Abstract
Fluorescence in situ hybridization (FISH) has been employed to identify microorganisms at the single cell level under a microscope. Extensive efforts have been made to improve and extend the FISH technique; however, the development of a widely applicable protocol is a continuing challenge. The present study evaluated the effects of divalent cations in the hybridization solution on the FISH-based detection of various species of bacteria and archaea with rRNA-targeted probes. A flow cytometric analysis after FISH with a standard hybridization buffer detected positive signals from less than 30% of Escherichia coli IAM 1264 cells. However, the number of cells with positive signals increased to more than 90% after the addition of calcium chloride to the hybridization buffer. Mn2+ also had positive effects, whereas Mg2+ did not. The positive effects of Ca2+ were similarly observed for bacteria belonging to Enterobacteriaceae, including Enterobacter sakazakii IAM 12660T, E. aerogenes IAM 12348, Klebsiella planticola IAM 14202, and Salmonella enterica subsp. enterica serovar Typhimurium strain LT2. These results indicate that the supplementation of Ca2+ to the hybridization buffer for FISH contributes to the efficient detection of Enterobacteriaceae cells.
Keywords: fluorescence in situ hybridization, Enterobacteriaceae, calcium ions, flow cytometry, Escherichia coli
Fluorescence in situ hybridization (FISH) was developed to quantitatively detect a specific group of microbes, including those yet to be cultivated, by microscopy (1, 3). FISH is currently one of the key techniques utilized in microbial ecology and environmental microbiology (24, 30, 41) as well as in public health (e.g., 20). FISH-based studies on microbiomes in humans and plants have recently attracted a great deal of attention (e.g., 4, 7, 16, 36). A large number of probes that target rRNA have been designed (14, 42), and a standard protocol is available (https://www.arb-silva.de/). FISH protocols continue to be modified to increase the intensity of fluorescence and expand its application to the targeting of genomic DNA and mRNA (5, 19, 22, 23, 31, 43, 44). Recent advances in rRNA-targeted FISH combined with other spectrometric imaging techniques have resulted in a clearer visualization of the microbial world (18, 27, 37, 39).
rRNA-targeted FISH occasionally fails to detect microbial cells even if rRNA gene sequence and oligonucleotide probes match completely. Previous methodological studies achieved improvements in the hybridization efficiency of probes to their targets (12, 13, 32, 45, 47). Cell wall structures also affect FISH results. Extensive efforts have been made to increase the probe permeability of microbial cells (6, 8–10, 28, 34). Nevertheless, an acceptable fluorescent signal intensity is not always obtained from all cells, even in laboratory-grown pure cultures. This phenomenon was traditionally considered to be due to a low cellular ribosome content (11, 26, 40). In the present study, we demonstrated that this phenomenon was a significant cause for concern, particularly when attempting to detect bacteria in the family Enterobacteriaceae, which includes medically important species. However, this issue was resolved with a slight modification to the standard FISH protocol, i.e., the addition of Ca2+ or Mn2+ to the hybridization solution.
Materials and Methods
Microbial strains and preparation of cells for FISH
Microorganisms (listed in Table 1) were obtained from public culture collections and cultivated according to their instructions. Cells were fixed with 3% paraformaldehyde pH 7.4, at 4°C for 20–24 h and stored in 99% ethanol:phosphate-buffered saline (PBS) solution (1:1 [v/v]) at −20°C as described by Wallner et al. (40). Prior to in situ hybridization, when indicated, the pretreatment of cells was conducted by an incubation in one of three solutions: 10 mg mL−1 of lysozyme at 37°C for 1 h, ethanol:formalin (9:1 [v/v]) at room temperature for 10 min, or 0.25 M HCl at room temperature for 30 min (8, 41).
Table 1.
