Abstract
The relationship among (i) the local incidence of cholera, (ii) the prevalence in the aquatic environment of Vibrio cholerae, and (iii) bacterial viruses that attack potentially virulent O1 and O139 serogroup strains of this organism (cholera phages) was studied in Dhaka, Bangladesh. Over nearly a 3-year period, we found that significantly more environmental water samples contained either a phage or a phage-susceptible V. cholerae strain than both (P < 0.00001). The number of cholera patients varied seasonally during this period and frequently coincided with the presence of pathogenic V. cholerae strains in water samples that otherwise lacked detectable cholera phages. Interepidemic periods were characterized by water samples containing cholera phages but no viable bacteria. Our data support the conclusion that cholera phages can influence cholera seasonality and may also play a role in emergence of new V. cholerae pandemic serogroups or clones.
Keywords: bacteriophage, seasonality, epidemiology, emergence, lysogeny
Epidemics of cholera caused by toxigenic Vibrio cholerae belonging to the O1 or O139 serogroups are a major public health problem in many developing countries of Asia, Africa, and Latin America (1). Cholera epidemics occur with seasonal regularity in the Ganges delta region of Bangladesh and India. Epidemics usually occur twice during a year, with the highest number of cases just after the monsoon during September to December. A somewhat smaller peak of cholera cases also is observed during the spring, between March and May. Although V. cholerae is a human pathogen, these bacteria constitute part of the normal aquatic flora in estuarine environments, and water is clearly a vehicle for transmission of V. cholerae. Although the seasonality of cholera in Bangladesh and elsewhere has been temporally associated with numerous physical and biological parameters (2), these associations do not directly cause epidemics, nor do they end them. More than a century of public health experience has shown that toxigenic O1 and O139 V. cholerae cells cause cholera epidemics and that the elimination of these cells from drinking water ends cholera epidemics. The parameters that directly modulate the level of viable cells belonging to the pathogenic clones of V. cholerae O1 and O139 in the Ganges delta aquatic environment remain unknown. Furthermore, the fact that pathogenic strains of V. cholerae are clonally distinct from environmental, nonpathogenic V. cholerae strains (1) undermines proposed mechanisms of seasonality and pandemic spread that are based on data from studies measuring the abundance of all Vibrio species in the aquatic environment (2).
Bacterial viruses (phages) are known to play a critical role in the evolution of pathogenic bacterial species, and V. cholerae in particular. For example, cholera toxin genes are transferred to nontoxigenic strains by means of a lysogenic filamentous phage, CTXΦ (3). Here we show that the presence of bacterial viruses acting on V. cholerae O1 or O139 (cholera phages or vibriophages) inversely correlates with the occurrence of viable V. cholerae in the aquatic environment and the number of locally reported cholera cases. We also demonstrate that some environmental V. cholerae strains of both epidemic and nonepidemic serogroups carry lysogenic phages and produce phage particles that kill epidemic strains. We identified at least one common O1 phage that is able to use several V. cholerae non-O1/non-O139 strains as alternative hosts. Such alternative hosts and lysogenic environmental V. cholerae strains may potentially act as cofactors in promoting cholera phage “blooms” within aquatic environments and thus negatively influence transmission of phage-sensitive, pathogenic V. cholerae strains by aquatic vehicles.
