Abstract
The characterization of microbial communities that promote or suppress soil-borne pathogens is important for controlling plant diseases. We compared prokaryotic communities in soil with or without the signs of tomato bacterial wilt caused by Ralstonia solanacearum. Soil samples were collected from a greenhouse at two different depths because this pathogen is present in deep soil. We used samples from sites in which we detected phcA, a key gene regulating R. solanacearum pathogenicity. The pyrosequencing of prokaryotic 16S rRNA sequences in four soil samples without disease symptoms but with phcA and in two soil samples with disease symptoms indicated that community richness was not significantly different between these two soils; however, microbial diversity in the lower soil layer was higher in soil samples without disease symptoms but with phcA. A difference in prokaryotic community structures between soil samples with and without bacterial wilt was only observed in the upper soil layer despite apparent similarities in the communities at the phylum level. Proteobacteria, Acidobacteria, Chloroflexi, Verrucomicrobia, and several Archaea were more abundant in soil samples without disease symptoms, whereas taxa in another eight phyla were more abundant in soil samples with disease symptoms. Furthermore, some prokaryotic taxa were abundant specifically in the lower layer of soil, regardless of whether disease was present. These prokaryotic taxa may suppress or accelerate the pathogenesis of bacterial wilt and are good targets for future studies on disease control.
Keywords: 454 pyrosequencing, lower soil layer, prokaryotic diversity, tomato bacterial wilt
Soil-borne pathogens cause various plant diseases such as take-all, damping off, crown rot, and wilting. Thus, pathogens pose a serious threat to crop production and food security. Bacterial wilt caused by the soil-borne pathogen Ralstonia solanacearum has been reported worldwide and is one of the most devastating plant diseases (26, 43). This pathogen infects more than 200 plant species, e.g., olive, tomato, tobacco, and eggplant, and causes great losses in agriculture and horticulture (18). It is mainly distributed in soil and enters roots through injured tissue or natural openings. Several approaches to control bacterial wilt, including soil amendments, crop rotation, and field sanitation (39), have been suggested; however, a definitive control method has not yet been developed.
Three elements are generally needed for the onset of plant disease: a susceptible host, the presence of a pathogen, and a conducive environment. A conducive environment comprises appropriate temperatures, soil chemical properties, soil types, and other factors. One of the most important factors in the outbreak of a soil-borne pathogen is soil microbial properties. Previous studies compared bacterial communities between soil samples with and without soil-borne disease symptoms, and showed that specific bacteria are involved in pathogen suppression (3, 21, 23, 25, 33, 35). Comparisons of microbial communities between soil samples with and without soil-borne disease pathogens are important for the selection of effective pathogen-controlling microorganisms and a clearer understanding of interactions between soil microbial communities and soil-borne pathogens. High-throughput sequencing technologies such as 454 pyrosequencing have been used in several recent studies that compared bacterial communities between soil samples with and without soil-borne disease pathogens, including Fusarium wilt, black root rot, and Rhizoctonia solani (20, 32, 45). R. solanacearum is present in soil at depths greater than 40 cm, and causes disease in tomato plants because their roots reach into deep soil (11). Therefore, an analysis of prokaryotic communities in this deep layer of soil is important. However, to the best of our knowledge, microbial communities have not yet been examined in this soil layer. Furthermore, soil samples from different fields with or without plant disease have been analyzed (20, 32, 33, 45). Difficulties are associated with identifying disease-associated microorganisms using comparisons between soil samples from different fields because members of indigenous soil microbial communities may markedly vary between sampling sites.
In the present study, we compared prokaryotic communities in soil samples with and without plants presenting tomato bacterial wilt using high-throughput sequencing and identified possible prokaryotic taxa associated with the promotion or suppression of this disease. In order to achieve this, we collected soil samples from a single greenhouse and investigated soil samples at depths less than and greater than 40 cm because the latter is also relevant to bacterial wilt.
Materials and Methods
Soil sampling and DNA extraction
Soil samples were collected from a greenhouse of tomato plants located in Kaizu City, Gifu Prefecture (35°23′ N, 136°63′ E; Fluvaquentic Haplosaprists) in late January 2015. The locations of plots in this greenhouse are described in Supplemental Fig. S1. Tomato plants have been continuously cultivated without serious plant diseases for 3 and 26 years in the northern and southern parts of the greenhouse, respectively. Plants in some areas of the northern part of the greenhouse coincidentally showed bacterial wilt symptoms at the time of sampling, whereas plants in other parts of the greenhouse remained disease-free. Therefore, two plots with plants showing signs of disease (plots N1 and N2) and four plots with no plants showing signs of disease (plots N3 to N6) in the northern part, and six plots with no plants showing signs of disease in the southern part (plots S1 to S6) were collected using a core sampler (Gauge Auger DIK-106B, Daiki Rika Kogyo, Saitama, Japan). We also collected soil samples from two different depths (20–30 cm and 40–50 cm) in each plot. A total of 24 samples were collected and stored at −20°C until used. Average soil pH (H2O) were 7.04 and 7.02 and electric conductivities (EC) were 0.053 and 0.042 mS cm−1 in the upper and lower layers, respectively. Soil pH and EC in the same layer were not significantly different between the soil sites with and without bacterial wilt symptoms (p<0.05, t-test).
DNA was extracted from 0.5 g of soil with an ISOIL for the Beads Beating kit (Nippongene, Tokyo, Japan) following the manufacturer’s instructions. DNA was quantified and its integrity was measured using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA) and by visualization on a 0.8% agarose gel in Tris-acetate-EDTA (TAE) buffer. The amount of DNA extracted from each soil sample is presented in Supplemental Table S1.
