Abstract
Tannerella forsythia is a periodontal pathogen expressing six secretory proteolytic enzymes with a unique multidomain structure referred to as KLIKK proteases. Two of these proteases, karilysin and mirolysin, were previously shown to protect the bacterium against complement-mediated bactericidal activity. The latter metalloprotease, however, was not characterized at the protein level. Therefore, we purified recombinant mirolysin and subjected it to detailed biochemical characterization. Mirolysin was obtained as a 66 kDa zymogen, which autoproteolytically processed itself into a 31 kDa active form via truncations at both the N- and C-termini. Further autodegradation was prevented by calcium. Substrate specificity was determined by the S1’ subsite of the substrate-binding pocket, which shows strong preference for Arg and Lys at the carbonyl side of a scissile peptide bond (P1’ residue). The protease cleaved an array of host proteins, including human fibronectin, fibrinogen, complement proteins C3, C4, and C5, and the antimicrobial peptide, LL-37. Degradation of LL-37 abolished not only the bactericidal activity of the peptide, but also its ability to bind lipopolysaccharide (LPS), thus quenching the endotoxin proinflammatory activity. Taken together, these results indicate that, through cleavage of LL-37 and complement proteins, mirolysin might be involved in evasion of the host immune response.
Keywords: antimicrobial peptide, metalloprotease, pathogenicity, periodontitis, proteolysis, virulence factor
Introduction
Periodontitis is arguably the most prevalent infection-driven chronic inflammatory disease of humankind. It is estimated that up to 15% of adults worldwide experience severe forms of periodontitis (Fox, 1992; Hugoson et al., 2008). In the United States alone, nearly 47% of people aged ≥30 (approximately 65 million adults) suffer from this disease (Eke et al., 2015). The progression of periodontitis is marked by gingival bleeding, suppuration, and progressive loss of alveolar bone, resulting in the formation of deep periodontal pockets. If left untreated, the disease can even lead to tooth loss (Armitage, 2004). Due to its chronic inflammatory and infectious character, periodontitis is associated with the progression and/or development of systemic diseases such as rheumatoid arthritis, cardiovascular diseases, diabetes, and aspiration pneumonia (Otomo-Corgel J et al., 2012; Benedyk M et al., 2016).
Because the etiology of periodontitis is multifactorial, it is impossible to implicate a single pathogen as responsible for development of the disease. Periodontitis begins with the formation of bacterial plaque in the subgingival space below the gum line. The plaque becomes pathogenic upon colonization by a consortium of bacteria: Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola, referred to as the “red complex” (Hajishengallis and Lamont, 2012; Wright et al., 2013). A common feature of bacteria belonging to the “red complex” is strong extracellular proteolytic activity, which is thought to be central to the virulence of these pathogens (Holt and Ebersole, 2005). The role of the secretory proteases of P. gingivalis and T. denticola in the pathogenesis of periodontitis has been well described (Guo et al., 2010; Ishihara, 2010). These enzymes are involved in several processes, including nutrient acquisition, immunomodulation, and corruption of the innate immune system (Potempa and Pike, 2009; Ishihara, 2010).
Until recently, our knowledge regarding the secretory proteases of T. forsythia has been very limited (Sharma, 2010). The only proteolytic enzyme that has been characterized in detail was PrtH, a cysteine protease of a caspase-like fold. PrtH detaches adherent cells from the substratum and increases IL-8 expression (Nakajima et al., 2006; Pei and Grishin, 2009). Our knowledge has been increased immensely with the discovery of a novel family of six secretory proteases in T. forsythia that includes three serine proteases (mirolase, miropsin-1, and miropsin-2) and three metalloproteases (karilysin, mirolysin, and forsilysin). These proteolytic enzymes are conserved in almost all clinical strains of T. forsythia but absent from the sequenced but not yet assembled genomes of 12 isolates of Tannerella BU063, which is a health-associated strain relative of T. forsythia. The enzymes are also capable of degrading elastin, a major component of gum connective tissue. Due to the presence of the sequential KL-I-K-K amino acids motif at the C-terminus, these proteases are referred to as KLIKK proteases. The KLIKK proteases possess a unique multidomain structure consisting of (beginning from the N-terminus) a signal peptide (SP) responsible for translocation across the inner membrane, an N-terminal profragment (NTP) responsible for enzyme latency, a catalytic domain (CD), and a C-terminal extension (CTE) encompassing a variable region flanked by two domains, which are nearly identical in all six KLIKK proteases (Ksiazek et al., 2015b). The domain at the C-terminus shares sequence similarity with the conserved C-terminal domain (CTD), which serves as a signal for the translocation of P. gingivalis and T. forsythia proteins across the outer membrane via a type IX secretion system (T9SS) (Sato et al., 2005; Nguyen et al., 2007; Tomek et al., 2014).
To date, only two KLIKK proteases, karilysin and mirolase, have been characterized in detail. Both proteases possess a unique mechanism responsible for their zymogenicity. Prokarilysin possesses the shortest propeptide described for metalloproteases, while in promirolase, a 19 kDa profragment forms a stable noncovalent complex with the CD after being cleaved off. Degradation of fibrinogen and the antimicrobial peptide LL-37 by karilysin, as well as synergistic inhibition of all complement activation pathways by both proteases, may contribute to the virulence of T. forsythia (Karim et al., 2010; Jusko et al., 2015; Ksiazek et al., 2015a; López-Pelegrín et al., 2015).
In contrast to karilysin, very little is known regarding the biochemical properties of mirolysin in the context of its putative role in the virulence of T. forsythia. Here, we characterized mirolysin, which exhibits a unique specificity for Xaa-Arg and Xaa-Lys peptide bonds and may contribute to the pathogenesis of periodontitis through inactivation of the biological activities of LL-37 and interference with complement functions.
Results
Mirolysin is a member of the pappalysin family (M43B) of metalloproteases
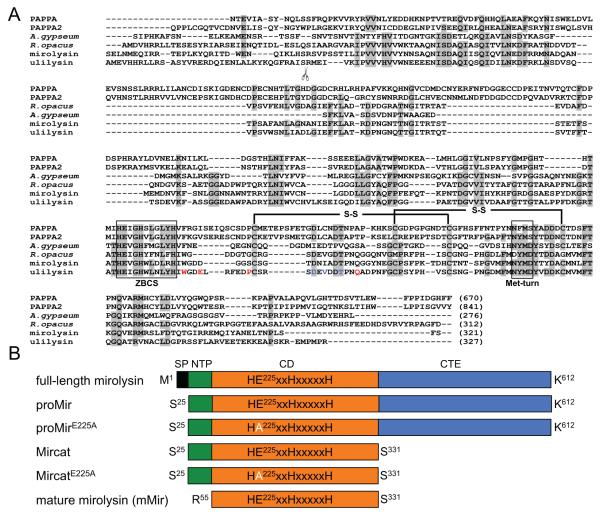
Bioinformatic analysis revealed that, according to the MEROPS database, mirolysin belongs to family M43, subfamily M43B, represented by pappalysin-1 (PAPPA) (Rawlings et al., 2016). Thus far, only one protease from subfamily M43B, ulilysin from Methanosarcina acetivorans, has been structurally characterized (Tallant et al., 2006). Due to its specificity, the metalloprotease was later renamed LysargiNase (Huesgen et al., 2015). Alignment of the CD sequence of mirolysin with five members of family M43B belonging to the three kingdoms of life revealed that mirolysin possesses both an extended zinc-binding consensus sequence (ZBCS), HEXXHXXGXXH/D, and a methionine-containing 1,4-β-turn called Met-turn. These findings identify mirolysin and other members of the M43B family as belonging to a group of metalloproteases called MetZincins, which includes astacins, adamalysins/ADAMs, serralysins, and matrixins. As in the case of ulilysin and PAPPA, mirolysin contains conserved cysteine residues and amino acids that are responsible for the binding of two calcium ions in ulilysin (Figure 1A; Overgaard et al., 2003; Tallant et al., 2006). Ulilysin, the CD of which shares almost 50% sequence identity with that of mirolysin, is synthesized as a 38 kDa zymogen that undergoes calcium-triggered autoactivation to the mature, 29 kDa protease. Autolytic activation entails removal of the N-terminal prodomain through cleavage of the Ser60-Arg peptide bond. The same peptide bond is conserved in mirolysin (Figure 1A), suggesting that the T. forsythia protease is also synthesized as a proenzyme that undergoes autoprocessing.
Figure 1.
