. J., Mayville, P., Muir, T. W. & Novick, R. P. (2000) Proc. Natl. Acad. Sci. USA 97, 13330–13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schenk, S. & Laddaga, R. A. (1992) FEMS Microbiol. Lett. 94, 133–138. [DOI] [PubMed] [Google Scholar]
- 24.Kreiswirth, B. N., Lofdahl, S., Betley, M. J., O'Reilly, M., Schlievert, P. M., Bergdoll, M. S. & Novick, R. P. (1983) Nature 305, 709–712. [DOI] [PubMed] [Google Scholar]
- 25.Bunce, C., Wheeler, L., Reed, G., Musser, J. & Barg, N. (1992) Infect. Immun. 60, 2636–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ford, C. W., Hamel, J. C., Stapert, D. & Yancey, R. J. (1989) J. Med. Microbiol. 28, 259–266. [DOI] [PubMed] [Google Scholar]
- 27.Lyon, G. J., Wright, J. S., Muir, T. W. & Novick, R. P. (2002) Biochemistry 41, 10095–10104. [DOI] [PubMed] [Google Scholar]
- 28.Wright, J. S., III, Lyon, G. J., George, E. A., Muir, T. W. & Novick, R. P. (2004) Proc. Natl. Acad. Sci. USA 101, 16168–16173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandenesch, F., Kornblum, J. & Novick, R. P. (1991) J. Bacteriol. 173, 6313–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novick, R. P. (1962) Biochem. J. 83, 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Contag, C. H., Contag, P. R., Mullins, J. I., Spilman, S. D., Stevenson, D. K. & Benaron, D. A. (1995) Mol. Microbiol. 18, 593–603. [DOI] [PubMed] [Google Scholar]
- 32.Molne, L., Verdrengh, M. & Tarkowski, A. (2000) Infect. Immun. 68, 6162–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gresham, H. D., Lowrance, J. H., Caver, T. E., Wilson, B. S., Cheung, A. L. & Lindberg, F. P. (2000) J. Immunol. 164, 3713–3722. [DOI] [PubMed] [Google Scholar]
- 34.Finlay-Jones, J. J., Hart, P. H., Spencer, L. K., Nulsen, M. F., Kenny, P. A. & McDonald, P. J. (1991) J. Med. Microbiol. 34, 73–81. [DOI] [PubMed] [Google Scholar]
- 35.Chan, P. F., Foster, S. J., Ingham, E. & Clements, M. O. (1998) J. Bacteriol. 180, 6082–6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bamberger, D. M., Herndon, B. L., Fitch, J., Florkowski, A. & Parkhurst, V. (2002) Antimicrob. Agents Chemother. 46, 2878–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vesga, O., Groeschel, M. C., Otten, M. F., Brar, D. W., Vann, J. M. & Proctor, R. A. (1996) J. Infect. Dis. 173, 739–742. [DOI] [PubMed] [Google Scholar]
- 38.Rothfork, J. M., Timmins, G. S., Harris, M. N., Chen, X., Lusis, A. J., Otto, M., Cheung, A. L. & Gresham, H. D. (2004) Proc. Natl. Acad. Sci. USA 101, 13867–13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker, J. T. (1924) J. Exp. Med. 40, 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haslinger, B., Strangfeld, K., Peters, G., Schulze-Osthoff, K. & Sinha, B. (2003) Cell. Microbiol. 5, 729–741. [DOI] [PubMed] [Google Scholar]
- 41.Hentzer, M. & Givskov, M. (2003) J. Clin. Invest. 112, 1300–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yarwood, J. M., McCormick, J. K., Paustian, M. L., Kapur, V. & Schlievert, P. M. (2002) J. Bacteriol. 184, 1095–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goerke, C., Campana, S., Bayer, M. G., Doring, G., Botzenhart, K. & Wolz, C. (2000) Infect. Immun. 68, 1304–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goerke, C., Fluckiger, U., Steinhuber, A., Zimmerli, W. & Wolz, C. (2001) Mol. Microbiol. 40, 1439–1447. [DOI] [PubMed] [Google Scholar]
- 45.Vandecasteele, S. J., Peetermans, W. E., Carbonez, A. & Van Eldere, J. (2004) J. Bacteriol. 186, 2236–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fast, D. J., Schlievert, P. M. & Nelson, R. D. (1988) J. Immunol. 140, 949–953. [PubMed] [Google Scholar]
- 47.Kreiswirth, B. N., Kravitz, G. R., Schlievert, P. M. & Novick, R. P. (1986) Ann. Intern. Med. 105, 704–707. [DOI] [PubMed] [Google Scholar]
- 48.Vojtov, N., Ross, H. F. & Novick, R. P. (2002) Proc. Natl. Acad. Sci. USA 99, 10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindsay, J. A., Ruzin, A., Ross, H. F., Kurepina, N. & Novick, R. P. (1998) Mol. Microbiol. 29, 527–543. [DOI] [PubMed] [Google Scholar]
- 50.Novick, R. P., Ross, H. F., Figueiredo, A. M. S., Abramochkin, G. & Muir, T. (2000) Science 287, 391a. [Google Scholar]
- 51.Novick, R. P. (1967) Fed. Proc. 26, 29–38. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.