Detection of R. solanacearum
The phcA gene, which plays a major role in the regulation of R. solanacearum pathogenicity (31), was amplified from DNA extracted from each soil sample. A two-step nested polymerase chain reaction (PCR) was performed to detect phcA using two primer sets (16): phcA2981f (5′-TGGATATCGGGCTGGCAA-3′) and phcA4741r (5′-CGCTTTTGCGCAAAGGGA-3′) for the first step and phcA3538f (5′-GTGCCACAGCATGTTCAGG-3′) and phcA4209r (5′-CCTAAAGCGCTTGAGCTCG-3′) for the second step. PCR was performed in a total volume of 50 μL in a 200-μL microtube that contained 0.5 μL of KAPA2G Robust HotStart DNA Polymerase (5 U μL−1) (KAPA Biosystems, Wilmington, MA, USA), 5 μL of PCR reaction buffer for a high GC content (KAPA Biosystems), 1.5 μL of each primer (3.2 pmol each), 4 μL of a dNTP mixture (10 mM), 2.5 μL of MgCl2 (25 mM), 1 μL of a DNA template, and 35.5 μL Milli-Q water. The following PCR amplification profile was used: 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 57.5°C for 30 s, and extension at 72°C for 60 s, with a final extension at 72°C for 5 min. One microliter of the PCR product from the first-step amplification was used as a template for the second-step PCR reaction. Second-step PCR conditions were the same as those in first-step PCR, except that the extension time was reduced to 30 s. Amplification was verified by gel electrophoresis (1.5% agarose in TAE buffer).
Tag-encoded amplicon pyrosequencing
PCR was performed with each soil sample in order to amplify the V4 variable region of 16S rRNA using the bacterial and archaeal universal primers 515F and 806R (6) coupled with Roche 454 Titanium sequencing adapters. The 515F primer (5′-GTGCCAGCMGCCGCGGTAA-3′) contained a 10-base pair (bp) bar-coded sequence with a Roche 454-A pyrosequencing adapter (Titanium Lib-L adapters), and a Roche 454-B adapter for the 806R primer (5′-GGACTACVSGGGTATCTAA-3′). PCR was performed in a 200-μL microtube with a total volume of 50 μL, consisting of 0.5 μL of each primer (50 pmol each), 5 μL of a 2.5 mM dNTP mixture, 5 μL of 10×Ex Taq DNA buffer (20 mM Mg2+; TaKaRa, Otsu, Japan), 0.25 μL of Ex Taq DNA polymerase (5 U μL−1) (TaKaRa), 1 μL of a DNA template, and 37.75 μL of Milli-Q water. The following PCR amplification profile was used: an initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 45 s, 50°C for 30 s, and 72°C for 90 s, and a final extension step of 72°C for 10 min. In 1.5% agarose gels showing two or more bands, PCR products of approximately 350 bp in length were excised from the agarose gel and purified using a MonoFas DNA purification kit (GL Sciences, Tokyo, Japan). Each PCR amplicon was cleaned twice using an Agencourt AMPure XP system (Beckman Coulter, Pasadena, CA, USA) to remove primers and short DNA fragments, and then quantified using a Qubit Fluorometer (Invitrogen, Carlsbad, CA, USA). The purified PCR amplicons were combined in equimolar ratios into a single tube for emulsion PCR (emPCR). An emPCR reaction was performed with an approximate ratio of 0.2:1 (amplicon:emPCR beads), and amplicon sequencing was performed following the manufacturer’s protocols (Roche Applied Science, Indianapolis, IN, USA) on a Roche 454 GS Junior Titanium sequencer using a Lib-L kit. Sequencing data were deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive under accession number DRA004754.
Data analysis
Raw standard flowgram format files were pre-processed in Quantitative Insights Into Microbial Ecology (QIIME) (5). Data from read sequences, quality, flows, and ancillary metadata were analyzed using the QIIME pipeline according to Campisaono et al. (4). Quality filtering consisted of discarding reads of <200 bp or >1,000 bp in length, excluding homopolymer runs of >6 bp and continuous ambiguous bases of >6, accepting one barcode correction and two primer mismatches. Moreover, reads with mean quality scores less than 25 were also removed. Singleton operational taxonomic units (OTUs) and chimeric sequences were then removed before the statistical analysis. Denoising was performed using the built-in Denoiser algorithm, and chimera removal and OTU picking were accomplished with USEARCH 61, based on pairwise identities of 0.97. Taxonomies were assigned with the Ribosomal Database Project (RDP) naïve Bayesian classifier with a minimum confidence of 0.8 against the Greengenes Database (October 2012 release). An OTU-based analysis was performed with pyrotag-based datasets to calculate richness and diversity using the phyloseq package in R (1.7.24) (24). The diversity within each individual sample was estimated using non-parametric Shannon and Simpson diversity indices. The Chao1 estimator and abundance-based coverage estimator (ACE) were calculated to estimate the richness of each sample. We used the Student’s t-test to assess differences in prokaryotic diversity and richness (p<0.05). A multivariate analysis of community structure and diversity were performed with pyrotag-based datasets using a weighted UniFrac dissimilarity matrix calculated in QIIME, jackknifing (1,000 reiterations), read abundance data at the deepest level possible (3,105 reads), and unconstrained ordination in a principal coordinate analysis (PCoA). Indicator values were then calculated using the indicspecies package in R (8), with the aim of identifying OTUs associated with soils without disease symptoms rather than soils with disease symptoms or vice versa (p<0.05). In order to calculate indicator values, 999 random permutation tests were performed.
Results and Discussion
Detection of pathogenic R. solanacearum
The presence of R. solanacearum in each soil sample was assessed by the detection of phcA, which plays a major role in the regulation of pathogenicity (31). The PCR products of phcA were obtained from all sites with disease, irrespective of soil depth. phcA was detected in four out of 10 sites without plant wilt disease (Table 1, Supplemental Fig. S2). In these four sites, pathologies associated with R. solanacearum were not observed. We classified soil samples into three types (Table 1): soil not showing the signs of disease but with phcA (ND-soil), soil with diseased tomato plants and phcA (D-soil), and soil not showing bacterial wilt or phcA (H-soil). We selected 4 ND-soil samples and 2 D-soil samples for subsequent experiments. The H-soil sites in this field were not included because we were unable to evaluate microbial communities for disease without the presence of the pathogen. Amplicons of phcA were detected in the lower layer (40–50 cm depth) of soil in all D-soil and ND-soil sites, except for one (plot S2), indicating that R. solanacearum was present in soil at a depth greater than 40 cm, as described in a previous study (11).