Summary of flow cytometry results after FISH
| Tested strains | Ratio of positive events (%)* | |
|---|---|---|
|
| ||
| w/Ca2+ | w/o Ca2+ | |
| Gammaproteobacteria (Enterobacteriaceae) | ||
| Escherichia coli IAM 1264 | 92.7 | 28.6 |
| Escherichia coli IAM 12119T | 66.7 | 5.0 |
| Proteus vulgaris JCM 1668T | 90.1 | 76.5 |
| Pantoea agglomerans IAM 12659 | 68.9 | 58.1 |
| Enterobacter sakazakii IAM 12660T | 68.9 | 10.9 |
| Enterobacter aerogenes IAM 12348T | 78.0 | 11.4 |
| Klebsiella planticola IAM 14202T | 90.3 | 41.4 |
| Klebsiella pneumoniae IAM 14200T | 95.9 | 58.5 |
| Citrobacter freundii NBRC 1268T | 39.0 | 9.3 |
| Salmonella enterica | ||
| subsp. enterica serovar Typhimurium LT2 | 80.1 | 38.4 |
| Gammaproteobacteria (others) | ||
| Pseudomonas aeruginosa JCM 14847 | 90.6 | 90.0 |
| Aeromonas salmonicida NBRC 13784T | 47.8 | 47.1 |
| Xanthomonas pisi NBRC 13556 | 33.6 | 25.9 |
| Vibrio orientalis NBRC 15638T | 96.5 | 96.3 |
| Shewanella putrefaciens NBRC 3908T | 92.7 | 93.6 |
| Alphaproteobacteria | ||
| Acetobacter pasteurianus IAM 1804 | 21.2 | 25.1 |
| Betaproteobacteria | ||
| Comamonas testosteroni NBRC 14951T | 87.2 | 76.6 |
| Chromobacterium violaceum NBRC 12614T | 86.5 | 86.2 |
| Firmicutes | ||
| Bacillus subtilis IAM 12118T** | 92.9 | 93.9 |
| Bacillus licheniformis IAM 13417** | 94.5 | 93.9 |
| Clostridium acetobutylicum NBRC 13948T** | 84.3 | 84.3 |
| Staphylococcus aureus IAM 1058** | 77.1 | 81.7 |
| Lactobacillus plantarum JCM 1149 | 63.9 | 62.7 |
| Lactococcuss lactis subsp. lactis IAM 1249 | 62.8 | 65.2 |
| Bacteroidetes | ||
| Bacteroides vulgatus NBRC 14291T | 95.8 | 94.8 |
| Actinobacteria | ||
| Bifidobacterium bifidum NBRC 100015T | 50.0 | 51.1 |
| Corynebacterium glutamicum NBRC 12168T** | 59.4 | 59.6 |
| Micrococcus luteus ATCC 398** | 68.0 | 68.1 |
| Euryarchaeota | ||
| Methanococcus voltae NBRC 100457T | 96.2 | 95.4 |
| Crenarchaeota | ||
| Acidianus brierleyi DSM 1651 | 56.9 | 58.0 |
Cells were harvested at the late exponential phase of growth and fixed with paraformaldehyde, as described in the Materials and Methods section.
The threshold for positive fluorescence was defined by <5% of total events observed in the negative control (i.e., without probe). Positive events % were assessed after FISH with the hybridization buffer containing 50 mg L−1 of calcium chloride (w/Ca2+) and not containing calcium chloride (w/o Ca2+). Average values from at least two independent experiments are shown.
The lysozyme treatment was conducted for these bacterial species before hybridization.
FISH and flow cytometry
FISH and flow cytometry were performed according to Amann et al. (2), Fuchs et al. (12), Manz et al. (25), and Yilmaz and Noguera (45). Fixed cells were suspended in standard hybridization buffer (0.9 M NaCl, 0.01% SDS, 20 mM Tris-HCl, pH 7.2 and 15% formamide) containing a fluorescein isothiocyanate (FITC)-labeled probe (EUB338) (1) or Alexa488-labeled probe (ARC915) (35) (0.5 pmol μL−1), as reported previously (1, 17, 35). In addition, the fluorescent probes, EC1153, EC1482, EC839S, EC839L, and EC1235, all of which have distinct target sequences (Escherichia coli 16S rRNA gene positions 1153–1172, 1482–1499, 839–856, 839–859, and 1235–1255, respectively) (12, 46), were used in experiments with E. coli. As a negative control, the fluorescent probe, non-EUB, which has a sequence complementary to that of EUB338, was used (48).
