Abstract
Betaine is an important osmoprotectant, synthesized by many plants in response to abiotic stresses. Almost all known biosynthetic pathways of betaine are two-step oxidations of choline. Recently, a biosynthetic pathway of betaine from glycine, catalyzed by two N-methyltransferase enzymes, was found. Here, the potential role of N-methyltransferase genes for betaine synthesis was examined in a freshwater cyanobacterium, Synechococcus sp. PCC 7942, and in Arabidopsis plants. It was found that the coexpression of N-methyltransferase genes in Synechococcus caused accumulation of a significant amount of betaine and conferred salt tolerance to a freshwater cyanobacterium sufficient for it to become capable of growth in seawater. Arabidopsis plants expressing N-methyltransferase genes also accumulated betaine to a high level in roots, stems, leaves, and flowers and improved seed yield under stress conditions. Betaine levels were higher than those produced by choline-oxidizing enzymes. These results demonstrate the usefulness of glycine N-methyltransferase genes for the improvement of abiotic stress tolerance in crop plants.
Keywords: cyanobacteria, methyltransferase, osmoprotectant, stress resistance
Today, ≈20% of the world's cultivated land and nearly half of all irrigated lands are affected by high salinity (1). High salinity causes ion imbalance and hyperosmotic stress in plants. Organisms that thrive in hypersaline environments possess specific mechanisms for the adjustment of their internal osmotic status. One such mechanism is the ability to accumulate low-molecular-weight organic-compatible solutes such as sugars, some amino acids, and quaternary ammonium compounds (2-4). Glycine betaine (N,N,N-trimethylglycine, hereafter betaine) is a major osmolyte (2-4). Another mechanism for adaptation to high salinity is the exclusion of the Na+ ion from sodium-sensitive sites (5). Genetic engineering techniques have been applied to improve the salt tolerance of plants (6-13). Considerable success has been demonstrated by manipulating the Na+/H+ antiporter genes (6-8). By contrast, the genetic engineering of betaine synthesis has been hampered by low accumulation levels of betaine (9-13). Most known biosynthetic pathways of betaine include a two-step oxidation of choline: choline → betaine aldehyde → betaine. The first step is catalyzed by choline monooxygenase (CMO) in plants (14), choline dehydrogenase (CDH) in animals and bacteria (15, 16), and choline oxidase in some bacteria (11, 17). The second step is catalyzed by NAD+-dependent betaine aldehyde dehydrogenase in all organisms (15, 18, 19), although in some bacteria, CDH and choline oxidase also catalyze the second step (15-17). Hitherto, all attempts at betaine synthesis have been carried out by using choline-oxidizing enzymes (9-13). The supply and transport of betaine precursors such as choline, ethanolamine, and serine to plastids may be of importance, because these precursors have been suggested to be limiting (12, 13).
Recently, we showed that a halotolerant cyanobacterium, Aphanothece halophytica, synthesizes betaine from glycine by three-step methylation (20). Two N-methyltransferase enzymes were involved in betaine synthesis. One enzyme [A. halophytica glycine sarcosine methyltransferase (ApGSMT)] catalyzed the methylation reactions of glycine to sarcosine and sarcosine to dimethylglycine, respectively, and the other enzyme [A. halophytica dimethylglycine methyltransferase (ApDMT)] catalyzed the specific methylation of dimethylglycine to betaine (20). It was shown that a reaction product, S-adenosyl-l-homocysteine (AdoHcy), a potent inhibitor of many methyltransferases, was a relatively weak inhibitor for betaine synthesis; and the final product, betaine, did not inhibit methylation activities even at a concentration of 1 M.
Here, we tested the potential role of genes that encode glycine N-methyltransferases (ApGSMT and ApDMT) for betaine synthesis in a freshwater cyanobacterium Synechococcus sp. PCC 7942 and in the higher plant, Arabidopsis. It was found that the coexpression of ApGSMT and ApDMT led to accumulation of significant amounts of betaine in Synechococcus cells and conferred sufficient salt tolerance that the cells were capable of growth in seawater. Arabidopsis plants expressing ApGSMT and ApDMT accumulated significant amounts of betaine in roots, stems, leaves, and flowers. Accumulation levels of betaine were higher than those of plants transformed with choline-oxidizing enzymes. These results demonstrated the usefulness of betaine synthesis from glycine for the construction of salt-tolerant crop plants.