Table 1.
Signs of disease and phcA gene amplification from each soil plot examined in this study.
| Sampling field | Plot number | Soil layer | Disease | phcA gene | Soil type |
|---|---|---|---|---|---|
| Northern part | N1 | Upper | + | + | D-soil |
| Lower | + | + | |||
| N2 | Upper | + | + | D-soil | |
| Lower | + | + | |||
| N3 | Upper | − | − | ND-soil | |
| Lower | − | + | |||
| N4 | Upper | − | − | ND-soil | |
| Lower | − | + | |||
| N5 | Upper | − | + | ND-soil | |
| Lower | − | + | |||
| N6 | Upper | − | − | H-soil | |
| Lower | − | − | |||
|
| |||||
| Southern part | S1 | Upper | − | − | H-soil |
| Lower | − | − | |||
| S2 | Upper | − | + | ND-soil | |
| Lower | − | − | |||
| S3 | Upper | − | − | H-soil | |
| Lower | − | − | |||
| S4 | Upper | − | − | H-soil | |
| Lower | − | − | |||
| S5 | Upper | − | − | H-soil | |
| Lower | − | − | |||
| S6 | Upper | − | − | H-soil | |
| Lower | − | − | |||
ND-soil indicates soil without disease but with phcA present; D-soil indicates soil with disease and phcA; and H-soil indicates soil without bacterial wilt or phcA.
In some soil environments, the incidence of the disease is known to be reduced in spite of the presence of pathogens, susceptible host plants, and climatic conditions favorable for disease development (2, 12, 13, 19). This soil, called disease-suppressive soil, has been reported for multiple soil-borne pathogens, including those causing Fusarium wilt (22), potato common scab (17), damping-off disease (15), tobacco black root rot (20), and bacterial wilt (33). However, in the present study, ND-soil with phcA was not defined as suppressive soil because we did not experimentally confirm its suppressive effects on bacterial wilt.
Richness, diversity, and microbial community structure
Pyrosequencing yielded a total of 105,967 high-quality sequences from the 12 samples. These sequences were clustered into 18,449 OTUs. A total of 1,452 and 1,206 OTUs were observed in the upper layers of ND- and D-soil, respectively, whereas 1,222 and 1,111 OTUs, respectively, were detected in the lower layers (Table 2). The number of OTUs and Chao1 and ACE richness indices were not significantly different between ND- and D-soil. However, the Shannon and Simpson diversity indices were significantly higher in D-than in ND-soil in the lower layer (each p<0.05). Previous studies indicated that the compositions of indigenous bacterial populations are simpler in conducive soil than in suppressive soil (32, 33). Therefore, some unique microbes may be involved in promoting or suppressing bacterial wilt disease.
Table 2.
Diversity and richness indices of prokaryotes in D- and ND-soil.
| Upper layer | Lower layer | |||
|---|---|---|---|---|
|
|
|
|||
| D-soil | ND-soil | D-soil | ND-soil | |
| Observed OTUs | 1452±136 | 1206±364 | 1222±150 | 1111±243 |
| Chao1 | 3609±736 | 3109±1306 | 3226±249 | 2994±743 |
| ACE | 3989±837 | 3254±1429 | 3323±129 | 3256±828 |
| Shannon | 4.35±0.17 | 4.46±0.08 | 4.15±0.14 | 4.01±0.13* |
| Simpson | 0.973±0.004 | 0.976±0.002 | 0.970±0.004 | 0.965±0.007* |
Asterisks represent pairs of means that are significantly different between D-soil (n=2) and ND-soil (n=4) in each layer (p<0.05).
DNA concentrations in the upper layer of soil were approximately three-to seven-fold higher than those in the lower layer (p<0.05) (Supplemental Table S1). Shannon and Simpson diversity indices were also higher in the upper layer (p<0.05). Watanabe et al. (40) indicated that the total DNA concentration in bulk soil was markedly reduced at depths greater than 30 cm because of the low carbon and nitrogen contents. The field in this study was plowed to a depth of 30 cm, and the use of an agrimotor may have consolidated the lower layer of soil. Thus, soil physicochemical properties may have differed between the upper and lower layers, and prokaryotes in the lower soil layer had a lower total microbial biomass and were less diverse than those in the upper layer (Table 2 and Supplemental Table S1).
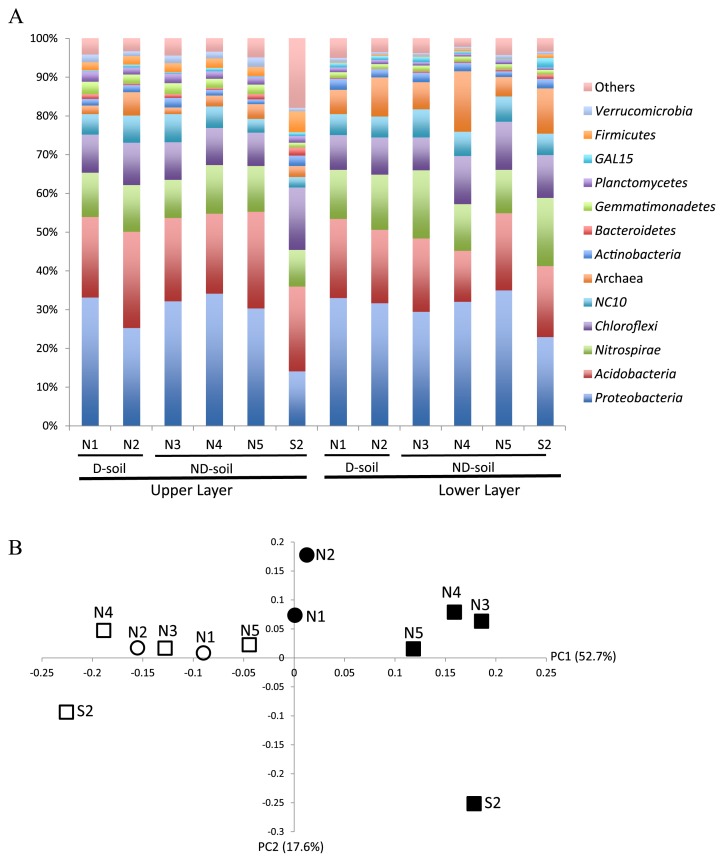
A total of 54 phyla were detected in the 12 soil samples. Proteobacteria and Acidobacteria accounted for 36% to 55% of the prokaryotic OTUs in all samples (Fig. 1A). Furthermore, Acidobacteria, Gemmatimonadetes, Planctomycetes, and Firmicutes were significantly more abundant in the upper layer than the lower layer, whereas Nitrospirae was less abundant (each p<0.05) (Fig. 1A). However, the relative abundance of each prokaryotic phylum was not significantly different between ND- and D-soil. These results indicate that prokaryotic community compositions were not significantly different at the phylum level between soil with and without disease.