Alignment of mirolysin with other members of the M43B protease family (A) and proteins used in this study (B). A) Sequence alignment of the catalytic domains of mirolysin (UniProt (UP) accession number: A0A0F7IPS1) and other pappalysins (family M43B) including human PAPPA (UP: Q13219) and PAPPA-2 (UP: Q9BXP8), ulilysin from Archaea (UP: Q8TL28), and proteases from the bacteria A. gypseum (UP: E5QZI4) and R. opacus (GenBank accession number: AII05686.1). Amino acid residues identical in a minimum of four out of six aligned sequenced are indicated by black letters on a gray background. Extended zinc-binding consensus sequences (ZBCS) and the Met-turn are boxed. Disulfide bonds identified for human pappalysin-1 (PAPPA) and ulilysin are marked with black lines. The scissors symbol indicates the site of autolytic cleavage during activation of ulilysin. The amino acid residues involved in the binding of two calcium ions by the protease are shown in red and blue fonts. B) Schematic drawing of the constructs and proteins expressed and purified in this study. SP – signal peptide; NTP – N-terminal profragment; CD – catalytic domain; CTE – C-terminal extension.
Expression, purification, and autoprocessing of mirolysin
To verify the zymogenicity of mirolysin, we cloned the coding sequence of mirolysin, without the putative SP (proMir), into the multicloning site of the pGEX-6P-1 vector for expression in E. coli BL21 (DE3) as a fusion protein with an N-terminal glutathione-S-transferase (GST) tag. Both the native form and a catalytically inactive mutein, in which the catalytic residue Glu225 was replaced by Ala (Figure 1B), were expressed. Both recombinant proteins were isolated by affinity chromatography on glutathione-Sepharose, with the GST tag removed by on-column cleavage with PreScission protease. The recombinant proteins were obtained as a 66 kDa tag-free proteins (Figure 2A, lane 3 in each panel). Of note, only a small amount (<0.3 mg/l of culture) was obtained, and all attempts to improve the expression yield were unsuccessful.
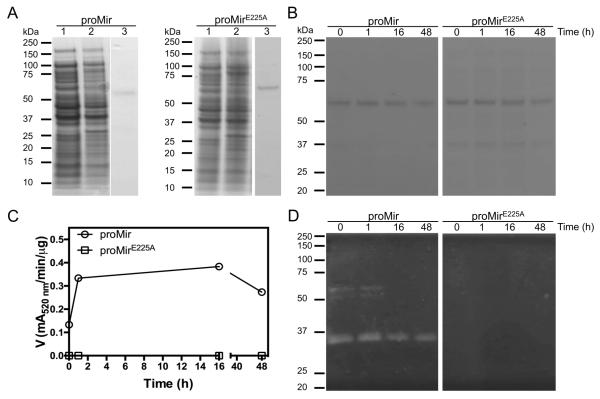
Figure 2.
Expression, purification, and autoprocessing of promirolysin. A) Both promirolysin (proMir) and the active-site-glutamic acid-to-alanine mutein (proMirE225A) were expressed in E. coli as a recombinant fusion protein with a GST and purified on a glutathione-Sepharose column with on-column removal of the tag. Lanes 1 and 2, E. coli extracts before and 16 h after induction of protein expression with IPTG, respectively; lane 3, the tag-free proMir and proMirE225A (66 kDa) proteins after purification by affinity chromatography with on-column removal of the GST by cleavage with PreScission protease. B-D) Proteolytic autoprocessing of mirolysin. ProMir and its active site mutein (proMirE225A) were incubated at 0.1 mg/ml in 50 mM Tris, pH 8.0, at 37°C. At defined time points, aliquots were taken for SDS-PAGE analysis (B) followed by measurement of activity employing Azocoll as a substrate (C) and zymography with gelatin as a substrate (D).
Other KLIKK proteases, including mirolase and karilysin, are synthesized as inactive zymogens that autoprocess into active enzymes. The same may apply to mirolysin, which possesses an apparent propeptide on the N-terminus that must be removed to activate the protease. To confirm this, we incubated both proMir and proMirE225A at 37°C for several hours. SDS-PAGE analysis revealed no change in the molecular mass of the proteins during incubation (Figure 2B), strongly suggesting that, under these conditions (50 mM Tris, pH 8.0), promirolysin is stable and does not undergo autoproteolysis. However, since the NTP encompasses only 30 amino acids residues, the change in molecular mass could be too small to be observed by SDS-PAGE. Therefore, we assayed the fractions after different periods of incubation for proteolytic activity using Azocoll as a substrate. Surprisingly, a significant amount of activity was detected even in the sample taken before incubation, and this increased approximately 3-fold after 1 h of incubation and stayed at that level after 16 h and 48 h of incubation (Figure 2C). As expected, no activity was detected in the proMirE225A samples. To study further the apparent activation of promirolysin, we performed zymography, which allows detection of the activity of zymogenic forms of prometalloproteases, which latency are exerted not only by “cysteine-switch” (Shimokawa et al., 2002), but also “aspartate-switch” (Karim et al., 2010; López-Pelegrín et al., 2015) mechanisms. Interestingly, in addition to a band with a molecular mass equivalent to 66 kDa (promirolysin), the analysis revealed an additional band just below 66 kDa (Figure 2D). These two bands were detected only in samples taken before incubation and after 1 h of incubation, while the strong ~31 kDa band corresponding to the fully processed CD of mirolysin was present in all analyzed samples. Taken together the increase of proteolytic activity observed during autoprocessing (Figure 2C) did not correlate with the results from SDS-PAGE (Figure 2B) indicating that promirolysin was stable during incubation at 37°C. This discrepancy can be explained by the presence of small fractions of active partially and fully processed protease in the 66 kDa promirolysin preparation.
Purification of mature mirolysin
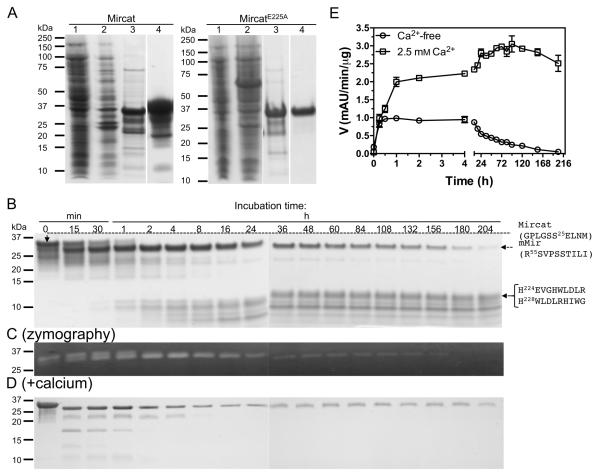
The findings presented above suggested that mirolysin, similar to karilysin and mirolase, is indeed synthesized as an inactive zymogen that autoprocesses into the mature form consisting of the CD alone (Karim et al., 2010; Ksiazek et al., 2015a). To further investigate the maturation of mirolysin, we expressed the protein without the CTE, but with the intact NTP (Figure 1B), which could act as a chaperone and thus be required for proper folding (Bryan, 2002). The recombinant proteins were purified by affinity chromatography on glutathione-Sepharose with simultaneous removal of the GST tag, followed by size exclusion chromatography (SEC) and ion exchange chromatography (only for MircatE225A). The procedure yielded more than 2 mg of purified protein per liter of culture (Figure 3A, lane 4 in each panel). Of note, in clear contrast to MircatE225A, lower bands were visible for Mircat, suggesting that this form of mirolysin is prone to autodegradation. To characterize the process in detail, Mircat was incubated at 37°C for 204 h in 50 mm Tris, 0.02% NaN3, pH 8.0, during which time aliquots were taken at specific time points for measurement of proteolytic activity using Azocoll and zymography, as well as SDS-PAGE analysis (Figure 3BCE). Mircat (35 kDa) completely processed itself into the mature 31 kDa form of mirolysin (mMir) within 2 h. This change in molecular mass correlated with a time-dependent increase in the proteolytic activity against Azocoll (Figure 3E) within the first 30 min of incubation. Similarly, zymographic analysis of the samples showed complete conversion of the upper band corresponding to Mircat to a lower band equivalent to that of mMir over 2 h (Figure 3C). This conversion was not associated with an increase in activity as measured by Azocoll degradation, apparently due to autodegradation of mMir that occurred after 1–2 h. After 4 h, the band corresponding to mMir gradually disappeared, and, by the end of the incubation period, had almost completely vanished. The gradual time-dependent decrease in the intensity of the mirolysin band correlated well with a decrease in proteolytic activity and accumulation of degradation products with molecular masses of 10 kDa and lower. The N-terminal sequence analysis of prominent bands at 10 kDa revealed that mirolysin cleaved itself at the Thr-His and Gly-His peptide bonds encompassing the Zn-chelating histidine residues in the conserved HEXXH motif (Figure 1B).
Figure 3.