Materials and Methods
Detection and Isolation of Phages. Environmental water samples were obtained in sterile containers, and initial processing of the samples to detect vibriophages and to culture for V. cholerae was done within 3 h of collection. For detection of phages, logarithmic-phase cells (500 μl) of each of 10 host bacterial strains in nutrient broth (Difco) were mixed with 3.5-ml aliquots of soft agar (nutrient broth containing 0.8% Bactoagar, Difco), and the mixtures were overlaid on nutrient agar plates. The 10 strains used for screening vibriophages were G-3669 (El Tor), G-7555 (El Tor), P-27457 (El Tor), AM-33363 (El Tor), AP-13550 (El Tor), AT-2352515 (El Tor), AL-30457 (El Tor), AI-885 (O139), AI 1852 (O139), and AL-11089 (O139). Aliquots of water (10–50 μl), which were prefiltered through 0.22-μm-pore filters (Millipore) to make them bacteria-free, were inoculated on the plates, and the plates were incubated for 16 h at 37°C. Aliquots of water samples mixed with serial dilutions of a control phage strain (10–104 particles per ml) were used as positive controls in all phage assays. A sample was scored positive for phages when a plaque was observed on the bacterial lawn in the plates. Plaques were counted to estimate the concentration of phage particles in the water. Phages from representative plaques were further purified and used for the production of high-titer stocks. High-titer phage stocks were used to prepare phage nucleic acids and to test for host range, morphology by electron microscopy, and DNA restriction patterns and hybridization (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
Culture of Environmental Samples. Water samples were analyzed for the presence of V. cholerae by methods described in ref. 4. Briefly, an aliquot (35 ml) of each water sample was centrifuged at 4,500 × g, and the pellet was resuspended in 3.5 ml of 10 mM PBS (pH 7.4). The suspension was vortexed to dislodge any bacteria adhering to solid particles and then centrifuged at low speed (1,000 × g) to precipitate particulate matter. One milliliter of this suspension was added to 10 ml of alkaline peptone water (APW) [peptone 1% (wt/vol)/NaCl 1% (wt/vol), pH 8.5] contained in 20-ml screw-cap glass tubes and incubated at 37°C with shaking for 6–8 h. Dilutions of this APW culture were streaked on taurocholate tellurite gelatin agar plates (5). Suspected Vibrio colonies were picked and subjected to biochemical and serological tests to identify V. cholerae belonging to the O1 and O139 serogroups.
Data Analysis. Frequency of isolation of vibriophages and V. cholerae from different water samples was summarized into tables for group comparisons. Statistical analyses were performed by using epi info 6.0 (USD, Stone Mountain, GA). The significance of difference in proportions was evaluated by the χ2 test.
Results
Influence of Environmental Vibriophages on Cholera. To study the prevalence of vibriophages and V. cholerae in the environment, we systematically analyzed water samples collected from two major rivers and a lake in Dhaka, the capital of Bangladesh, where cholera epidemics occur every year. Sampling was done monthly during a period between January 2001 and November 2003. A panel of 10 different V. cholerae clinical isolates from recent and previous epidemic outbreaks of cholera was used as potential indicator strains to detect the presence of vibriophages in water. All water samples also were analyzed for the presence of V. cholerae by standard enrichment culture techniques and for the presence of O1 and O139 serogroup strains by serological methods.
We found that the majority of water samples showed an inverse correlation between the presence of vibriophages capable of lysing a given serogroup of V. cholerae and the presence of a strain of that same serogroup (Table 1). Of a total of 221 water samples analyzed from January 2001 to November 2003, 114 samples (51.5%) contained either a vibriophage or an epidemic V. cholerae, whereas only 15 samples (7.2%) contained both. Of these 15 samples, 10 samples contained V. cholerae that were resistant to the phage present in the same sample. Overall, the number of water samples containing both a vibriophage and an epidemic V. cholerae strain susceptible to coisolated phages was significantly less than that predicted by chance alone based on the frequency of samples that contained either vibriophages or phage-susceptible V. cholerae (P < 0.00001). Thus, if a phage lytic to epidemic V. cholerae O1 was present in a water sample, the water was free of phage-susceptible V. cholerae O1 strains (Table 3, which is published as supporting information on the PNAS web site). Conversely, a majority of those water samples that contained either V. cholerae O1 or O139 did not contain a detectable phage that lysed the corresponding serogroup strain. Many samples were obtained that carried an O1 strain but only O139-specific phages (e.g., Gulshan Lake in July 2001, August 2002, January 2003, and March 2003 and Buriganga River in October 2001 and October 2003). Similarly, other samples contained an O139 strain but only O1-specific phages (e.g., Gulshan Lake in May 2002). If a V. cholerae strain of a specific serogroup was present in a water sample together with a phage that recognized the same serogroup, in most of these cases (73%) the actual strain present was resistant to the coincidentally isolated phage. To address the possibility that V. cholerae present in the water sample were simply killed during processing of water samples by phages present in samples, we performed control experiments on “spiked” water samples containing known concentrations of V. cholerae O1 and O1 phages (Table 4, which is published as supporting information on the PNAS web site). These experiments supported the conclusions noted above in that spiking water samples with phages only modestly reduced the titers of viable V. cholerae and then only when phages were added at levels >100 plaque-forming units/ml.