As supplemental cations, CaCl2, MnSO4, or MgCl2 was added to the hybridization buffer at appropriate concentrations. After incubating at 46°C for 20–24 h, the buffer was replaced with a washing solution (375 mM NaCl, 0.01% SDS, 20 mM Tris-HCl at pH 7.2 and 5 mM EDTA), and the sample was incubated at 46°C for 20 min. Cells were then suspended in PBS and analyzed on the same day. Fluorescently hybridized cells were analyzed with the flow cytometer, Epics Altra (Beckman Coulter, Miami, FL, USA). The experimental conditions used for flow cytometry included deionized particle-free water (Milli-Q water filtered through a membrane with a pore size of 0.1 μm) as sheath fluid and an argon laser at a wavelength of 488 nm (power level, 15 mW). Side scattering with a 488-nm band pass filter was used for the discrimination of bacterial cells from other particles. The green fluorescence of FITC and Alexa488 was assessed using the PMT2 channel (515 to 525 nm). EXPO32 (Beckman Coulter) was used for data analyses. A total of 50,000 events were analyzed in each sample at a data rate of <3,000 events s−1. The threshold for positive fluorescence was defined as a fluorescence signal intensity of less than 5% of the total events observed in the negative control (i.e., without a fluorescent probe). Epifluorescence microscopic observations were performed after samples had been stained with 4′,6-diamidino-2-phenylindole (DAPI) using Axioskop2 (Carl Zeiss, Jena, Germany).
Results and Discussion
Effects of divalent cations on FISH for E. coli detection
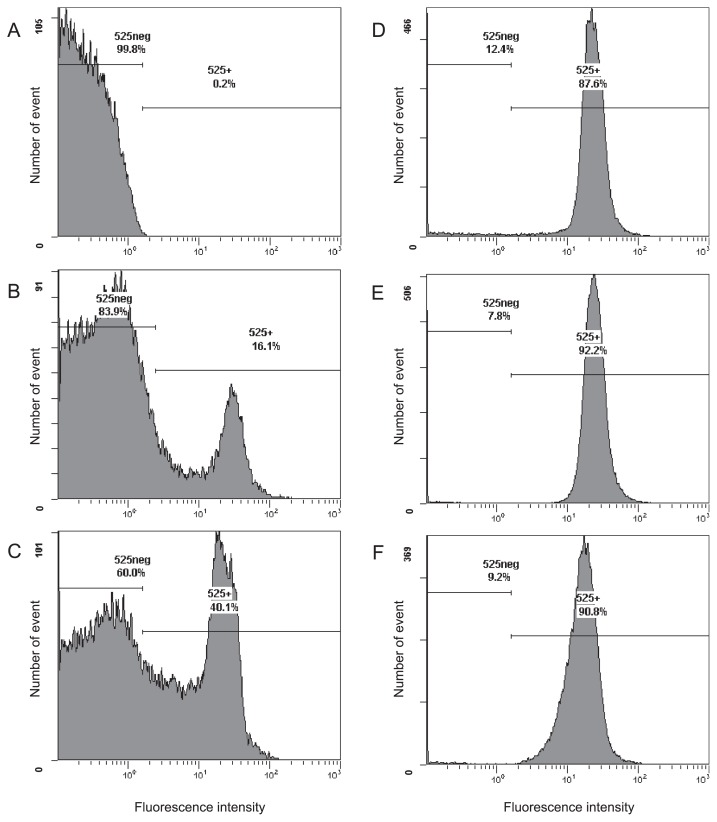
Following standard protocols, E. coli IAM 1264 cells harvested at the stationary phase of growth were fixed with paraformaldehyde, and hybridized with the fluorescent-labeled probe EUB338 for 20 h (2, 12). Flow cytometry results showed that 16.1% of all cells exhibited a positive signal, i.e., 83.9% of cells showed low fluorescence signal intensity (Fig. 1B; the positive signal percentage was 13.5±2.1% in three independent experiments using different batches of E. coli cultures), indistinguishable from cells incubated without the fluorescent probe (Fig. 1A). This low positive signal percentage may be caused by a decrease in the membrane permeability of cells in the stationary phase (21). The positive signal percentage was slightly increased by the addition of 0.5 mg L−1 of calcium chloride into the hybridization buffer (Fig. 1C) and markedly increased to more than 90% following the addition of 5 mg L−1 or more of calcium chloride (Fig. 1D, E, and F). A microscopic analysis of the hybridized cells also revealed that the ratio of fluorescently-labeled cells to total cells was clearly increased by the addition of calcium chloride (Fig. S1). As shown in Fig. 1D, E, and F, the fluorescence signal intensity from cells was not markedly affected by Ca2+. A similar effect of calcium ions was observed in E. coli IAM 1264 cells under multiple conditions; harvested in the exponential phase of growth, fixed with paraformaldehyde for a shorter time (4–6 h), and hybridized with EC1153, EC1482, EC839S, EC839L, and EC1235 probes targeting 16S rRNA (12, 46) (data not shown).