Fig. 1.
Relative abundances of prokaryotic phyla (A) and a UniFrac-weighted principal component analysis of prokaryotic communities in D- and ND-soil (B). (B) Closed circle: D-soil, upper layer. Open circle: D-soil, lower layer. Closed square: ND-soil, upper layer. Open square: ND-soil, lower layer.
A weighted UniFrac analysis and PCoA showed that the prokaryotic communities in soil samples were classifiable into two groups according to soil depth (Fig. 1B). The community structures in the upper soil layer were also distinct between ND- and D-soil, whereas those in the lower soil layer were similar (Fig. 1B). Therefore, a difference in prokaryotic community structures between ND- and D-soil was only observed in the upper layer of soil collected from a single greenhouse; however, this difference was not apparent at the bacterial phylum level. The prokaryotic communities of soil sample S2, collected from the southern part of the field, were distantly related to samples collected from the northern part, both in the upper and lower layers, suggesting that the prokaryotic community structure differed according to soil management practices rather than the observation of bacterial wilt.
Specific OTUs in ND-soil
Twenty-five OTUs were unique or significantly more abundant in ND-soil than in D-soil in their respective layers (Table 3). Among them, seven OTUs were commonly detected in the upper and lower layers, whereas eight and 10 OTUs were detected in one of the layers only. OTUs belonged to the following phyla: Acidobacteria (8 OTUs), Proteobacteria (8 OTUs), Chloroflexi (4 OTUs), Aenigmarchaeota (1 OTU), Euryarchaeota (1 OTU), Crenarchaeota (1 OTU), Woesearchaeota (1 OTU), and Verrucomicrobia (1 OTU).
Table 3.
Operational taxonomic units (OTUs) abundant in ND-soil.
| Soil layer | OTU number | Phylum | Closest relative | D-soil | ND-soil | D-soil | ND-soil |
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Upper layer | Lower layer | ||||||
| Upper layer | OTU239890 | Acidobacteria | Acidobacteria Gp2 | 0 | 2.53 E-04 | 0 | 0 |
| OTU17668 | Acidobacteria | Acidobacteria Gp3 | 4.76 E-04 | 2.43 E-03 | 4.82 E-05 | 3.09 E-05 | |
| OTU4234 | Chloroflexi | Anaerolinae | 0 | 6.32 E-05 | 0 | 0 | |
| OTU4724 | Chloroflexi | Anaerolinae | 1.43 E-04 | 8.22 E-04 | 0 | 0 | |
| OTU723819 | Crenarchaeota | Thermoportei | 4.76 E-05 | 2.43 E-03 | 4.82 E-05 | 3.09 E-05 | |
| OTU952203 | Proteobacteria | Pseudodugonella sp. | 9.52 E-05 | 4.11 E-04 | 0 | 0 | |
| OTU21359 | Proteobacteria | Cystobacterineae | 0 | 1.90 E-04 | 0 | 0 | |
| OTU2761428 | Proteobacteria | Dyella | 0 | 2.85 E-04 | 0 | 0 | |
|
| |||||||
| Lower layer | OTU8713 | Acidobacteria | ‘Candidatus Koribacter’ | 4.76 E-04 | 0 | 0 | 3.09 E-04 |
| OTU14103 | Acidobacteria | Acidobacteria Gp12 | 0 | 0 | 0 | 2.47 E-04 | |
| OTU4584 | Acidobacteria | Acidobacteria Gp13 | 0 | 0 | 9.64 E-05 | 2.47 E-04 | |
| OTU1107276 | Acidobacteria | Acidobacteria Gp2 | 4.76 E-04 | 0 | 4.82 E-05 | 2.17 E-04 | |
| OTU18194 | Aenigmarchaeota | ‘Candidatus Aenigmarchaeum’ | 4.76 E-04 | 0 | 0 | 2.47 E-04 | |
| OTU19114 | Chloroflexi | Anaerolineaceae | 0 | 0 | 4.82 E-05 | 1.24 E-04 | |
| OTU889 | Euryarchaeota | Methanomassiliicoccus | 0 | 0 | 9.64 E-05 | 4.33 E-04 | |
| OTU16969 | Proteobacteria | Azospirillum sp. EP3-3L | 4.76 E-04 | 0 | 0 | 3.09 E-04 | |
| OTU14184 | Proteobacteria | Desulfurmonadales | 0 | 0 | 1.45 E-04 | 4.64 E-04 | |
| OTU21385 | Woesearchaeota | Woesearchaeota Incertae Sedis AR15 | 4.76 E-04 | 0 | 0 | 1.86 E-04 | |
|
| |||||||
| Upper and lower layers | OTU2240 | Acidobacteria | Acidobacteria Gp1 | 0 | 2.40 E-03 | 0 | 6.19 E-05 |
| OTU663880 | Acidobacteria | Acidobacteria Gp4 | 0 | 1.90 E-04 | 9.64 E-05 | 9.90 E-04 | |
| OTU23191 | Chloroflexi | Anaerolineaceae | 0 | 6.32 E-05 | 9.64 E-05 | 3.71 E-04 | |
| OTU142261 | Proteobacteria | Hephaestia sp. | 4.76 E-04 | 7.27 E-04 | 0 | 6.19 E-05 | |
| OTU10519 | Proteobacteria | Nitrosospira sp. APG3 | 0 | 2.85 E-04 | 0 | 3.09 E-05 | |
| OTU21362 | Proteobacteria | Denitratisoma sp. | 0 | 5.38 E-04 | 0 | 2.47 E-04 | |
| OTU2781 | Verrucomicrobia | Spartobacteria sp. | 0 | 5.38 E-04 | 0 | 2.47 E-04 | |
The relative abundances of OTUs are shown in the table. Each OTU was identified using an indicspecies analysis (p<0.05).