Expression, purification, and the effect of calcium on autoprocessing of mirolysin without C-terminal extension (CTE). A) Catalytic domain preceded by the prodomain of mirolysin and its active site mutein (Mircat and MircatE225A, respectively) were obtained as recombinant proteins and purified by affinity chromatography on glutathione-Sepharose with simultaneous removal of the GST tag by PreScission cleavage followed by size exclusion chromatography (SEC) and ion exchange chromatography (MircatE225A only). Lanes 1 and 2, E. coli extracts before and 8 h after induction of protein expression with IPTG, respectively; lanes 3 and 4, proteins after purification by affinity chromatography and SEC followed by ion exchange chromatography for MircatE225A only, respectively. B-E) Mircat was incubated at 0.5 mg/ml in 50 mM Tris, 0.02% NaN3 pH 8.0, alone (B) or supplemented with 2.5 mM CaCl2 (D). At specific time points, aliquots were taken and analyzed by SDS-PAGE (B, D) followed by zymography (C, for incubation in calcium-free buffer only) and measurement of residual activity against Azocoll as a substrate (E). N-terminal sequences determined by Edman degradation are shown on the right.
The CD (without the CTE) of mirolysin shares almost 50% sequence identity with ulilysin, including conservation of almost all of the residues required to chelate calcium (Figure 1A). This strongly suggested that mirolysin should also bind calcium, which may affect protein stability and autoproteolysis. To verify this, we monitored the processing of mirolysin in the presence of 2.5 mm CaCl2, which corresponds to the calcium concentration in human serum and gingival crevicular fluid (Koregol et al., 2011; Fijorek et al., 2014). Under these conditions, autoprocessing of mirolysin was much more rapid (Figure 3D). According to SDS-PAGE analysis, complete conversion of Mircat into mMir occurred within 15 min, although the activity increased for up to 1 h in a time-dependent manner (Figure 3E). This may be associated with degradation of the initial cleavage products, including the released NTP. Importantly, there was no accumulation of fragments with masses of 10 kDa or lower and the proteolytic activity of mirolysin was stable. After 16 h of incubation, only one band corresponding to mMir was visible by SDS-PAGE, and its intensity did not change over 204 h of incubation. This indicated that mMir is very stable in the presence of calcium (Figure 3D). Based on this finding, the homogenous, 31 kDa mMir was obtained from the Mircat sample, incubated overnight for 12–16 h, and subjected to dialysis to remove small molecular mass degradation products. Of note, in all experiments described above, MircatE225A was used as a negative control, as it did not undergo autoprocessing and had no proteolytic activity against Azocoll or as detected by zymographic analysis (data not shown).
Biochemical characterization
Consistent with its similarity to the M43B family of metalloproteases, mirolysin activity was inhibited by treatment with the diagnostic metalloprotease inhibitor, o-phenanthroline, and by EDTA, a general divalent cation-chelating compound. As predicted, mirolysin activity was not affected by specific inhibitors of cysteine proteases, E-64 and was only slightly inhibited by the serine protease inhibitor Pefabloc. Also, other inhibitors of these catalytic classes of proteases, leupeptin, iodoacetic acid (IAA), and TLCK, had no effect on mirolysin activity (Table 1).
Table 1.
The effect of inhibitors, reducing agents, and divalent cations on the proteolytic activity of mirolysin.
| Inhibitors and metal ions | Concentration | % of control |
|---|---|---|
| EDTA | 1 mM | 0 |
| 10 mM | 0 | |
| Pefabloc | 10 μM | 91 |
| 100 μM | 82 | |
| 1,10-Phenanthroline | 1 mM | 2 |
| 10 mM | 2 | |
| Iodoacetic acid | 5 mM | 130 |
| 10 mM | 147 | |
| E-64 | 5 μM | 115 |
| 10 μM | 115 | |
| ZnCl2 | 0.01 mM | 112 |
| 0.1 mM | 96 | |
| CaCl2 | 1 mM | 24 |
| 10 mM | 0 | |
| 0.1 mM | 116 | |
| 1 mM | 218 | |
| 5 mM | 230 | |
| TLCK | 10 mM | 247 |
| 5 mM | 91 | |
| 10 mM | 100 | |
| Leupeptin L-cysteine |
50 μM | 112 |
| 1 mM | 109 | |
| 10 mM | 91 | |
| DTT | 1 mM | 98 |
| 10 mM | 74 |
Mirolysin activity determined at 37°C in 50 mM Tris-HCl, pH 8.0, using Azocoll as a substrate, was set at 100%.
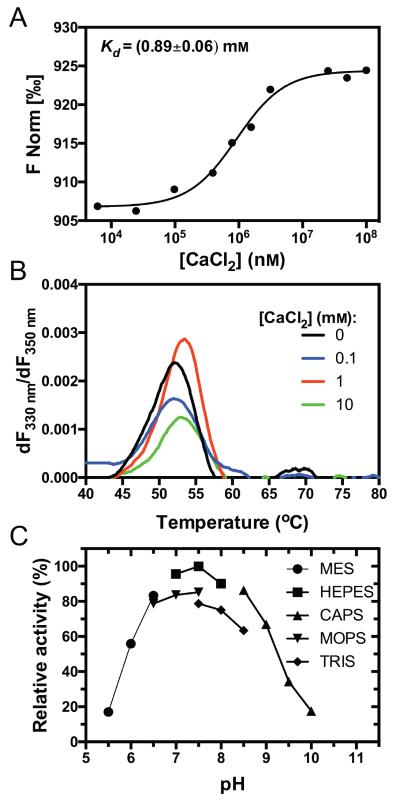
Alignment of mirolysin with other members of the M43B family revealed that all of the conserved cysteine residues involved in the formation of disulfide bridges in pappalysin and ulilysin are also conserved in mirolysin (Figure 1A). Nevertheless, preincubation with reducing agents had almost no effect on enzyme activity (Table 1). We also examined the effect of divalent cations on mirolysin activity. At a high concentration (10 mM), Zn2+ inhibited enzyme activity, while Ca2+ at concentrations ranging from 1–10 mM enhanced the activity 2-fold (Table 1). This increase in activity corroborated the results from analysis of mirolysin processing. To investigate calcium binding by mirolysin in more detail, we determined the dissociation constant (Kd) for the interaction between mirolysin and Ca2+ and studied the effect of calcium on enzyme stability using microscale thermophoresis (MST) (Jerabek-Willemsen M et al., 2011) and differential scanning fluorimetry (DSF) techniques, respectively. The Kd of the interaction was determined to be at 0.89 mM (Figure 4A), and melting temperatures (Tm) were calculated to demonstrate the effect of Ca2+ ions on MircatE225A stability. As shown in Figure 4B, calcium increased the Tm in a concentration-dependent manner, but the observed changes were negligible, with only a 1.5°C increase in the Tm in the presence of 10 mM CaCl2, suggesting that Ca2+ exerts no major effect on the structural stability of mirolysin.
Figure 4.
Determination of the dissociation constant (Kd) of mirolysin in the presence of calcium, the effect of calcium on thermal stability, and the optimum pH. A) The dissociation constant (Kd) was determined using microscale thermophoresis. MircatE225A was titrated with increasing concentrations of CaCl2. The binding data were fitted, and the Kd values were determined. The results are presented as the mean±SD from three experiments. B) The effect of increasing concentrations of calcium on the T of MircatE225A m was evaluated by differential scanning fluorimetry. Briefly, protein samples in the presence of increasing concentrations of CaCl2 were gradually heated and fluorescence was recorded at two emission wavelengths, 330 and 350 nM. The Tm was calculated based on the plots of first derivative of the ratio of two measured fluorescence values against temperature. C) Activity of mirolysin (mMir) was determined in buffers with different pH values. The activity measured in 50 mM HEPES pH 7.5 was set to 100%.
Mirolysin was active at pH values ranging from 6 to 9, and activity was dependent on the buffer composition, despite using buffers with the same molarity (50 mM; Figure 4C). This could be explained by differences in the ionic strength of the buffers, as calculated using software developed by Bob Beynon (https://www.liverpool.ac.uk/buffers/buffercalc.html). For example, the ionic strength of buffers at pH 7.5 is 0.027 for HEPES, 0.035 for MOPS, and 0.042 for Tris, which correlates with the observed differences in activity.