Table 1. Presence of specific vibriophages and susceptible V. cholerae strains belonging to the O1 and O139 serogroups in two major rivers and a lake in Dhaka, Bangladesh, between January 2001 and November 2003.
| No. of water samples (%)
|
||||||
|---|---|---|---|---|---|---|
| Source of water | No. of water samples analyzed* (A) | Phage-positive and V. cholerae-negative (B) | Phage-negative and V. cholerae-positive (C) | Samples positive for either phage or V. cholerae (B + C) | Both phage- and susceptible V. cholerae-positive (D) | P value† |
| Gulshan Lake | 63 | 26 (41.2) | 19 (30.1) | 45 (71.4) | 1 (1.5) | 0.0001 |
| Buriganga River | 62 | 30 (48.3) | 12 (19.3) | 42 (67.7) | 2 (3.2) | 0.004 |
| Turag River | 96 | 31 (32.2) | 18 (18.7) | 49 (51.0) | 2 (2.0) | 0.02 |
| Total samples | 221 | 87 (39.3) | 49 (22.1) | 136 (61.5) | 5 (2.2) | <0.00001 |
Samples were collected from two different points along each of the Gulshan Lake and Buriganga River and from three different points along the Turag River in Dhaka, Bangladesh. Samples were scored as phage-positive if they contained phages that plaqued on O1 or O139 V. cholerae indicator strains. Samples were scored as V. cholerae-positive if they contained O1 or O139 strains. Fifteen samples contained both phages and V. cholerae, but 10 of these strains were resistant to the coisolated phage and, accordingly, were eliminated from column D in the analysis.
Significance of D being lower numerically than expected by chance alone given A number of samples scored as B, C, or D. Statistical analyses were performed by using epi info 6.0. The significance of difference in proportions was evaluated by the χ2 test.
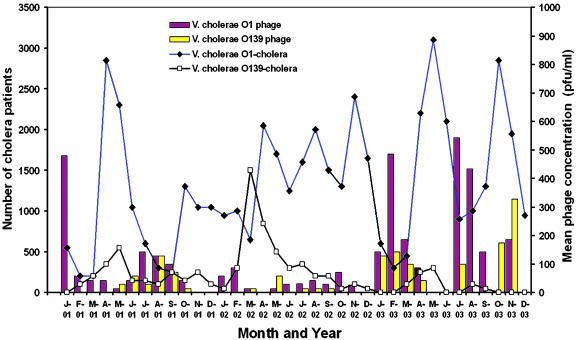
Our finding of an inverse association between the presence of virulent phages and phage-susceptible V. cholerae O1 or O139 in environmental water led us to examine whether incidence of cholera due to these two serogroups could also be correlated with these findings. We investigated this possibility by using a diarrhea surveillance program of the International Centre for Diarrhoeal Disease Research in Dhaka, Bangladesh (ICDDR, B) (6). Diarrhea patients in and around Dhaka city are usually treated in the Dhaka diarrhea hospital of ICDDR, B. In the Dhaka hospital, stool samples from 2% of all diarrhea patients presenting for treatment are analyzed for the presence of all known diarrheal pathogens, including V. cholerae. Monthly numbers of cholera patients extrapolated from this 2% surveillance system were compared with the monthly phage isolation data during the same period. As shown in Fig. 1, cholera epidemics due to either the O1 or O139 serogroup strains generally coincided with a marked reduction in the prevalence of vibriophages specific for the serogroup dominating a particular epidemic wave. For example, the early upward slope of epidemic curves most often corresponded to periods of low vibriophage isolation (e.g., see the O139-dominated epidemic occurring from February to April of 2002 and the corresponding bars for isolation of O139-specific bacteriophages in Fig. 1). This inverse relationship between the prevalence of a serogroup-specific vibriophage and the incidence of cholera due to that serogroup of V. cholerae was seen on both sides of the epidemic wave. Temporal blooms of serogroup-specific phages usually coincided with the downward