Acidobacteria was predominant in ND-soil. Acidobacteria subdivisions 1, 3, 4, and 6 are the most abundant bacteria in agricultural soil (17). In this study, OTUs belonging to subdivisions 1, 2, 3, 4, 12, and 13 were specifically abundant in ND-soil. Previous studies showed that Acidobacteria subdivisions 4 and 6 were more abundant in soil that suppresses potato common scab and Fusarium wilt (29, 32). Moreover, the abundance of Acidobacteria subdivisions 1 and 3 negatively correlated with that of R. solanacearum (41). However, previous studies also demonstrated that some members of this phylum are more abundant in soil with plants showing signs of disease due to several soil-borne pathogens (21, 30, 32, 41). These bacteria may play a role in preventing bacterial wilt; however, the ecology of Acidobacteria has not yet been defined in detail.
The proteobacterial genera Hephaestia, Cystobacterineae, Azospirillum, Nitrosospira, Denitratisoma, Desulfuromonas, Pseudoduganella, and Dyella were abundant in ND-soil. Azospirillum and Dyella were previously detected in disease-suppressive soil (4, 20). The detection of Azospirillum in ND-soil is interesting because they are known as plant growth-promoting bacteria and for their nitrogen-fixing ability (34, 46). Nitrosospira, Denitratisoma, and Dyella are also involved in the nitrogen cycle in soil environments (9, 36, 37). The suppressive effect on plant disease has been discussed for bacteria that participate in the nitrogen cycle and the nitrogen they release has been found to affect microbial community structures in soil (7, 14). Therefore, bacteria that play a role in the nitrogen cycle in soil may be involved in the control of bacterial wilt.
Anaerolineae, belonging to Chloroflexi, were also abundant in ND-soil. These bacteria are neutrophilic, strictly anaerobic chemo-organotrophs that have the ability to utilize sugars and polysaccharides (28). They inhabit extremely diverse environments, including sediments, subsurface habitats, anaerobic-dechlorinating environments, hot springs, deep hot aquifers, and anaerobic wastewater sludges (44). They have relatively long doubling times (45–100 h) in cultures; therefore, they are difficult to isolate. We detected not only bacteria, but also Archaea in the phyla Aenigmarchaeota, Euryarchaeota, Crenarchaeota, and Woesearchaeota in ND-soil. To the best of our knowledge, we are the first to have detected Archaea in suppressive soil. We are also the first to have detected and analyzed Chloroflexi and Archaea in ND-soil associated with tomato bacterial wilt using pyrosequencing methods.
Woesearchaeota, Euryarchaeota, Aenigmarchaeota, Acidobacteria subdivision 12 and 13, Desulfuromonadales, and Azospirillum were specifically detected in the lower layer of ND-soil. These bacteria are found in oligotrophic and anaerobic environments (27, 38). Due to these conditions, which are specific to the lower layer of soil, unique microbes were detected in the lower soil layer. Moreover, prokaryotes were less diverse in ND-than in D-soil (Table 2). Therefore, these indicator bacteria may have important roles in the suppression of bacterial wilt because tomato roots are found in deep (<40 cm) soil.
Specific OTUs in D-soil
In D-soil, 37 indicator OTUs belonging to eight phyla were detected as significantly abundant OTUs (Table 4). OTUs belonging to Actinobacteria and Planctomycetes were specifically detected in D-soil rather than in ND-soil. The OTUs of Acidobacteria (7 OTUs) were dominant in the upper layer, whereas those of Proteobacteria (6 OTUs) and Acidobacteria (6 OTUs) were mainly detected in the lower layer of D-soil. We specifically detected OTUs belonging to Acidobacteria subdivisions 7 and 25 and Holophaga subdivision 8 in D-soil. Moreover, unique OTUs belonging to subdivisions 2, 3, 12, and 13 were found in ND- and D-soil. Members of Acidobacteria, Proteobacteria, and Actinobacteria were more abundant in soil with the appearance of disease (20, 21, 32). OTUs belonging to Planctomycetes were more abundant in D-soil, which is in contrast to previous findings indicating that they are frequency observed in soil and are aerobic heterotrophs (1, 22, 30, 42). Moreover, Planctomycetes bacteria have a unique metabolism and the ability to oxidize ammonia under anaerobic conditions (10). The abundance of these bacteria correlated with the appearance of disease; therefore, they may be associated with bacterial wilt.
Table 4.