Determination of mirolysin specificity
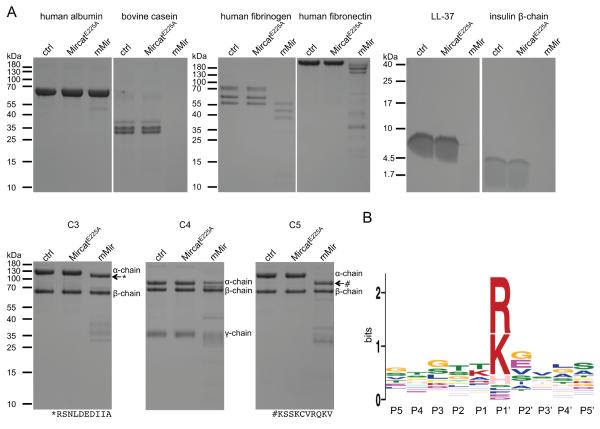
As mentioned previously, the name of the archaeal metalloprotease ulilysin was changed to LysargiNase in order to better describe the specificity of the enzyme, which is restricted to Xaa-Lys and Xaa-Arg peptide bonds (Huesgen et al., 2015). To determine whether mirolysin shows the same specificity, mMir was incubated with the following proteinaceous substrates: bovine casein and human albumin, fibrinogen, fibronectin, complement proteins (factors C3, C4, and C5), and the peptides human cathelicidin LL-37 and bovine insulin β-chain. The reaction was stopped by boiling in SDS-PAGE sample buffer, and the digestion products were separated by SDS-PAGE (Figure 5A). Mirolysin cleaved all of the analyzed substrates except albumin. Two types of proteolysis were identified: complete and limited. The latter was observed only for complement proteins. To determine mirolysin specificity, peptides generated by the enzyme were identified by mass spectrometry, thus allowing the determination of cleavage sites within the polypeptide chains of the substrates. Based on these results, we mapped the enzymatic specificity of mirolysin using Weblogo (Figure 5B). Like LysargiNase, mirolysin cleaved preferentially peptide bonds with arginine and lysine at the P1’ position. Interestingly, unlike LysargiNase, mirolysin also cleaved Xaa-His peptide bonds.
Figure 5.
Mirolysin hydrolyzes a broad range of substrates and cleaves peptide bonds at arginine and lysine. A) The proteins human albumin, fibrinogen, fibronectin, and bovine casein, the human complement proteins C3, C4, and C5 (10 μg), and the peptides human cathelicidin LL-37 and bovine insulin β-chain (4 μg) were incubated in 50 mM Tris, 150 mM NaCl, 2.5 mM CaCl2, 0.02% NaN3, pH 7.5, at 37°C for 8 h alone (control, ctrl) or with mMir at a substrate:enzyme weight ratio of 1:20 (complement proteins), 1:100 (other proteinaceous substrates), and 1:1 000 (peptides). Substrates incubated with MircatE225A served as a negative control. The identity of the bands indicated by arrows was determined by N-terminal sequencing. The obtained sequences are shown below the gels. B) The digestive peptides resulting from cleavage of substrates by mirolysin were identified by mass spectrometry, thus allowing the determination of cleavage sites within the substrate polypeptide chains. Based on these results, the mirolysin cleavage motif was generated employing MEME (Multiple Em for Motif Elicitation).
The limited proteolysis of the α-chains of C3 and C5 could lead to release of anaphylatoxins. To examine this, we determined the N-terminal sequence of truncated α-chains of C3 and C5 (Figure 5B). The analysis revealed that, indeed, both proteins were cleaved at the N-terminus. However, the cleavage of C3 led to the formation of C3a desArg (without C-terminal arginine), while C5 was truncated 127 residues downstream of the C5a peptide. The efficient cleavage of complement factors by mMir corroborates the observation of efficient inactivation of complement functions by mirolysin (Jusko et al., 2015).
Mirolysin abolishes the biological activities of LL-37
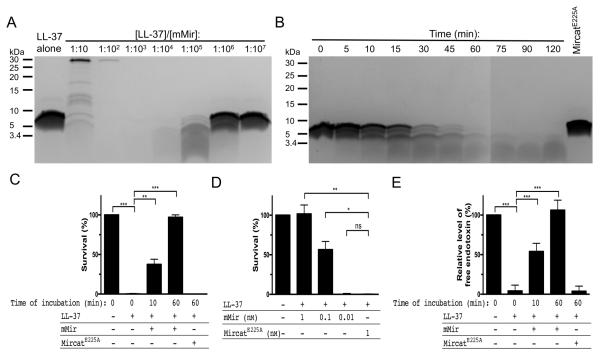
LL-37 is thought to play a crucial role in maintaining homeostasis in the periodontium (Pütsep K et al., 2002). Therefore, we monitored the concentration- and time-dependent cleavage of LL-37 by mirolysin. Mirolysin cleaved LL-37 very efficiently, and, after 1 h of incubation at a 1:100 000 molar ratio of substrate/enzyme, no native LL-37 remained in the sample (Figure 6A). Accumulated product(s) with lower molecular mass peptides (<3.4 kDa) (Figure 6B) was completely degraded during incubation for 1 h at a 1:1 000 (substrate/enzyme) molar ratio (Figure 6A). LL-37 was not affected by the catalytically inactive mirolysin mutein, MircatE225A.
Figure 6.
Mirolysin degrades LL-37 in a concentration- and time-dependent manner, abolishing its antimicrobial activity. A, B) LL-37 was incubated with mature mirolysin (mMir) at different substrate:enzyme molar ratios (concentration-dependent proteolysis) for 1 h (A) or at a constant 100 000:1 substrate:enzyme molar ratio for different time intervals (time-dependent proteolysis) (B) in 50 mM Tris, 150 mM NaCl, 2.5 mM CaCl2, 0.02% NaN3, pH 7.6, at 37°C. The reaction was terminated by boiling with SDS-PAGE sample buffer, and samples were subjected to SDS-PAGE. LL-37 incubated with MircatE225A served as a negative control. C) LL-37 was incubated alone or with mirolysin (mMir) at a molar ratio of 100 000:1 for 10 and 60 min, then mixed with bacteria (E. coli ATTCC 25922), incubated for 2 h at 37°C, and plated onto agar plates to determine the numbers of CFU. Percentage survival was calculated and compared to bacteria grown without antimicrobial peptide LL-37 (100% survival). D) Bacteria were mixed with mMir at concentrations of 0.01, 0.1, and 1 nM or MircatE225A (control), then LL-37 was added, and after incubation for 2 h at 37°C the mixtures were plated onto agar plates to determine the numbers of CFU. Percentage survival was calculated and compared to bacteria treated with MircatE225A (no survival). E) LL-37, intact or cleaved by mMir, was mixed and then incubated for 5 min at room temperature with endotoxin from E. coli 0111:B4. Intact LL-37 incubated with MircatE225A was used as a negative control. The level of the Limulus test reactive endotoxin was determined and compared to endotoxin without LL-37 (100%). *p<0.05; **p<0.01; ***p<0.001.
To assess the bactericidal potency of LL-37 after incubation with mirolysin, we performed an antimicrobial assay employing E. coli ATCC 33694 as a target. After just 10 min of incubation with mirolysin, the bactericidal activity of LL-37 against E. coli was reduced by 60%, and was entirely gone after 1 h (Figure 6C). Of note, the inactive mirolysin mutein MircatE225A exerted no effect on the antimicrobial activity of the peptide. T. forsythia is naturally resistant to LL-37, but the dental plaque is also inhabited by other species of bacteria, many of which are susceptible to LL-37 (Ouhara et al., 2005). Therefore, we hypothesized that secreted mirolysin could protect bacteria dwelling together with T. forsythia in the dental plaque. To verify this, mirolysin at different concentrations was added to a suspension of E. coli, which was then immediately treated with LL-37. MircatE225A served as a negative control. Mirolysin at a 1 nM concentration completely protected E. coli against the antimicrobial activity of LL-37. At a concentration of 0.1 nM, mirolysin increased E. coli survival to approximately 50%, but, at lower concentrations (0.01 nM), the enzyme had no protective effect. The inactive form of the enzyme had no effect on E. coli viability at any concentration (Figure 6D).
In addition to the antimicrobial activity, LL-37 exerts potent anti-inflammatory activity, which is mediated through direct binding to lipopolysaccharide (LPS). To determine how cleavage of LL-37 by mirolysin affects the ability of the peptide to neutralize LPS, we used a commercially available kit based on Limulus amebocyte lysate. Intact LL-37 prevented LPS from activation of a protease zymogen, Factor C, which is the first step in clotting induction. This ability was completely lost after preincubation of the peptide with mirolysin, which led to degradation of the peptide. Abrogation of the ability of LL-37 to neutralize LPS was dependent on mirolysin activity, since the catalytically inactive mutein of mirolysin had no effect (Figure 6E).
Discussion
Peptidases (proteases) are widely distributed among prokaryotes, so it is not surprising that bacterial proteases are involved in processes such as nutrient acquisition, degradation of misfolded proteins, regulation of gene expression, and post-translational modification of proteins. Proteolysis is irreversible, so bacterial proteases must be tightly controlled, in both a spatial and temporal manner. The first is achieved by compartmentalization of synthesized proteins, which are directed to the periplasm or extracellular milieu by different secretory pathways. In the case of prepromirolysin, a SP directs the protein to the SEC translocon. During translocation to the inner membrane, the SP is cleaved off and promirolase folds in the periplasm. The conserved CTD serves as a signal for transport by the T9SS (Ksiazek et al., 2015b), which moves mirolysin across the outer membrane to the extracellular milieu. During the journey through the periplasm, mirolysin is maintained as an inactive zymogen by the N-terminal profragment (NTP). The NTP, however, seems to provide stable latency only in the absence of calcium.