slope of epidemic curves dominated by the corresponding serogroup of V. cholerae (e.g., see May–September 2001, January–April 2003, and July–August 2003 in Fig. 1). Additionally, on one occasion these dynamic changes in serogroup-specific cholera incidence occurred out of temporal register for O1 and O139. From January to March of 2002, when the incidence of O1 cases was falling, the incidence of O139 cases was dramatically increasing (Fig. 1). Thus, environmental factors (2) that might be conducive or inhibitory to cholera epidemics (temperature, salinity, pH, UV light, aquatic carbon, copepod, or plankton levels) either occasionally affect the two serogroups of cholera differently, or, alternatively, some other serogroup-specific parameter modulates temporal incidence of the disease. If the presence of phages in the aquatic environment has an adverse effect on cholera transmission, then serogroup-specific phage predation provides a feasible parameter that might modulate the occasional temporal discordance between epidemics caused by the two serogroups O1 and O139.
Fig. 1.
Mean concentration of lytic vibriophages in the aquatic environment of Dhaka, Bangladesh, and the estimated number of cholera cases reporting to the International Centre for Diarrhoeal Disease Research hospital in Dhaka from 2001 to 2003. Number of cholera cases is extrapolated from a 2% surveillance sample of all patients presenting for treatment. pfu, plaque-forming units.
Diversity of Phages and Their V. cholerae Hosts. We characterized vibriophages isolated from the environmental surveillance for their genetic and phenotypic diversity (Table 2) and observed their morphology by electron microscopy (Fig. 2 and Table 5, which is published as supporting information on the PNAS web site). During this period, we observed only six phage types, which were designated JSF1, JSF2, JSF3, JSF4, JSF5, and JSF6 based on the restriction patterns of their genomes. Whereas JSF1, JSF4, and JSF5 genomes cross-hybridized, those of JSF2, JSF3, and JSF6 were unique and did not cross-hybridize with any of the other phages. Thus, the phages represented four genetically unrelated groups. Phage JSF3 was specific for V. cholerae O139 strains, whereas all other phages were specific for V. cholerae O1 strains. However, JSF6 could use, in addition to O1 strains, a number of V. cholerae non-O1/non-O139 strains as alternative hosts; JSF6 phages produced from these non-O1/non-O139 host strains were virulent against V. cholerae O1 strains (Fig. 3).
Table 2. Characteristics of vibriophages isolated from surface water samples in Bangladesh between January 2001 and November 2003.
| Phage designation | Host specificity | Plaque type | Isolation of lysogens* | Phage production by lysogens* | Crossimmunity | DNA homology |
|---|---|---|---|---|---|---|
| JSF1 | V. cholerae O1 | Clear | - | - | JSF4, JSF5 | JSF4, JSF5 |
| JSF2 | V. cholerae O1 | Turbid | + | + | None | None |
| JSF3 | V. cholerae O139 | Clear | +† | +† | None | None |
| JSF4 | V. cholerae O1 | Clear | +† | +† | JSF1, JSF5 | JSF1, JSF5 |
| JSF5 | V. cholerae O1 | Clear | - | - | JSF1, JSF4 | JSF1, JSF4 |
| JSF6 | V. cholerae O1 and some non-O1 | Clear | - | - | None | None |
Minus sign indicates that the phage designated was negative for the property being scored; plus sign indicates that the phage was positive for the property being scored.