Fig. 1.
The distribution of the fluorescence signal intensity of cells hybridized in hybridization buffer supplemented with CaCl2. E. coli IAM 1264 cells were harvested at the stationary phase of growth and fixed with paraformaldehyde, as described in the Materials and Methods section. After in situ hybridization with the FITC-labeled probe, cells were analyzed with a flow cytometer. The x-axis and y-axis represent the fluorescence intensity (at 525 nm) and number of detected events, respectively. A, negative control (without probe); B, no divalent cation supplementation; C, 0.5 mg L−1 CaCl2; D, 5 mg L−1 CaCl2; E, 50 mg L−1 CaCl2; and F, 500 mg L−1 CaCl2. 525 neg, fraction of negative signals and 525+, fraction of positive signals.
The effects of other divalent cations were evaluated (Fig. S2). Although supplementation with 50 mg L−1 of manganese sulfate produced no effect (Fig. S2A), the addition of 500 mg L−1 of manganese sulfate to the hybridization buffer mildly increased the ratio of hybridized cells (Fig. S2B). In contrast, supplementation with magnesium chloride reduced the number of positive events (Fig. S2C and D).
Divalent cations have stabilizing effects on the hybridization of nucleotides. However, the low concentrations of divalent cations used in this study were not expected to increase hybridization efficiency and reduce probe specificity in FISH. It has been theoretically and experimentally demonstrated that less than 0.5 mM of divalent cations (i.e., below the effective concentrations of Ca2+ and Mn2+ in this study) has a negligible effect on the melting temperatures (Tm) of oligonucleotides in the presence of high concentrations of Na+ greater than 0.1 M (29, 38). We confirmed that the supplemental addition of calcium chloride did not cause false positive signals when the probe non-EUB was applied as a negative control (data not shown).
Hanahan reported that divalent cations had a marked effect on the genetic transformation efficiency of E. coli by plasmids, and Ca2+ and Mn2+ were found to be more effective than Mg2+ (15). Hanahan also suggested that divalent cations affected the phosphate moieties of DNA, lipid bilayers, and lipopolysaccharides, resulting in an increase in the permeability of E. coli cells to oligonucleotide probes (15).
The pretreatment of cells prior to FISH has been shown to increase probe permeability (3, 34). Ootsubo et al. treated fixed enteric bacterial cells including E. coli, with a non-ionic detergent before hybridization (33). In the present study, we found that positive signal percentages after ethanol:formalin and HCl treatments of E. coli IAM 1264 cells were 59.8% and 26.9%, respectively, and the lysozyme treatment was as effective (the number of positive signals out of all events, 93.8%) as the addition of Ca2+ to the hybridization buffer. However, the supplementation of Ca2+ to the hybridization buffer is a simple solution that does not alter cell morphology for the efficient detection of E. coli.
Effects of Ca2+ on FISH detection of a number of species of bacteria and archaea
The effects of Ca2+ on additional bacterial and archaeal species were evaluated. FISH and flow cytometry were completed on 27 species of bacteria and two species of archaea using the EUB338 probe for bacteria and the ARC915 probe for archaea. The ratio of positive events to the total number of events was assessed for each species using cells harvested at the late exponential phase of growth (Table 1). As observed in E. coli IAM 1264, the ratio of positive cells in E. coli IAM 12119T was low under standard conditions; however, the number of positive cells increased after the addition of calcium chloride to the hybridization buffer. Similarly, a low positive signal ratio was observed for Enterobacter spp., Klebsiella planticola IAM 14202T, Citrobacter freundii NBRC 1268T, and Salmonella enterica subsp. enterica serovar Typhimurium strain LT2 in the family Enterobacteriaceae in Gammaproteobacteria; however, the supplementation of calcium chloride to the hybridization buffer increased the number of positive cells. Other species in Enterobacteriaceae showed a relatively high percentage of positive cells under standard conditions, such as Proteus vulgaris JCM 1668T, Pantoea agglomerans IAM 12659, and K. pneumoniae IAM 14200T, and the effects of calcium chloride were still positive (Table 1). However, as shown in Table 1, calcium chloride did not markedly alter the FISH results obtained for bacterial and archaeal species other than those for Enterobacteriaceae.