In bacteria, similar to eukaryotes, calcium concentrations within the cell are tightly regulated. Although there are no data available for T. forsythia, calcium levels were measured in the model bacteria, E. coli. In E. coli, as well as in other tested bacteria, the cytoplasmic Ca2+ concentration is kept constant at 1 μm. The periplasmic concentration may vary depending on the extracellular concentration, but, nevertheless, it is maintained in the range of 60–300 μm (Jones et al., 2002; Dominguez, 2004). The determined Kd value for interaction of mirolysin with calcium is 890 μm, which suggested that mirolysin could not bind calcium in subcellular compartments of T. forsythia (cytoplasm and periplasm), but only after secretion into the extracellular milieu, gingival crevicular fluid, where the calcium concentration is around 2.5 mm. Thus, such regulation could prevent unwanted activation of secretory proteases in the periplasm. Mirolysin is the third KLIKK protease to be characterized; the first two were karilysin and mirolase. All of these proteases bind calcium, but, interestingly, Ca2+ exerts a different effect on each enzyme. In karilysin, Ca2+ increases thermal stability, in mirolase, it is essential for enzyme activity, and, in mirolysin, it facilitates autoactivation of promirolysin and prevents autoproteolytic degradation of the mature enzyme. As a result, calcium concentration within the subcellular compartments may be a global regulator of KLIKK protease activity.
Despite almost 50% identity in of the amino acid sequence with the CD of ulilysin, mirolysin possesses some unique features. First, both proteases bind calcium, but this binding results in different effects. Ca2+ is essential for ulilysin activity, while it only increases the activity of mirolase and provides resistance to autoproteolysis. Second, although both ulilysin and mirolysin show strong preference for lysine or arginine at the P1’ position, ulilysin, similar to trypsin, slightly prefers lysine (52%) to arginine (40%). In mirolysin, the opposite is true: 49% of cleavage occurred before arginine and only 33% in front of lysine. Moreover, mirolysin also cleaved Xaa-His peptide bonds (8%). However, it needs to be kept in mind that mirolysin specificity was determined based on only 39 cleavage sites compared to 1,917 unique cleavage sites determined for ulilysin (Huesgen et al., 2015).
Two disulfide bridges spanning the CD are conserved in PAPPA and ulilysin (Tallant et al., 2006). Those cysteine residues are also present in mirolysin (Figure 1A), but they are either not engaged in disulfide bridges or these bridges are not structurally important, since protein reduction with DTT has little effect on mirolysin activity. The former assumption is corroborated by the fact that M43B protease from Rhodococcus opacus lacks the cysteine residues that form these disulfide bridges (Figure 1A). Furthermore, despite the presence of numerous cysteine residues, disulfide bridges are absent in T. forsythia and P. gingivalis proteins with structures known at atomic resolution. This is likely a structural and functional adaptation to the anaerobic, reducing environment niche occupied by periodontal pathogens.
The human cathelicidin LL-37 is considered as a crucial antimicrobial peptide responsible for the maintenance of homeostasis in the periodontium (Hosokawa et al., 2006). Levels of LL-37 are increased in patients suffering from periodontitis compared to healthy controls (Türkoğlu et al., 2009), and genetic deficiency of LL-37 results in rapid development of severe periodontitis (Pütsep K et al., 2002). Thus, hydrolysis of LL-37 by mirolysin may contribute to the natural resistance of T. forsythia to this peptide. During LL-37 degradation, mirolysin may act synergistically with karilysin and mirolase, which also cleave LL-37. Mirolysin alone, at concentrations as low as 1 nM, fully protected E. coli against LL-37. Thus, it is tempting to speculate that mirolysin could provide protection to LL-37 against periodontal pathogens inhabiting subgingival biofilm with T. forsythia.
In addition to its bactericidal activity, LL-37 also possesses potent immunoregulatory properties mediated by binding and neutralizing the proinflammatory activity of LPS (endotoxin) (Golec, 2007). LPS is a component of the outer membrane of Gram-negative bacteria, which are the most predominant bacteria in dental plaques. Therefore, degradation of LL-37 by mirolysin, which abolishes the ability of the peptide to bind LPS, may result in increased inflammation at the site of infection, thus contributing to development of periodontitis.
In dental plaques, which are bathed in gingival crevicular fluid (GCF) composed of almost 70% human serum, T. forsythia is exposed to the bactericidal activity of the complement system. Complement activation contributes to bacterial death, either directly by assembly of a membrane attacking complex (MAC) on the bacterial surface, or indirectly through the release of potent anaphylatoxins (C3a and C5a), which recruit and activate neutrophils and macrophages to kill bacteria (Potempa M and Potempa J, 2012). Anaphylatoxin activity is regulated by serum carboxypeptidase, which removes the C-terminal arginine, resulting in the formation of desArg forms of these potent mediators. This leads to the loss of almost 90% and 100% of the biologic activity of C3a and C5a, respectively (Bajic et al., 2013). For many years, C3a was considered a proinflammatory mediator. However, it has been recently shown that C3a possesses both pro- and anti-inflammatory properties, the latter being exerted through blocking the mobilization of neutrophils from bone marrow into the blood stream and suppressing the production of proinflammatory cytokines by these cells (Coulthard et al., 2015). In this context, it is important to emphasize that mirolysin efficiently generated C3a desArg devoid of anti-inflammatory properties, which cannot counterbalance the activity of C5a released by T. forsythia proteases (Jusko et al., 2015). This may contribute to excessive, sustained inflammation at the site of infection, the environment in which periodontal pathogens thrive (Hajishengallis, 2010).
Apart from the proteolysis-mediated disruption of two major bactericidal systems present in the oral cavity, mirolysin is also able to efficiently degrade other physiologically relevant human proteins, including fibrinogen and fibronectin. Thus, mirolysin could interfere with clotting at the site of infection, but this is more likely to facilitate the degradation of proteins abundant in GCF, which generates a pool of peptides indispensable for the growth of asaccharolytic bacteria (Tanner and Izard, 2006).
Taken together, our findings regarding the expression and biochemical characterization of mirolysin identify it as a specific protease that preferentially hydrolyzes Xaa-Arg and Xaa-Lys peptide bonds. Based on its high level of proteolytic activity and cleavage of complement proteins and LL-37, mirolysin may not only provide nutrients for the asaccharolytic consortium of periodontal pathogens, but also protect itself, as well as other pathogens in dysbiotic subgingival biofilm, against the human innate immune system. Confirmation of this hypothesis is currently the subject of ongoing research in our laboratory.
Materials and methods
Chemicals and reagents
The restriction endonucleases BamHI and XhoI, T4 DNA ligase, dNTP, GeneJET™ Gel Extraction Kit, GeneJET™ PCR Purification Kit, Phusion DNA Polymerase, and GeneJET™ Plasmid Miniprep Kit were acquired from Thermo Scientific (Waltham, MA, USA). The QuikChange Lightning Site-Directed Mutagenesis Kit was purchased from Stratagene (La Jolla, CA, USA). All primers used in the study were synthesized by Genomed (Warsaw, Poland). The expression vector pGEX-6P-1, glutathione-Sepharose 4 Fast Flow, 3C protease (PreScission protease), HiLoad 16/600 Superdex 75 pg SEC column, and Mono Q 4.6/100 PE ion exchange column were obtained from GE Healthcare Life Sciences (Little Chalfont, UK). The Pierce Protein Concentrators 9K MWCO (7 ml and 20 ml) were from Thermo Scientific (Rockford, Il, USA). The protein molecular weight standards Precision Plus Protein Dual Xtra and Pageruler Unstained Low Range Protein Ladder were purchased from Bio-Rad (Hercules, CA, USA) and Thermo Scientific, respectively. Polyvinylidene difluoride (PVDF) membranes were from Bio-Rad. Spectra/Por 4 12,000 to 14,000 Dalton MWCO dialysis membranes were acquired from Spectrum Laboratories (Rancho Dominguez, CA, USA). Calbiochem Azocoll was purchased from Merck (Darmstadt, Germany). Human fibrinogen, human fibronectin, bovine casein, bovine insulin β-chain, human albumin, RPMI, and the Hank’s balanced salts protease inhibitors leupeptin (N-acetyl-L-leucyl-Leucyl-L-argininal hemisulfate salt), E-64 (trans-epoxy-succinyl-l-leucyl-amido (4-guanidino) butane), IAA, Pefabloc (4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride), EDTA, and 1,10-phenanthroline (orthophenantrholine) were from Sigma (St. Louis, MO, USA). The human complement proteins C3, C4, and C5 were acquired from Complement Technology (Tyler, TX, USA). Synthetic LL-37 (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES) was purchased from GenScript (Piscataway, NJ, USA). All other chemical reagents were obtained from BioShop Canada (Burlington, ON, Canada).