JSF3 and JSF4 phages appear to be primarily lytic but can occasionally persistently infect a host strain. Such persistently infected cells shed virus and display viral immunity but do not show typical chromosomal junction fragments consistent with stable integration of the viral genome into the bacterial chromosome.
Fig. 2.
Electron micrograph of vibriophages isolated from environmental waters in Bangladesh. (A–F) Morphology of phages JSF1–JSF6. (Scale bar: 50 nm.)
Fig. 3.
Plaque morphology of different vibriophages on lawns of susceptible V. cholerae strains. Description of phages and host strains are as follows: JSF1 phage plated on a V. cholerae O1 clinical strain isolated from the recent epidemic (A), JSF2 phage isolated from water (B) and from a lysogenic non-O1/non-O139 strain (G) plated on an epidemic V. cholerae O1 strain, JSF3 phage plated on a V. cholerae O139 clinical strain (C), JSF6 phage plated on a V. cholerae O1 strain (F) and an environmental non-O1/non-O139 strain (H), JSF6 phage produced from an environmental non-O1/non-O139 strain and plated on a clinical V. cholerae O1 strain (E), and JSF4 phage plated on a V. cholerae O1 strain (D).
Production of Phages by Lysogenic V. cholerae. All phages except JSF2 produced clear plaques, suggesting that these phages were lytic and highly virulent for V. cholerae. JSF2 was a temperate phage related to the previously described κ group of phages (7, 8), as revealed by electron microscopy (hexagonal isometric head of 52.3 ± 1.7 nm and tail of 89.7 ± 2.2 nm × 15.9 ± 1.2 nm, with cross-striations), and produced characteristic turbid plaques on susceptible V. cholerae O1 strains. Temperate vibriophages exemplified by the κ group of vibriophages can grow lytically (killing the host cell and producing numerous progeny phage particles) or form lysogenic V. cholerae cells (9, 10). Lysogenic cells are immune to κ phages but carry dormant prophages that can be induced to replicate lytically through signals that typically cause DNA damage (e.g., UV irradiation). Furthermore, one highly conserved gene (glo) encoded by κ vibriophages has been shown to provide resistance to superinfection by other κ phages through the process of surface exclusion (11). Thus, we reasoned that lysogenic bacterial cells might on occasion shed viable phage particles to which they as a population were clearly immune. These phages in turn could amplify quickly if the aquatic environment contained sufficient susceptible V. cholerae cells, thus interrupting an ongoing cholera epidemic by reducing the concentration of viable V. cholerae cells in water and their transmission to new victims.
To evaluate this hypothesis, we analyzed all V. cholerae strains isolated from the environment during the study period for the presence of lysogenic phages by DNA hybridization using probes derived from the genome of each of the six phage types isolated. Several strains belonging to the O1, O139, or non-O1/non-O139 serogroups were found to carry lysogenic phages related to JSF2, JSF3, or JSF4. Probing of Southern blots with labeled phage DNA revealed patterns of hybridizing fragments derived from these strains that matched those derived from the native phage genomes (see Fig. 5, which is published as supporting information on the PNAS web site). Probe-positive V. cholerae strains were further tested for phage production in their culture supernatant. These strains spontaneously produced significant titers of the respective phages (between 102 and 104 particles per ml) when freshly isolated. Furthermore, when treated with the DNA damaging agent mitomycin C, most strains produced even higher levels of phage particles. The phages produced by these lysogens were active against V. cholerae O1 or O139 strains. However, consistent with their lysogenic nature, the environmental strains of V. cholerae that produced phages after induction were resistant to superinfection by the phage they elaborated.