Conclusions
The addition of Ca2+ to the hybridization buffer for FISH increased the number of positive signals from E. coli. This effect was independent of the probe sequence and appears to have been caused by an increase in membrane permeability for oligonucleotide probes. Ca2+ was similarly effective for bacteria in Enterobacteriaceae including S. enterica subsp. enterica serovar Typhimurium strain LT2. These Enterobacteriaceae cells may have a distinctive cell wall structure. Calcium chloride at a concentration of less than 50 mg L−1 did not produce negative effects in any of the microbes examined in this study (Table 1). As a simple application of our results, bottled hard water (>20 mg L−1 of Ca2+ and <100 mg L−1 of Mg2+) may be used to prepare hybridization solutions rather than the pure water typically used in laboratory settings. The protocol introduced in this study is useful for analyzing intestinal bacterial flora and detecting pathogenic bacteria including E. coli and Salmonella in medical and public health fields.
Supplementary material
Acknowledgements
We thank Tomoko Yamamoto and Akiko Takaya of Chiba University for their kind preparation of S. enterica cells, and Bernhard Maximilian Fuchs and Rudolf Amann for their helpful suggestions.
References
- 1.Amann R.I., Binder B.J., Olson R.J., Chisholm S.W., Devereux R., Stahl D.A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R.I., Krumholz L., Stahl D. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R., Fuchs B.M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Microbiol. 2008;6:339–348. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- 4.Attar N. FISHing in the oral microbiota. Nat Rev Microbiol. 2016;14:132–133. doi: 10.1038/nrmicro.2016.21. [DOI] [PubMed] [Google Scholar]
- 5.Barrero-Canosa J., Moraru C., Zeugner L., Fuchs B.M., Amann R. Direct-geneFISH: a simplified protocol for the simultaneous detection and quantification of genes and rRNA in microorganisms. Environ Microbiol. 2017;19:70–82. doi: 10.1111/1462-2920.13432. [DOI] [PubMed] [Google Scholar]
- 6.Beimfohr C., Krause A., Amann R., Ludwig W., Schleifer K.-H. In situ identification of lactococci, enterococci and streptococci. Syst Appl Microbiol. 1993;16:450–456. [Google Scholar]
- 7.Berg G., Grube M., Schloter M., Smalla K. Unraveling the plant microbiome: looking back and future perspective. Front Microbiol. 2014;5:148. doi: 10.3389/fmicb.2014.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun-Howland E.B., Danielsen S.A., Nierzwicki-Bauer S.A. Development of a rapid method for detecting bacterial cells in situ using 16S rRNA-targeted probes. Biotechniques. 1992;13:928–934. [PubMed] [Google Scholar]
- 9.Burggraf S., Mayer T., Amann R., Schadhauser S., Woese C.R., Stetter K.O. Identifying members of the domain Archaea with rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1994;60:3112–3119. doi: 10.1128/aem.60.9.3112-3119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr E.L., Eales K., Soddell J., Seviour R.J. Improved permeabilization protocols for fluorescence in situ hybridization (FISH) of mycolic-acid-containing bacteria found in foams. J Microbiol Methods. 2005;61:47–54. doi: 10.1016/j.mimet.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 11.Franks A.H., Harmsen H.J.M., Raangs G.C., Jansen G.J., Schut F., Welling G.W. Variations of bacterial populations in human feces measured by fluorescence in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs B.M., Wallner G., Beisker W., Schwippl I., Ludwig W., Amann R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 1998;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs B.M., Glockner O.F., Wulf J., Amann R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 2000;66:3603–3607. doi: 10.1128/aem.66.8.3603-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greuter D., Loy A., Horn M. probeBase--an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016. Nucleic Acids Res. 2016;44:D586–589. doi: 10.1093/nar/gkv1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 16.Harmsen H.J.M., Gibson G.R., Elfferich P., Raangs G.C., Wildeboer-Veloo A.C., Argaiz A., Roberfroid M.B., Welling G.W. Comparison of viable cell counts and fluorescence in situ hybridization using specific rRNA-based probes for the quantification of human fecal bacteria. FEMS Microbiol Lett. 2000;183:125–129. doi: 10.1111/j.1574-6968.2000.tb08945.x. [DOI] [PubMed] [Google Scholar]