Cloning and mutant construction
Genomic DNA from T. forsythia was isolated from strain ATCC 43037. The coding sequences of the full-length mirolysin gene, proMir (Ser25-Lys612, BFO_2661; GenBank accession number: AEW21083.1), and mirolysin CD with NTP, Mircat (Ser25-Ser331), both without the nucleotide sequence that encodes the SP, were amplified by PCR using the following primers (restriction sites are underlined): proMir_F: 5′-ACAGGATCCTCTGAGTTGAATATGGAAC-3′ proMir_R: 5′-CGGCTCGAGTTACTTCTTAATCAATTTCTGC-3′ Mircat_F: 5′-ACAGGATCCTCTGAGTTGAATATGGAACAAATCC-3′ Mircat_R: 5′-CGGCTCGAGTTAGGAGAAAGAAAGTGGATTCG-3′
The PCR products were purified and cloned into the pGEX-6P-1 vector, which provides the sequence for an N-terminal GST tag and inserts five additional residues (Gly-Pro-Leu-Gly-Ser) in the N-terminus of mirolysin. The wild-type (WT) constructs (pGEX-6P-1_proMir, pGEX-6P1_Mircat) were used to obtain E225A variants (proMirE225A and MircatE225A) in which the active site Glu-225 is substituted by alanine (Figure 1B). This was achieved with the QuikChange Lightning Site-Directed Mutagenesis Kit and the following mutagenic primers: E225A-F (5′ GGACAGCGACGCATGCAGTAGGGCATTGGTTAG 3′) and E225A-R (5′ CTAACCAATGCCCTACTGCATGCGTCGCTGTCC 3′). All obtained plasmids were confirmed by DNA sequencing and then transformed into the Escherichia coli strain BL21 (DE3) (New England Biolabs, Ipswich, MA, USA), which allowed expression under the control of the T7 promoter.
Recombinant protein expression and purification
Transformed E. coli BL21 (DE3) were grown in LB Lennox media at 37°C to an optical density (OD600 nm) ranging from 0.75 to 1.0 and cooled for 30 min at 4°C. Expression of recombinant proteins was then induced by the addition of 0.1 mm isopropyl β-D-1-galactopyranoside (IPTG). After 6–8 h of cultivation at 20°C, cells were harvested by centrifugation (6 000 g, 15 min, 4°C) and resuspended in PBS supplemented with 0.02% NaN3 (15 ml per pellet from 1 l of culture). They were then lysed by sonication (35 × 0.5 s pulses at an amplitude of 70%) using a Branson Digital Sonifier 450 (Branson Ultrasonics, Danbury, CT, USA). The cell lysates were clarified by centrifugation (50 000 g, 50 min, 4°C) followed by filtration through a 0.45 μm syringe filter and then applied at 4°C to a glutathione-Sepharose 4 Fast Flow column (bed volume = 5 ml) equilibrated with PBS supplemented with 0.02% NaN3. After extensive washing, 10 ml of PBS supplemented with 0.02% NaN3 containing 100 μl of PreScission protease stock solution (1 U/ml) was applied to the column and incubated for 40 h at 4°C. The final purification of recombinant protein (Mircat and MircatE225A) was accomplished by SEC on a Superdex 75 pre-equilibrated with 50 mm Tris-HCL (pH 8.0) and, for MircatE225A only, ion exchange chromatography employing Mono Q, and two buffers: 20 mm Tris-HCl (pH 8.0) and 20 mm Tris-HCl, 1 m NaCl (pH 8.0). The total amount of purified protein was determined by measuring the absorbance at 280 nm using a Spectrophotometer NanoDrop 1000 and BCA assay (Thermo Scientific). Purity of the obtained protein was verified by SDS-PAGE.
Gel electrophoresis and zymography
Mirolysin purification, autocatalytic processing, and protein substrate degradation were monitored by SDS-PAGE using NuPAGE Novex 4–12% and 4–20% gels (only for degradation of LL-37) and Bis-Tris Protein Gels with MES buffer (Thermo Scientific). Gels were stained with SimplyBlue SafeStain (Thermo Scientific) and destained in water.
Zymographic analysis was performed on samples mixed 1:1 with sample buffer (0.125 m Tris-HCl, pH 7.8, 20% glycerol, 4% SDS, 0.1% Bromophenol Blue) for 15 min at 20°C and electrophoresed on 12% SDS-PAGE gels (T:C, 27.5:1) containing gelatin at a final concentration of 0.1 mg/ml, a substrate for proteases. Gels were incubated twice in 2.5% Triton X-100 for 30 min, followed by incubation in developing buffer (0.2 M Tris-HCl, 5 mm CaCl2, 1 mm DTT, pH 7.8) for 3 h at 37°C. Finally, the gels were incubated in destaining/fixing solution (methanol/acetic acid/water (30:10:60)), stained with 0.1% amido black in 10% acetic acid for 1 h, and then destained successively in destaining/fixing solution containing 10% acetic acid and 1% acetic acid, which revealed clear zones of substrate hydrolysis on a blue background.
Proteolytic processing and activation of mirolysin
Mirolysin (proMir and Mircat) was incubated in 50 mm Tris, 0.02% NaN3, pH 8.0, at 37°C for up to 204 h. At specific time points (0, 1, 16, 48 h for proMir and 0, 0.25, 0.5, 1, 2, 4, 8, 16, 24, 36, 48, 60, 72, 84, 96, 120, 156, 204 h for Mircat), aliquots were taken to measure proteolytic activity using Azocoll as a substrate (see the Enzymatic activity assays section), zymography, and SDS-PAGE analysis. Enzymatically inactive muteins (E225A) of both proMir and Mircat served as controls.
Purification of mature mirolysin (mMir)
Mircat at a concentration of 1 mg/ml was incubated in 50 mM Tris, 2.5 mm CaCl2, 0.02% NaN3, pH 8.0, at 37°C, as described above, and processing was monitored by SDS-PAGE. When only one band of molecular weight 31 kDa was visible on the gels, the processing was stopped. The samples were then extensively dialyzed against buffer containing 5 mm Tris, 50 mm NaCl, 1 mm CaCl2, 0.02% NaN3, pH 8.0, and concentrated to a concentration of 1 mg/ml. The obtained mMir was aliquoted and stored at −20°C.
N-terminal sequence analysis
Samples were resolved on 4–12% NuPAGE gels and then electrotransferred in 10 mM CAPS, 10% methanol, pH 11, onto a PVDF membrane using a mini trans-blot module (Bio-Rad). Protein bands were visualized by CBB G-250 staining, excised, and analyzed by automated Edman degradation using a Procise 494HT amino acid sequencer (Applied Biosystems, Carlsbad, CA, USA).
Determination of the Kd by microscale thermophoresis (MST)
A Monolith NT.LabelFree instrument (NanoTemper Technologies GmbH, Munich, Germany) was used to analyze the binding interactions between recombinant mirolysin and calcium (Ca2+). Calcium at concentrations ranging from 6.1 to 10 000 μm was incubated with 375 nm mirolysin (MircatE225A) in assay buffer (5 mm Tris, 50 mm NaCl, 0.02% NaN3, 0.05% Pluronic F-127, pH 7.5) for 5 min at 20°C. The samples were loaded into Monolith NT.LabelFree Standard Treated glass capillaries, and an initial fluorescence measurement was taken followed by thermophoresis measurements using 20% of power of a light-emitting diode (LED) and 20% MST power, respectively. Kd values were calculated using MO.Affinity Analysis software. The experiment was performed in triplicate.
Differential scanning fluorimetry
Differential scanning fluorimetry employing the Prometheus NT.48 apparatus (NanoTemper) was used to investigate the effect of Ca2+ concentration on MircatE225A thermal stability. MircatE225A (375 nM) in 50 mm Tris, pH 8.0, was incubated with increasing concentrations of CaCl2 (0.01, 0.1, 1, and 10 mm) for 15 min at 20°C. The samples were then loaded into nanoDSF standard grade capillaries and then heated from 40°C to 95°C at a rate of 1°C per min. Protein unfolding was monitored by measuring the fluorescence intensity at two emission wavelengths at 330 nm and 350 nm. Analysis of the obtained data was performed using the control software (NanoTemper), which allowed calculation of the protein Tm based on the first derivative of the ratio of fluorescence intensity measured at 350 and 330 nm.