Territorialism and V. cholerae Evolution. Together, these data suggest a model for how environmental phages might influence the occurrence of epidemics, cholera seasonality, and changes in serogroup prevalence (Fig. 4). The inverse correlation between the environmental concentration of vibriophages and the presence of susceptible V. cholerae strains in water suggests that epidemics would most likely begin in periods of low phage concentration (after floods and the monsoon season). Predictably, this inverse correlation would not hold for V. cholerae strains that were either phage-resistant (due to serogroup type) or lysogenic for prevalent phages and thus immune to lysis by these phages. However, such lysogenic cells might, after exposure to DNA damaging agents such as sunlight (12), serve as a source of phages that could attack nonlysogenic cells. If both lysogenic strains and nonlysogenic strains competed for a similar environmental niche, this competition would lead to a selective pressure for strains to become phage-resistant through lysogeny while simultaneously providing the lysogenized bacterium with a “vibriocide” (i.e., phage) that could destroy nonlysogenic competing strains. Jouravleva et al. (13) speculated that such territorialism might be in play, given the differences in the lysogeny and sensitivity of clinical isolates to V. cholerae filamentous vibriophage 493. Our data showing that disease incidence tracks inversely with the prevalence of lytic phages in the environment also suggests that V. cholerae cells lysogenic for phages might modulate epidemics through phage-related territorialism.
Fig. 4.
Schematic model for the influence of phage on cholera seasonality and serogroup emergence. Seasonal cholera epidemics (increased cases over time) occur in waves, with different serogroups (e.g., O1 or O139) dominating. Corresponding conditions that exist in the aquatic environment are diagramed in the circles shown. The absence of yellow phage specific for yellow oval cells provides an opportunity for that serogroup to begin the seasonal epidemic and transmit efficiently. However, yellow phages eventually amplify in the environment and attack this serogroup, ending that epidemic. A different serogroup (red oval cells) is resistant to the yellow phage. In the case shown, red cells actually carry a prophage (yellow intracellular circle) and thus shed yellow phage. A second epidemic wave due to the red serogroup follows and runs its course until red phages bloom and prevent environmental transmission of this serogroup. The interepidemic period is dominated by sporadic disease due to other serogroups (blue cells) resistant to both phages. These strains usually lack typical virulence factors but are more environmentally adapted than virulent strains. However, blue cells may harbor prophages that kill virulent serogroups and may acquire virulence determinants by horizontal gene transfer. Thus, blue cells may eventually emerge to become a new epidemic serogroup.
Other than CTXΦ, the most common phage released by toxigenic El Tor O1 strains are κ-like (7,10). The κ-like JSF2 phage was frequently isolated from water samples in this study, together with a V. cholerae O1 strain that was lysogenized with this phage (Table 3). In the case of κ phage lysogens, territorialism would be a highly successful strategy if κ lysogens could also become resistant to other O1 phages. Indeed, several non-O1 serogroups of V. cholerae (among them, O139 and O141) curiously carry κ prophages and produce particles that lyse only O1 strains (8, 14). It was previously hypothesized that these non-O1 strains were derived from precursor O1 strains that happened to be lysogenized by κ before their conversion to a new non-O1 serogroup. In light of the data presented in this report, we propose that these precursor O1 κ lysogens might have switched to non-O1 serogroups to escape other non-κ, O1-specific phages amplified during O1 epidemics (e.g., JSF1, JSF4, JSF5, and JSF6). This dynamic could have led to the emergence of globally distributed pathogenic strains of both the O139 and O141 serogroups as well as numerous other pathogenic non-O1/non-O139 strains that carry prophages with O1 specificity (Fig. 4). Finally, phages JSF1, JSF4, and JSF5 lysed 9 of 10 strains of the classical biotype examined. It is well known that the seventh pandemic O1 clone of the El Tor biotype globally displaced O1 strains of the classical biotype within a decade of its introduction into South Asia (1). The sensitivity of O1 classical strains to phages currently prevalent in the Bangladesh aquatic environment suggests that these phage types may have played a role in the elimination of the previously highly successful classical biotype.