Operational taxonomic units (OTUs) abundant in D-soil.
| Soil layer | OTU number | Phylum | Closest relative | D-soil | ND-soil | D-soil | ND-soil |
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Upper layer | Lower layer | ||||||
| Upper layer | OTU15038 | Acidobacteria | ‘Candidatus Koribacter’ | 2.4E-04 | 0 | 0 | 0 |
| OTU17589 | Acidobacteria | Acidobacteria_Gp13 | 1.4E-04 | 0 | 0 | 0 | |
| OTU16659 | Acidobacteria | Acidobacteria_Gp2 | 1.9E-04 | 0 | 0 | 0 | |
| OTU4398 | Acidobacteria | Acidobacteria_Gp2 | 2.9E-04 | 3.2E-05 | 0 | 0 | |
| OTU16427 | Acidobacteria | Acidobacteria_Gp3 | 1.4E-04 | 0 | 0 | 0 | |
| OTU1216 | Acidobacteria | Acidobacteria_Gp7 | 2.4E-04 | 6.3E-05 | 0 | 0 | |
| OTU14433 | Acidobacteria | Acidobacteria_Gp7 | 4.3E-04 | 3.2E-05 | 0 | 6.2E-05 | |
| OTU10268 | Planctomycetes | Pirellula | 2.4E-04 | 0 | 0 | 0 | |
| OTU7125 | Planctomycetes | Planctomycetaceae | 3.3E-04 | 3.2E-05 | 0 | 0 | |
| OTU9710 | Proteobacteria | Alphaproteobacteria | 9.5E-05 | 0 | 1.1E-03 | 1.5E-04 | |
| OTU21086 | Proteobacteria | Betaproteobacteria | 1.4E-04 | 3.2E-05 | 0 | 0 | |
| OTU1323 | Verrucomicrobia | Spartobacteria | 9.5E-05 | 0 | 0 | 0 | |
| OTU17053 | Verrucomicrobia | Spartobacteria_genera_incertae_sedis | 2.9E-04 | 0 | 0 | 3.1E-05 | |
|
| |||||||
| Lower layer | OTU18877 | Acidobacteria | Acidobacteria_Gp12 | 0 | 0 | 4.3E-04 | 6.2E-05 |
| OTU1053 | Acidobacteria | Acidobacteria_Gp13 | 0 | 0 | 1.9E-04 | 3.1E-05 | |
| OTU2699 | Acidobacteria | Acidobacteria_Gp25 | 0 | 0 | 6.7E-04 | 1.2E-04 | |
| OTU15771 | Acidobacteria | Acidobacteria_Gp3 | 0 | 0 | 2.4E-04 | 0 | |
| OTU7061 | Acidobacteria | Acidobacteria_Gp3 | 0 | 6.3E-05 | 2.4E-04 | 0 | |
| OTU8173 | Acidobacteria | Holophaga | 0 | 0 | 2.4E-04 | 6.2E-05 | |
| OTU5670 | Actinobacteria | Actinobacteria | 0 | 0 | 9.6E-05 | 0 | |
| OTU2126 | Actinobacteria | Actinobacteria | 0 | 0 | 6.7E-04 | 9.3E-05 | |
| OTU21537 | Pacearchaeota | Pacearchaeota Incertae Sedis AR13 | 0 | 0 | 9.6E-05 | 0 | |
| OTU2970 | Woesearchaeota | UnclassifiedWoesearchaeota | 0 | 0 | 6.7E-04 | 1.5E-04 | |
| OTU15020 | Chloroflexi | Anaerolineaceae | 0 | 0 | 9.6E-05 | 0 | |
| OTU24535 | Proteobacteria | Rhodospirillales | 0 | 3.2E-05 | 2.9E-04 | 3.1E-05 | |
| OTU80 | Proteobacteria | Deltaproteobacteria | 0 | 0 | 1.4E-04 | 0 | |
| OTU6774 | Proteobacteria | Geobacter | 0 | 0 | 1.9E-04 | 0 | |
| OTU3473 | Proteobacteria | Nannocystineae | 0 | 0 | 2.4E-04 | 0 | |
| OTU23743 | Proteobacteria | Methylococcaceae | 0 | 0 | 1.9E-04 | 0 | |
| OTU9686 | Proteobacteria | Gammaproteobacteria | 5.2E-04 | 1.3E-04 | 3.9E-04 | 0 | |
|
| |||||||
| Upper and lower layers | OTU15704 | Acidobacteria | Acidobacteria_Gp25 | 9.5E-05 | 0 | 9.6E-05 | 0 |
| OTU9423 | Armatimonadetes | Chthonomonas | 4.8E-05 | 3.2E-05 | 1.9E-04 | 0 | |
| OTU19666 | Chloroflexi | Anaerolineaceae | 1.4E-04 | 0 | 1.4E-04 | 3.1E-05 | |
| OTU15429 | Planctomycetes | ‘Candidatus Kuenenia’ | 4.8E-05 | 0 | 4.8E-04 | 0 | |
| OTU19754 | Proteobacteria | Betaproteobacteria | 5.7E-03 | 4.7E-04 | 1.1E-02 | 1.9E-03 | |
| OTU6110 | Proteobacteria | Deltaproteobacteria | 5.2E-04 | 1.9E-04 | 5.8E-04 | 1.2E-04 | |
| OTU377 | Proteobacteria | Gammaproteobacteria | 2.4E-04 | 3.2E-05 | 9.6E-05 | 3.1E-05 | |
The relative abundances of OTUs are shown in the table. Each OTU was identified using an indicspecies analysis (p<0.05).
Conclusions
In the present study, we compared prokaryotic communities in upper and lower layers of soil with or without bacterial wilt using a 454 pyrosequencing analysis. We classified ND- and D-soil according to the appearance of a plant pathogen in the field depending on the detection of phcA in both soil types. In the lower layer of soil, prokaryotic diversity was less in ND-than in D-soil. A difference in prokaryotic community structures between ND- and D-soil was only observed in the upper layer of soil, despite an apparent similarity at the phylum level, and this may have been because soil was sampled from a single greenhouse. Twenty-five and 37 indicator OTUs were found to be more abundant in ND- and D-soil, respectively. Some bacteria and Archaea were specifically detected in the lower layer of ND-soil. The specific microbes detected in ND-soil may play important roles in suppressing the soilborne pathogens of bacterial wilt, and future studies are needed in order to clarify the roles of the prokaryotes detected.
Supplementary material
Acknowledgements
This work was supported by the Council for Science, Technology, and Innovation (CSTI), Cross-ministerial Strategic Innovation Promotion Program (SIP), and “Technologies for creating next-generation agriculture, forestry, and fisheries” (funding agency: Bio-oriented Technology Research Advancement Institution, NARO). This work was also partially supported by the RIKEN Competitive Program for Creative Science and Technology.