Enzymatic activity assays
Mirolysin activity was routinely determined in 50 mm Tris (pH 8.0) using Azocoll as a substrate. Briefly, 125 μl of 15 mg/ml suspension of substrate in assay buffer was added to the same volume of the enzyme solution in Eppendorf tubes and the mixtures were incubated for 1–2 h at 37°C with shaking. Undigested Azocoll was removed by centrifugation (16 100 g, 5 min, 4°C), and the absorbance at 520 nm was measured using a SpectraMAX microplate reader (Molecular Devices, Sunnyvale, CA, USA).
The optimum pH was determined using the following buffers at a concentration of 50 mm: MES (pH 5.5, 6.0, and 6.5), MOPS (pH 6.5, 7.0, and 7.5), HEPES (pH 7.0, 7.5, and 8.0), Tris (pH 7.5, 8.0, and 8.5), and CAPS (pH 8.5, 9.0, 9.5, and 10.0). To test the effect of inhibitors, reducing agents, and divalent metal ions on mirolysin activity, mMir was preincubated with each compound (listed in Table 1) in 50 mm Tris (pH 8.0) for 15 min at room temperature and the residual activity was measured using Azocoll as a substrate.
Human albumin, fibrinogen, fibronectin, bovine casein, the human complement proteins C3, C4, and C5 (10 μg), and the peptides human cathelicidin LL-37 and bovine insulin β-chain (4 μg) were incubated with mMir at the enzyme/substrate weight ratios 1:20 (complement proteins), 1:100 (all other proteins), and 1:1 000 (LL-37 and insulin β-chain) for 8 h at 37°C in 50 mm Tris, 150 mm NaCl, 5 mm CaCl2, 0.02% NaN3, pH 8.0. Obtained samples were resolved by SDS-PAGE. MircatE225A was used as a negative control.
Determination of mirolysin cleavage specificity
To determine mirolysin cleavage specificity, different mammalian proteins and peptides cleaved by mirolysin were examined. Samples were mixed 1:1 (v/v) with 15% metaphosphoric acid, incubated in ice for 30 min, and centrifuged (10 min, 16 100 g, 4°C). Supernatants, free of undigested proteins, were gently collected and lyophilized. The samples were then analyzed by NanoLC-MS/MS using an EASY-nLC II system (Thermo Scientific) connected to a TripleTOF 5600 mass spectrometer (AB SCIEX; Framingham, MA, USA). The mirolysin-digested samples were dissolved in 0.1% formic acid and loaded onto a 0.1 × 20 nm C18 trap column and a 0.075 × 150 nm C18 analytical column. The peptides were eluted and electrosprayed directly into the mass spectrometer using a 20 min gradient from 5% to 35% solvent B (0.1% formic acid in 90% acetonitrile) at a flow rate of 250 nl/min. An information-dependent acquisition method was employed to automatically run experiments acquiring up to 25 MS/MS spectra per cycle using a 0.9 s cycle time and an exclusion window of 6 s. The raw data were converted to MGF files and searched against the SWISS-Prot mammal database (Version 2015-07) using an in-house Mascot search engine. No enzyme was specified, and oxidation of methionine was included as a variable modification. Peptide mass tolerance and fragment mass tolerance were set at 10 ppm and 0.2 Da, respectively, and the significance threshold (p) was set at 0.01. Identified peptide sequences were used to reveal the cleavage sites (P1-P1’) within substrates digested by mirolysin. The 10-long sequences (P5-P5’) were then used to determine mirolysin cleavage specificity using the MEME Suite server (Bailey et al., 2009).
Time- and concentration-dependent LL-37 cleavage by mirolysin
LL-37 (4 μg) was incubated with mirolysin (mMir) at peptide/enzyme molar ratios varying from 10:1 to 10,000,000:1 in 20 μl of 50 mm Tris, 150 mm NaCl, 2.5 mm CaCl2, pH 8.0, for 1 h at 37°C. For time-dependent analysis, LL-37 was exposed to mirolysin for times ranging from 0 to 120 min at a molar ratio of 100 000:1 under the same conditions. The reaction was terminated by the addition of boiling SDS-PAGE sample buffer. After denaturation (5 min, 95°C), samples were subjected to SDS-PAGE analysis. LL-37 incubated with MircatE225A was used as a control.
Antimicrobial assay
To evaluate the antimicrobial activity of LL-37 cleaved by mirolysin, a colony reduction assay using E. coli ATCC 33694 grown in LB was performed. E. coli cells were collected by centrifugation (5 000 g, 8 min), washed three times with PBS, and resuspended in RPMI supplemented with 10% Hank’s balanced salt solution) to a final concentration of a 1.1 × 106 colony-forming units (CFU)/ml. LL-37 (20 μm), either intact or cleaved by mirolysin, was prepared in 50 mm Tris, 150 mm NaCl, 2.5 mm CaCl2, pH 7.5, and 180 μl of each bacterial suspension was added to Eppendorf tubes. After 1 h of incubation at 37°C, aliquots were diluted in 10% HBSS with RPMI to a final concentration of 103 CFU/ml and 100 μl aliquots of each sample were plated onto LB agar plates. Alternatively, aliquots of bacterial suspension were mixed with different concentrations (1, 0.1, and 0.01 nM) of mMir and MircatE225A, and the intact LL-37 was then added to a final concentration of 2 μM. After 1 h of incubation, aliquots were plated onto LB agar plates. All experiments were performed in triplicate.
Determination of endotoxin level
LL-37 (2 μm), intact or cleaved by mirolysin (mMir), was mixed 1:1 (v/v) and then incubated for 5 min at RT with endotoxin (0.5 EU/ml) from E. coli 0111:B4. Intact LL-37 incubated with MircatE225A was used as a negative control. The level of free endotoxin was determined by QCL-1000 assay (Lonza, Basel, Switzerland) per the manufacturer’s instructions.
Data analysis
All graphing and statistical analysis (Student’s t test) was performed using GraphPad Prism. Differences were considered significant when p<0.05.
Acknowledgements
This study was financially supported in part by grants K/DSC/003700 (decision no. BMN 31/2016, Faculty of Biochemistry, Biophysics and Biotechnology of Jagiellonian University), UMO-2015/17/B/NZ1/00666 and UMO-2012/04/A/NZ1/00051 (National Science Centre Poland), DE 09761 and DE 022597 from US National Institutes of Health (NIH). MK has obtained a scholarship from Ministry of Science and Higher Education (1306/MOB/IV/2015/0, “Mobilność Plus”). The Faculty of Biochemistry, Biophysics, and Biotechnology of Jagiellonian University is a partner of the Leading National Research Center (KNOW) supported by the Ministry of Science and Higher Education.