- 17.Iino T., Tamaki H., Tamazawa S., Ueno Y., Ohkuma M., Suzuki K., Igarashi Y., Haruta S. Candidatus Methanogranum caenicola: a novel methanogen from the anaerobic digested sludge, and proposal of Methanomassiliicoccaceae fam. nov. and Methanomassiliicoccales ord. nov., for a methanogenic lineage of the class Thermoplasmata. Microbes Environ. 2013;28:244–250. doi: 10.1264/jsme2.ME12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaltenpoth M., Strupat K., Svatos A. Linking metabolite production to taxonomic identity in environmental samples by (MA) LDI-FISH. ISME J. 2016;10:527–531. doi: 10.1038/ismej.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawakami S., Hasegawa T., Imachi H., Yamaguchi T., Harada H., Ohashi A., Kubota K. Detection of single-copy functional genes in prokaryotic cells by two-pass TSA-FISH with polynucleotide probes. J Microbiol Methods. 2012;88:218–223. doi: 10.1016/j.mimet.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Kenzaka T., Utrarachkij F., Suthienkul O., Nasu M. Rapid monitoring of Escherichia coli in Southeast Asian urban canals by fluorescent-bacteriophage assay. J Health Sci. 2006;52:666–671. [Google Scholar]
- 21.Kolter R., Siegele D.A., Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 22.Kubota K. CARD-FISH for environmental microorganisms: Technical advancement and future applications. Microbes Environ. 2013;28:3–12. doi: 10.1264/jsme2.ME12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubota K., Morono Y., Ito M., Terada T., Itezono S., Harada H., Inagaki F. Gold-ISH: a nano-size gold particle-based phylogenetic identification compatible with NanoSIMS. Syst Appl Microbiol. 2014;37:261–266. doi: 10.1016/j.syapm.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence J., Korber D., Neu T. Analytical Imaging and Microscopy Techniques. In: Hurst C., Crawford R., Garland J., Lipson D., Mills A., Stetzenbach L., editors. Manual of Environmental Microbiology. 3rd ed. ASM Press; Washington DC: 2007. pp. 40–68. [Google Scholar]
- 25.Manz W., Amann R., Ludwig W., Wagner M. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 26.Manz W., Szewzyk U., Ericsson P., Amann R., Schleifer K.H., Stenström T.A. In situ identification of bacteria in drinking water and adjoining biofilms by hybridization with 16S and 23S rRNA-directed fluorescent oligonucleotide probes. Appl Environ Microbiol. 1993;59:2293–2298. doi: 10.1128/aem.59.7.2293-2298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musat N., Stryhanyuk H., Bombach P., Adrian L., Audinot J.N., Richnow H.H. The effect of FISH and CARD-FISH on the isotopic composition of 13C- and 15N-labeled Pseudomonas putida cells measured by nanoSIMS. Syst Appl Microbiol. 2014;37:267–276. doi: 10.1016/j.syapm.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura K., Terada T., Sekiguchi Y., Shinzato N., Meng X.Y., Enoki M., Kamagata Y. Application of pseudomurein endoisopeptidase to fluorescence in situ hybridization of methanogens within the family Methanobacteriaceae. Appl Environ Microbiol. 2006;72:6907–6913. doi: 10.1128/AEM.01499-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakano S., Fujimoto M., Hara N., Sugimoto N. Nucleic acid duplex stability: influence of base composition on cation effects. Nucleic Acids Res. 1999;27:2957–2965. doi: 10.1093/nar/27.14.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen P.H., Daims H., Lemmer H. FISH Handbook for Biological Wastewater Treatment: Identification and Quantification of Microorganisms in Activated Sludge and Biofilms by FISH. IWA Publishing; New York: 2009. [Google Scholar]
- 31.Noguera D.R. Could in situ DNA-hybridization chain reaction enable simple and effective detection of identify and function in whole cell hybridization? Environ Microbiol. 2015;17:2559–2561. doi: 10.1111/1462-2920.12969. [DOI] [PubMed] [Google Scholar]