References
- 1.Asakawa S., Kimura M. Comparison of bacterial community structures at main habitats in paddy field ecosystem based on DGGE analysis. Soil Biol Biochem. 2008;40:1322–1329. [Google Scholar]
- 2.Baker K.F., Cook R.J. Biological control of plant pathogens. WH Freeman; San Francisco: 1974. p. 433. [Google Scholar]
- 3.Bernard E., Larkin R.P., Tabantzis S., Erich M.S., Alyokhin A., Sewell G., Lannan A., Gross S.D. Compost, rapeseed rotation, and biocontrol agents significantly impact soil microbial communities in organic and conventional potato production systems. Appl Soil Ecol. 2012;52:29–41. [Google Scholar]
- 4.Campisaono A., Antonielli L., Pancher M., Yousaf S., Pindo M., Pertot I. Bacterial endophytic communities in the grapevine depend on pest management. PLoS One. 2014;9:e112763. doi: 10.1371/journal.pone.0112763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caporaso J.G., Kuczynski J., Stombaugh J., et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., Fierer N., Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequence per sample. Proc Natl Acad Sci USA. 2011;108:S4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen M.F., Yamasaki H., Mazzola M. Brassica napus seed meal soil amendment modifies microbial community structure, nitric oxide production and incidence of Rhizoctonia root rot. Soil Biol Biochem. 2005;37:1215–1227. [Google Scholar]
- 8.De Cáceres M., Legendre P. Associations between species and groups of sites: indices and statistical inference. Ecology. 2009;90:3566–3574. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
- 9.Fahrbach M., Kuever J., Meinke R., Kämpfer P., Hollender J. Denitrisoma oestradiolicum gen. nov., sp. nov., a 17β-oestradiol-degrading, denitrifying betaproteobacterium. Int J Syst Evol Microbiol. 2006;56:1547–1552. doi: 10.1099/ijs.0.63672-0. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst J.A., Sagulenko E. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol. 2011;9:403–413. doi: 10.1038/nrmicro2578. [DOI] [PubMed] [Google Scholar]
- 11.Graham J., Lloyd A.B. Survival of potato strain (race 3) of Pseudomonas solanacearum in the deeper soil layers. Aust J Agric Res. 1979;30:489–496. [Google Scholar]
- 12.Haas D., Défago G. Biological control of soil-borne pathogens by fluorescent Pseudomonas. Nat Rev Microbiol. 2005;3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 13.Ho W.C., Cherm L.L., Ko W.H. Pseudomonas solanacearum- suppressive soils in Taiwan. Soil Biol Biochem. 1988;20:489–492. [Google Scholar]
- 14.Hossain S., Bergkvist G., Glinwood R., Berglund K., Måretensson A., Hallin S., Persson P. Brassicaceae cover crops reduce Aphanomyces pea root rot without suppressing genetic potential of microbial nitrogen cycling. Plant Soil. 2015;392:227–238. [Google Scholar]
- 15.Hunter P.J., Petch G.M., Calvo-Bado L.A., Pettitt T.R., Parsons N.R., Morgan J.A., Whipps J.M. Difference in microbial activity and microbial populations of peat associated with suppression of damping-off disease caused by Pythium sylvaticum. Appl Environ Microbiol. 2006;72:6452–6460. doi: 10.1128/AEM.00313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue Y., Nakaho K. Sensitive quantitative detection of Ralstonia solanacearum in soil by the most probable number-polymerase chain reaction (MPN-PCR) method. Appl Microbiol Biotechnol. 2014;98:4169–4177. doi: 10.1007/s00253-014-5604-z. [DOI] [PubMed] [Google Scholar]
- 17.Keilak A., Pijl A.S., van Veen J.A., Kowalchuk G.A. Phylogenetic diversity of Acidobacteria in a former agricultural soil. ISME J. 2009;3:378–382. doi: 10.1038/ismej.2008.113. [DOI] [PubMed] [Google Scholar]
- 18.Kelman A. One hundred and one years of research on bacterial wilt. In: Prior P.H., Allen C., Elphinstone J., editors. Bacterial Wilt Disease: Molecular and Ecological Aspects. Springer; Heidelberg: 1998. pp. 1–5. [Google Scholar]
- 19.Koga K., Hara H., Tanaka H. Suppressive soils to bacterial wilt of tobacco in Japan and population dynamics of Pseudomonas solanacearum in these soils. Ann Phytopathol Soc Jpn. 1997;63:304–308. [Google Scholar]
- 20.Kyselková M., Kopecky J., Frapolli M., Défago G., Ságová-Marecková M., Grundmann G.L., Moënne-Loccoz Y. Comparison of rhizobacterial community composition in soil suppressive or conductive to tobacco black root rot disease. ISME J. 2009;3:1127–1138. doi: 10.1038/ismej.2009.61. [DOI] [PubMed] [Google Scholar]
- 21.Li G.J., Ren G.D., Jia Z.J., Dong Y.H. Composition and activity of rhizosphere microbial community associated with healthy and diseased greenhouse tomatoes. Plant Soil. 2014;380:337–347. [Google Scholar]
- 22.Li X., Zhang Y., Ding C., Jia Z., He Z., Zhang T., Wang X. Declined soil suppressiveness to Fusarium oxysporum by rhizosphere microflora of cotton in soil sickness. Biol Fertil Soils. 2015;51:935–946. [Google Scholar]
- 23.Luo J., Ran W., Hu J., Yang X., Xu Y., Shen Q. Application of bio-organic fertilizer significantly affected fungal diversity of soils. Soil Sci Soc Am J. 2010;74:2039–2048. [Google Scholar]