References
- Armitage GC. Periodontal diagnoses and classification of periodontal diseases. Periodontol. 2000. 2004;34:9–21. doi: 10.1046/j.0906-6713.2002.003421.x. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic. Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic G, Yatime L, Klos A, Andersen GR. Human C3a and C3a desArg anaphylatoxins have conserved structures, in contrast to C5a and C5a desArg. Protein Sci. 2013;22:204–212. doi: 10.1002/pro.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedyk M, Mydel PM, Delaleu N, Płaza K, Gawron K, Milewska A, Maresz K, Koziel J, Pyrc K, Potempa J. Gingipains: critical factors in the development of aspiration pneumonia caused by Porphyromonas gingivalis. J. Innate Immun. 2016;8:185–198. doi: 10.1159/000441724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan PN. Prodomains and protein folding catalysis. Chem. Rev. 2002;102:4805–4816. doi: 10.1021/cr010190b. [DOI] [PubMed] [Google Scholar]
- Coulthard LG, Woodruff TM. Is the complement activation product C3a a proinflammatory molecule? Re-evaluating the evidence and the myth. J. Immunol. 2015;194:3542–3548. doi: 10.4049/jimmunol.1403068. [DOI] [PubMed] [Google Scholar]
- Dominguez DC. Calcium signalling in bacteria. Mol. Microbiol. 2004;54:291–297. doi: 10.1111/j.1365-2958.2004.04276.x. [DOI] [PubMed] [Google Scholar]
- Eke PI, Dye BA, Wei L, Slade GD, Thornton-Evans GO, Borgnakke WS, Taylor GW, Page RC, Beck JD, Genco RJ. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J. Periodontol. 2015;86:611–622. doi: 10.1902/jop.2015.140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijorek K, Püsküllüoğlu M, Tomaszewska D, Tomaszewski R, Glinka A, Polak S. Serum potassium, sodium and calcium levels in healthy individuals - literature review and data analysis. Folia Med Cracov. 2014;54:53–70. [PubMed] [Google Scholar]
- Fox CH. New considerations in the prevalence of periodontal disease. Curr. Opin. Dent. 1992;2:5–11. [PubMed] [Google Scholar]
- Golec M. Cathelicidin LL-37: LPS-neutralizing, pleiotropic peptide. Ann. Agric. Environ. Med. 2007;14:1–4. [PubMed] [Google Scholar]
- Guo Y, Nguyen KA, Potempa J. Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon's knife to a meat chopper-like brutal degradation of proteins. Periodontol. 2000. 2010;54:15–44. doi: 10.1111/j.1600-0757.2010.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Complement and periodontitis. Biochem Pharmacol. 2010;80:1992–2001. doi: 10.1016/j.bcp.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000. 2005;38:72–112. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Hosokawa I, Hosokawa Y, Komatsuzawa H, Goncalves RB, Karimbux N, Napimoga MH, Seki M, Ouhara K, Sugai M, Taubman MA, et al. Innate immune peptide LL-37 displays distinct expression pattern from beta-defensins in inflamed gingival tissue. Clin. Exp. Immunol. 2006;146:218–225. doi: 10.1111/j.1365-2249.2006.03200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesgen PF, Lange PF, Rogers LD, Solis N, Eckhard U, Kleifeld O, Goulas T, Gomis-Rüth FX, Overall CM. LysargiNase mirrors trypsin for protein C-terminal and methylation-site identification. Nat. Methods. 2015;12:55–58. doi: 10.1038/nmeth.3177. [DOI] [PubMed] [Google Scholar]
- Hugoson A, Sjödin B, Norderyd O. Trends over 30 years, 1973-2003, in the prevalence and severity of periodontal disease. J. Clin. Periodontol. 2008;35:405–414. doi: 10.1111/j.1600-051X.2008.01225.x. [DOI] [PubMed] [Google Scholar]
- Ishihara K. Virulence factors of Treponema denticola. Periodontol. 2000. 2010;54:117–135. doi: 10.1111/j.1600-0757.2009.00345.x. [DOI] [PubMed] [Google Scholar]
- Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011;9:342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HE, Holland IB, Campbell AK. Direct measurement of free Ca(2+) shows different regulation of Ca(2+) between the periplasm and the cytosol of Escherichia coli. Cell Calcium. 2002;32:183–192. doi: 10.1016/s0143416002001537. [DOI] [PubMed] [Google Scholar]
- Jusko M, Potempa J, Mizgalska D, Bielecka E, Ksiazek M, Riesbeck K, Garred P, Eick S, Blom AM. A Metalloproteinase Mirolysin of Tannerella forsythia Inhibits All Pathways of the Complement System. J. Immunol. 2015;195:2231–2240. doi: 10.4049/jimmunol.1402892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim AY, Kulczycka M, Kantyka T, Dubin G, Jabaiah A, Daugherty PS, Thogersen IB, Enghild JJ, Nguyen KA, Potempa J. A novel matrix metalloprotease-like enzyme (karilysin) of the periodontal pathogen Tannerella forsythia ATCC 43037. Biol. Chem. 2010;391:105–117. doi: 10.1515/BC.2010.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koregol AC, More SP, Nainegali S, Kalburgi N, Verma S. Analysis of inorganic ions in gingival crevicular fluid as indicators of periodontal disease activity: A clinico-biochemical study. Contemp Clin Dent. 2011;2:278–282. doi: 10.4103/0976-237X.91788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek M, Karim AY, Bryzek D, Enghild JJ, Thøgersen IB, Koziel J, Potempa J. Mirolase, a novel subtilisin-like serine protease from the periodontopathogen Tannerella forsythia. Biol. Chem. 2015a;396:261–275. doi: 10.1515/hsz-2014-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek M, Mizgalska D, Eick S, Thøgersen IB, Enghild JJ, Potempa J. KLIKK proteases of Tannerella forsythia: putative virulence factors with a unique domain structure. Front Microbiol. 2015b;6:312. doi: 10.3389/fmicb.2015.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Pelegrín M, Ksiazek M, Karim AY, Guevara T, Arolas JL, Potempa J, Gomis-Rüth FX. A novel mechanism of latency in matrix metalloproteinases. J. Biol. Chem. 2015;290:4728–4740. doi: 10.1074/jbc.M114.605956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Tomi N, Fukuyo Y, Ishikura H, Ohno Y, Arvind R, Arai T, Ishikawa I, Arakawa S. Isolation and identification of a cytopathic activity in Tannerella forsythia. Biochem. Biophys. Res. Commun. 2006;351:133–139. doi: 10.1016/j.bbrc.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Nguyen KA, Travis J, Potempa J. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-Negative bacteria? J. Bacteriol. 2007;189:833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo-Corgel J, Pucher JJ, Rethman MP, Reynolds MA. State of the science: chronic periodontitis and systemic health. J. Evid. Based Dent. Pract. 2012;12:20–28. doi: 10.1016/S1532-3382(12)70006-4. [DOI] [PubMed] [Google Scholar]
- Ouhara K, Komatsuzawa H, Yamada S, Shiba H, Fujiwara T, Ohara M, Sayama K, Hashimoto K, Kurihara H, Sugai M. Susceptibilities of periodontopathogenic and cariogenic bacteria to antibacterial peptides, {beta}-defensins and LL37, produced by human epithelial cells. J. Antimicrob. Chemother. 2005;55:888–896. doi: 10.1093/jac/dki103. [DOI] [PubMed] [Google Scholar]
- Overgaard MT, Sorensen ES, Stachowiak D, Boldt HB, Kristensen L, Sottrup-Jensen L, Oxvig C. Complex of pregnancy-associated plasma protein-A and the proform of eosinophil major basic protein. Disulfide structure and carbohydrate attachment. J. Biol. Chem. 2003;278:2106–2117. doi: 10.1074/jbc.M208777200. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. Prediction of a caspase-like fold in Tannerella forsythia virulence factor PrtH. Cell Cycle. 2009;8:1453–1455. doi: 10.4161/cc.8.9.8243. [DOI] [PubMed] [Google Scholar]
- Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J. Innate Immun. 2009;1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa M, Potempa J. Protease-dependent mechanisms of complement evasion by bacterial pathogens. Biol. Chem. 2012;393:873–888. doi: 10.1515/hsz-2012-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pütsep K, Carlsson G, Boman HG, Andersson M. Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. Lancet. 2002;360:1144–1149. doi: 10.1016/S0140-6736(02)11201-3. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Finn RD. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016;44:D343–D350. doi: 10.1093/nar/gkv1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sakai E, Veith PD, Shoji M, Kikuchi Y, Yukitake H, Ohara N, Naito M, Okamoto K, Reynolds EC, et al. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J. Biol. Chem. 2005;280:8668–8677. doi: 10.1074/jbc.M413544200. [DOI] [PubMed] [Google Scholar]
- Sharma A. Virulence mechanisms of Tannerella forsythia. Periodontol. 2000. 2010;54:106–116. doi: 10.1111/j.1600-0757.2009.00332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa Ki.K., Katayama M, Matsuda Y, Takahashi H, Hara I, Sato H, Kaneko S. Matrix metalloproteinase (MMP)-2 and MMP-9 activities in human seminal plasma. Mol. Hum. Reprod. 2002;8:32–6. doi: 10.1093/molehr/8.1.32. [DOI] [PubMed] [Google Scholar]
- Staniec D, Ksiazek M, Thøgersen IB, Enghild JJ, Sroka A, Bryzek D, Bogyo M, Abrahamson M, Potempa J. Calcium regulates the activity and structural stability of Tpr, a bacterial calpain-like peptidase. J. Biol. Chem. 2015;290:27248–27260. doi: 10.1074/jbc.M115.648782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallant C, García-Castellanos R, Seco J, Baumann U, Gomis-Rüth FX. Molecular analysis of ulilysin, the structural prototype of a new family of metzincin metalloproteases. J. Biol. Chem. 2006;281:17920–17928. doi: 10.1074/jbc.M600907200. [DOI] [PubMed] [Google Scholar]
- Tanner AC, Izard J. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol. 2000. 2006;42:88–113. doi: 10.1111/j.1600-0757.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- Tomek MB, Neumann L, Nimeth I, Koerdt A, Andesner P, Messner P, Mach L, Potempa JS, Schäffer C. The S-layer proteins of Tannerella forsythia are secreted via a type IX secretion system that is decoupled from protein O-glycosylation. Mol. Oral Microbiol. 2014;29:307–320. doi: 10.1111/omi.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Türkoğlu O, Emingil G, Kütükçüler N, Atilla G. Gingival crevicular fluid levels of cathelicidin LL-37 and interleukin-18 in patients with chronic periodontitis. J. Periodontol. 2009;80:969–976. doi: 10.1902/jop.2009.080532. [DOI] [PubMed] [Google Scholar]
- Wright CJ, Burns LH, Jack AA, Jack AA, Back CR, Dutton LC, Nobbs AH, Lamont RJ, Jenkinson HF. Microbial interactions in building of communities. Mol. Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]