Similarly, phage JSF6 grows on several non-O1/non-O139 hosts but also lyses O1 strains. It is apparent that the concentration of permissive hosts for JSF6 in the aquatic environment might lead to higher concentrations of JSF6 particles and render the water unsuitable for propagation of an O1 epidemic. Thus, environmental non-O1/non-O139 V. cholerae strains could influence the epidemiology of O1 and O139 strains by seeding the environment with phages that kill O1 and O139 strains (Fig. 4). The low incidence of cholera in the interepidemic period could be explained by such an ecological role for non-O1/non-O139 environmental V. cholerae strains as lytic or lysogenic hosts for phages that also kill O1 and O139 strains.
It has been previously reported that the abundance of other aquatically adapted bacterial genera, such as Prochlorococcus and Synechococcus, was inversely correlated with phage abundance in open ocean waters (15). This variation of abundance of phage and host species was thought to be reflective of phage sensitivity of host cells, production of phages by lysogenic cells, and the host range of different lytic and temperate phages (15). Earlier studies have documented seasonal variation in lysogeny for Synechococcus (16) and shown that this variation might be influenced by nutritional stresses such as phosphate limitation (17). Thus, Vibrio, as an esturine-adapted genus, might also participate in dynamic population variations based in part on phage predation and lysogeny cycles that could be influenced by physical and nutritional aquatic parameters that vary seasonally. The prevalence of pathogenic V. cholerae clones in aquatic environments (and thus the potential for water-borne epidemics of cholera) could, in turn, be influenced by complex seasonal cycles that affect lysogeny of nonpathogenic, environmental vibrios that serve as alternate cholera phage hosts as well as by blooms of phages driven by contamination of the environment by V. cholerae shed by cholera victims.
Conclusions and Implications. About one and a half centuries ago, the British physician John Snow epidemiologically linked Thames River water from the Broad Street pump to cholera cases in London (18). Since then, numerous studies have shown that V. cholerae belong to a group of organisms whose major habitats are aquatic ecosystems, and the role of water sources in the spread of cholera has been well documented (19–21). Interactions of aquatic microorganisms and V. cholerae have also been documented, leading to the suggestion that environmental concentration of Vibrio species might increase in response to zooplankton or phytoplankton blooms driven by global or local aquatic conditions (2, 22, 23). Our data suggest that phages in the environment are an additional factor that causes the V. cholerae population in aquatic environments to wax or wane in a recurrent cycle. We propose that phages play their predominant role in ending cholera epidemics. But their removal by conditions such as severe flooding might also contribute to making water more conducive to V. cholerae human-to-human transfer. Thus, the absence of phages in the aquatic environment might promote cholera epidemics, particularly when V. cholerae is introduced for the first time. Such a concept can account for the explosive nature of cholera epidemics that occurred when this organism was recently introduced into previously cholera-free areas, such as Latin America and Africa (1). Finally, the phage cycle described here might explain much of the enigmatic seasonality of endemic cholera, as well. In countries where cholera exhibits a seasonal behavior characterized by fluctuations in incidence, environmental surveillance for vibriophages could be useful in tracking outbreaks (24), predicting epidemics, and anticipating emergence of new serogroups. Vibriophages might even be employed as biological control agents in cholera endemic areas.
Supplementary Material
Acknowledgments
This work was funded by Ellison Medical Foundation Grant No. ID-T-0007-01 under a subagreement between Harvard Medical School and the International Centre for Diarrhoeal Disease Research in Dhaka, Bangladesh (ICDDR,B), and by National Institutes of Health Grant GM-068851. ICDDR,B is supported by countries and agencies that share its concern for the health problems of developing countries.