- 32.Okten H.E., Yilmaz L.S., Noguera D.R. Exploring the in situ accessibility of small subunit ribosomal RNA of members of the domains Bacteria and Eukarya to oligonucleotide probes. Syst Appl Microbiol. 2012;35:485–495. doi: 10.1016/j.syapm.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Ootsubo M., Shimizu T., Tanaka R., Sawabe T., Tajima K., Yoshimizu M., Ezura Y., Ezaki T., Oyaizu H. Oligonucleotide probe for detecting Enterobacteriaceae by in situ hybridization. J Appl Microbiol. 2002;93:60–68. doi: 10.1046/j.1365-2672.2002.01668.x. [DOI] [PubMed] [Google Scholar]
- 34.Pernthaler A., Pernthaler J., Amann R. Sensitive multicolor in situ hybridization for the identification of environmental microorganisms. In: Kowlchuk G., de Bruijn F.J., Head I.M., Akkermans A.D., van Elsas J.D., editors. Molecular Microbial Ecology Manual. Kluwer Academic; Dordrecht: 2004. pp. 711–726. [Google Scholar]
- 35.Raskin L., Rittmann B.E., Stahl D.A. Competition and coexistence of sulfate-reducing and methanogenic populations in anaerobic biofilms. Appl Environ Microbiol. 1996;62:3847–3857. doi: 10.1128/aem.62.10.3847-3857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohde A., Hammeri J.A., Appel B., Dieckmann R., Al Dahouk S. FISHing for bacteria in food—A promising tool for the reliable detection of pathogenic bacteria? Food Microbiol. 2015;46:395–407. doi: 10.1016/j.fm.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt H., Eickhorst T., Mussmann M. Gold-FISH: a new approach for the in situ detection of single microbial cells combining fluorescence and scanning electron microscopy. Syst Appl Microbiol. 2012;35:518–525. doi: 10.1016/j.syapm.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Tan Z.-J., Chen S.-J. RNA helix stability in mixed Na+/Mg2+ solution. Biophysic J. 2007;92:3615–3632. doi: 10.1529/biophysj.106.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner M. Single-cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Annu Rev Microbiol. 2009;63:411–429. doi: 10.1146/annurev.micro.091208.073233. [DOI] [PubMed] [Google Scholar]
- 40.Wallner G., Amann R., Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 41.Wendeberg A. Fluorescence in situ hybridization for the identification of environmental microbes. Cold Spring Harb Protoc. 2010;1 doi: 10.1101/pdb.prot5366. pdb.prot5366. [DOI] [PubMed] [Google Scholar]
- 42.Wright E.S., Yilmaz L.S., Corcoran A.M., Ökten H.E., Noguera D.R. Automated design of probes for rRNA-targeted fluorescence in situ hybridization reveals the advantages of using dual probes for accurate identification. Appl Environ Microbiol. 2014;80:5124–5133. doi: 10.1128/AEM.01685-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi T., Kawakami S., Hatamoto M., Imachi H., Takahashi M., Araki N., Yamaguchi T., Kubota K. In situ DNA-hybridization chain reaction (HCR): a facilitated in situ HCR system for the detection of environmental microorganisms. Environ Microbiol. 2015;17:2532–2541. doi: 10.1111/1462-2920.12745. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi T., Fuchs B.M., Amann R., Kawakami S., Kubota K., Hatamoto M., Yamaguchi T. Rapid and sensitive identification of marine bacteria by an improved in situ DNA hybridization chain reaction (quickHCR-FISH) Syst Appl Microbiol. 2015;38:400–405. doi: 10.1016/j.syapm.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 45.Yilmaz L.S., Noguera D.R. Mechanistic approach to the problem of hybridization efficiency in fluorescent in situ hybridization. Appl Environ Microbiol. 2004;70:7126–7139. doi: 10.1128/AEM.70.12.7126-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yilmaz L., Okten H.E., Noguera D.R. Making all parts of the 16S rRNA of Escherichia coli accessible in situ to single DNA oligonucleotides. Appl Environ Microbiol. 2006;72:733–744. doi: 10.1128/AEM.72.1.733-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yilmaz L.S., Parnerkar S., Noguera D.R. mathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl Environ Microbiol. 2011;77:1118–1122. doi: 10.1128/AEM.01733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zarda B., Amann R., Wallner G., Schleifer K.H. Identification of single bacterial cells using digoxigenin-labelled, rRNA-targeted oligonucleotides. J Gen Microbiol. 1991;137:2823–2830. doi: 10.1099/00221287-137-12-2823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.