- 24.McMurdie P.J., Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendes R., Kruijt M., de Bruijin I., et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 26.Nishiyama M., Shiomi Y., Suzuki S., Marumoto T. Suppression of growth of Ralstonia solanacearum, tomato bacterial wilt agent, on/in tomato seedlings cultivated in a suppressive soil. Soil Sci Plant Nutr. 1999;45:79–87. [Google Scholar]
- 27.Ortiz-Alvarez R., Casamayor E.O. High occurrence of Pacearchaeota and Woesearchaeota (Archaea superphylum DPANN) in the surface waters of oligotrophic high-altitude lakes. Environ Microbiol Rep. 2016;8:210–217. doi: 10.1111/1758-2229.12370. [DOI] [PubMed] [Google Scholar]
- 28.Podosokorskaya O.A., Bonch-Osmolovskaya E.A., Novikov A.A., Kolganova T.V., Kublanov I.V. Ornatilinea apprima gen. nov., sp. nov., a cellulolytic representative of the class Anaerolineae. Int J Syst Evol Microbiol. 2013;63:86–92. doi: 10.1099/ijs.0.041012-0. [DOI] [PubMed] [Google Scholar]
- 29.Rosenzweig N., Tiedje J.M., Quensen J.F., III, Meng Q., Hao J.J. Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses. Plant Dis. 2012;96:718–725. doi: 10.1094/PDIS-07-11-0571. [DOI] [PubMed] [Google Scholar]
- 30.Sanguin H., Sarniguet A., Gazengel K., Moënner-Loccoz Y., Grundamann G.L. Rhizosphere bacterial communities associated with disease suppressiveness stages of take-all decline in wheat monoculture. New Phytol. 2009;184:694–707. doi: 10.1111/j.1469-8137.2009.03010.x. [DOI] [PubMed] [Google Scholar]
- 31.Schell M.A. Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu Rev Phytopathol. 2000;38:263–292. doi: 10.1146/annurev.phyto.38.1.263. [DOI] [PubMed] [Google Scholar]
- 32.Shen Z., Ruan Y., Chao X., Zhang J., Li R., Shen Q. Rhizosphere microbial community manipulated by 2 years of consecutive biofertilizer application associated with banana Fusarium wilt disease suppression. Biol Fertil Soils. 2015;51:553–562. [Google Scholar]
- 33.Shiomi Y., Nishiyama M., Onizuka T., Marumoto T. Comparison of bacterial community structure in the rhizoplane of tomato plants grown in soils suppressive and conductive towards bacterial wilt. Appl Environ Microbiol. 1999;65:3996–4001. doi: 10.1128/aem.65.9.3996-4001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Somers E., Ptacek D., Gysegom P., Srinivasan M., Vanderleyden J. Azospirillum brasilense produces the auxin-like phenylacetic acid by using the key enzyme for indole-3-acetic acid biosynthesis. Appl Environ Microbiol. 2005;71:1803–1810. doi: 10.1128/AEM.71.4.1803-1810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stutz E.W., Défago G., Kern H. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology. 1986;76:181–185. [Google Scholar]
- 36.Tago K., Ishii S., Nishizawa T., Otsuka S., Senoo K. Phylogenetic and functional diversity of denitrifying bacteria isolated from various rice paddy and rice-soybean rotation field. Microbes Environ. 2011;26:30–35. doi: 10.1264/jsme2.me10167. [DOI] [PubMed] [Google Scholar]
- 37.Taylor A.E., Bottomley P.J. Nitrite production by Nitrosomonas europeae and Nitrosospira sp. AV in soils at different solution concentrations of ammonium. Soil Biol Biochem. 2006;38:828–836. [Google Scholar]
- 38.Thrash J.C., Coates J.D. Phylum XVII. Acidobacteria phyl. Nov. In: Krieg N.R., Staley J.T., Brown D.R., Hedlund B.P., Paster B.J., Ward N.J., Ludwig W., Whitman B.W., editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Vol. 4. Springer; New York: 2011. pp. 725–735. [Google Scholar]
- 39.Wang J.F., Lin C.H. Integrated management of tomato bacterial wilt. AVRDC-The World Vegetable Center; Taiwan: 2005. [Google Scholar]
- 40.Watanabe T., Wang G., Taki K., Ohashi Y., Kimura M., Asakawa S. Vertical changes in bacterial and archaeal communities with soil depth in Japanese paddy fields. Soil Sci Plant Nutr. 2010;56:705–715. [Google Scholar]
- 41.Wu B., Wang X., Yang L., et al. Effects of Bacillus amyloliquefaciens ZM9 on bacterial wilt and rhizosphere microbial communities of tobacco. Appl Soil Environ. 2016;103:1–12. [Google Scholar]
- 42.Xiong W., Zhao Q., Zhao J., Xun W., Li R., Zhang R., Wu H., Shen Q. Different continuous cropping spans significantly affect microbial community membership and structure in a vanilla-grown soil as revealed by deep pyrosequencing. Microb Ecol. 2015;70:209–218. doi: 10.1007/s00248-014-0516-0. [DOI] [PubMed] [Google Scholar]
- 43.Yabuuchi E., Kosako Y., Yano I., Hotta H., Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]
- 44.Yamada T., Sekiguchi Y. Cultivation of uncultured Chloroflexi subphyla: significance and ecophysiology of formerly uncultured Chloroflexi ‘Subphylum I’ with natural and biotechnological relevance. Microbes Environ. 2009;24:205–216. doi: 10.1264/jsme2.me09151s. [DOI] [PubMed] [Google Scholar]
- 45.Yin C., Hulbert S.H., Schroeder K.L., Mavrodi O., Mavrodi D., Dhingra A., Schillinger W.F., Paulitz T.C. Role of bacterial communities in the natural suppression of Rhizoctonia solani bare patch disease of wheat (Triticum aestivum L.) Appl Environ Microbiol. 2013;79:7428–7438. doi: 10.1128/AEM.01610-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou D., Huang X.F., Chaparro J.M., Badri D.V., Manter D.K., Vivanco J.M., Guo J. Root and bacterial secretions regulate the interaction between plants and PGPR leading to distinct plant growth promotion effects. Plant Soil. 2016;401:259–272. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.