References
- 1.Faruque, S. M., Albert, M. J. & Mekalanos, J. J. (1998) Microbiol. Mol. Biol. Rev. 62, 1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipp, E. K., Huq, A. & Colwell, R. R. (2002) Clin. Microbiol. Rev. 15, 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waldor, M. K. & Mekalanos, J. J. (1996) Science 272, 1910–1914. [DOI] [PubMed] [Google Scholar]
- 4.Faruque, S. M., Chowdhury, N., Kamruzzaman, M., Dziejman, M., Rahman, M. H., Sack, D. A., Nair, G. B. & Mekalanos, J. J. (2004) Proc. Natl. Acad. Sci. USA 101, 2123–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monsur, K. A. (1961) Trans. R. Soc. Trop. Med. Hyg. 55, 440–442. [DOI] [PubMed] [Google Scholar]
- 6.Greenough, W. B., III (2004) J. Clin. Invest. 113, 334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeya, K. (1974) in Cholera, eds. Barua, D. & Burrows, W. (Saunders, Philadelphia), pp. 74–83.
- 8.Reidl, J. & Mekalanos, J. J. (1995) Mol. Microbiol. 18, 685–701. [DOI] [PubMed] [Google Scholar]
- 9.Brige, E. A. (2000) in Bacterial and Bacteriophage Genetics (Springer, New York), 4th Ed., pp. 253–292.
- 10.Guidolin, A. & Manning, P. A. (1987) Micriobiol. Rev. 51, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nesper, J., Blass, J., Fountoulakis, M. & Reidl, J. (1999) J. Bacteriol. 181, 2902–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faruque, S. M., Asadulghani, Rahman, M. M., Waldor, M. K. & Sack, D. A. (2000) Infect. Immun. 68, 4795–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jouravleva, E. A., McDonald, G. A., Gaon, C. F., Boesman-Finkelstein, M. & Finkelstein, R. A. (1998) Microbiology 144, 315–324. [DOI] [PubMed] [Google Scholar]
- 14.Kapfhammer, D., Blass, J., Evers, S. & Reidl, J. (2002) J. Bacteriol. 184, 6592–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan, M. B., Waterbury, J. B. & Chisholm, S. W. (2003) Nature 424, 1047–1051. [DOI] [PubMed] [Google Scholar]
- 16.McDaniel, L., Houchin, L., Williamson, S. J. & Paul, J. H. (2002) Nature 415, 496. [DOI] [PubMed] [Google Scholar]
- 17.Williamson, S. J., Houchin, L. A., McDaniel, L. & Paul, J. H. (2002) Appl. Environ. Microbiol. 68, 4307–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snow, J. (1855) in On the Mode of Communication of Cholera (J. Churchill, London), 2nd Ed., pp. 1–162.
- 19.Glass, R. I. & Black, R. E. (1992) in Cholera, eds. Barua, D. & Greenough, W. B., III (Plenum, New York), pp. 129–154.
- 20.Colwell, R. R. & Huq, A. (1994) in Vibrio cholerae and Cholera: Molecular to Global Perspectives, eds. Wachsmuth, I. K., Blake, P. A. & Olsvik, O. (Am. Soc. Microbiol., Washington, DC), pp. 117–133.
- 21.Islam, M. S., Rahim, Z., Alam, M. J., Begum, S., Moniruzzaman, S. M., Umeda, A., Amako, K., Albert, M. J., Sack, R. B., Huq, A. & Colwell, R. R. (1999) Trans. R. Soc. Trop. Med. Hyg. 93, 36–40. [DOI] [PubMed] [Google Scholar]
- 22.Pascual, M., Rodo, X., Ellner, S. P., Colwell, R. R. & Bouma, M. J. (2000) Science 289, 1766–1769. [DOI] [PubMed] [Google Scholar]
- 23.Colwell, R. R. (1996) Science 274, 2025–2031. [DOI] [PubMed] [Google Scholar]
- 24.Madico, G., Checkley, W., Gilman, R. H., Bravo, N., Cabrera, L., Calderon, M. & Ceballos, A. (1996) J. Clin. Microbiol. 34, 2968–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.