Materials and Methods
Strains and Culture Conditions. Synechococcus sp. PCC 7942 cells were grown at 30°C under continuous fluorescent white light, 40 μE·m-2·s-1; E, einstein; 1 einstein = 1 mol of photons) in BG11 liquid medium and bubbled with 3% CO2. The Synechococcus cells expressing ApGSMT and ApDMT and Escherichia coli bet-cluster genes were grown in the same conditions as wild-type cells but supplemented with 10 μg·ml-1 streptomycin. The growth of cyanobacterial cells was monitored by measuring absorbance at 730 nm with a Shimadzu UV-160A spectrophotometer.
Construction of the Expression Vectors for ApGSMT and ApDMT in Cyanobacteria. The plasmids pUC303-Bm (21, 22) and pUC303-Bet (13, 23) were used to transform control cells and for the expression of E. coli bet-cluster genes coding for the choline oxidation enzymes CDH and betaine aldehyde dehydrogenase, respectively. The plasmid pUC303-ApNhaP was used for the expression of the Na+/H+ antiporter gene from A. halophytica (ApNhaP1) (23). For the construction of a coexpression vector, the region containing both the promoter and the coding region of ApGSMT was amplified from genomic DNA of A. halophytica by PCR by using the primer set, GSMTbamF, 5′-ATGGATCCTTCAATGTAAGGGGTTGCT-3′ and GSMTbamR, 5′-CGGGATCC TTAATCTTTTTTCGCAAC-3′. The corresponding region for ApDMT was amplified by using the primer set, DMTsalF, 5′-GCGTCGACCTAGGGT TTGTGGAAC TT-3′ and DMTsalR, 5′-ACGTCGACACCAAGAATCTGCTTAGT T-3′. PCR products were subcloned into pBSK+ at the EcoRV restriction site and then sequenced. DNA fragments covering the promoter and the coding region of ApGSMT and ApDMT were prepared by digestion with BamHI and SalI, respectively. Each fragment was ligated into the corresponding site of the shuttle vector, pUC303. The generated plasmid pUC303-ApGSMT/DMT was transferred to Synechococcus cells, as described (23). The transformants were selected on BG11 agar containing 10 μg·ml-1 streptomycin.
Construction of Transgenic Arabidopsis Expressing ApGSMT and Ap-DMT. The plasmids pApGSMT-SK+ and pApDMT-SK+, which contain the coding regions of ApGSMT and ApDMT in pBSK+, respectively (20), were digested with NcoI and BamHI. The resulting fragments were ligated into the NcoI and BglII sites of pCAMBIA1301, generating pApGSMT-CAMBIA and pApDMT-CAMBIA, respectively. The fragment containing the cauliflower mosaic virus 35S promoter and the coding region of ApDMT was prepared by double digestion of pApDMT-CAMBIA with SmaI and DraI, then ligated into the SmaI site of pApGSMT-CAMBIA. The resulting plasmid pApDMT/GSMT-CAMBIA was transferred to Agrobacterium tumefaciens strain LBA4404. Arabidopsis thaliana (ecotype Columbia) plants were transformed by using the floral-dip method (24). Transgenic plants were screened on Murashige and Skoog (MS) agar medium containing 40 μg·ml-1 hygromycin. Seedlings were grown on MS medium in a growth chamber (Sanyo MLR-350HT, Tokyo) with a cycle of 16 h of light at 200 μE m-2·s-1 and 8 h dark, at 60% relative humidity. Two homozygous transformants showing high-level expression of the introduced genes were used for analysis of stress tolerance. CMO transgenic Arabidopsis plants were constructed as described (13).
Stress Treatment for Plants. For stress treatment during germination, surface-sterilized seeds were soaked, plated on MS agar medium, and incubated in the growth chamber. For salt and oxidative stresses, seeds were plated on MS agar medium containing various concentrations of NaCl or CuSO4, respectively. For low-temperature treatment during imbibition, sterilized seeds were allowed to imbibe at 0°C for the indicated times and then inoculated on MS plates at 22°C. For substrate supplement experiments, 1-5 mM glycine, sarcosine, or dimethylglycine and 50-100 μM S-adenosylmethionine (SAM) were added to the growth medium. For salt-stress treatment during the reproductive stage, plants grown under normal conditions on vermiculite were irrigated with 1/10 MS plus 200 mM NaCl just before the primary inflorescence appeared.
To measure the quantum yield of photosystem II under various abiotic stresses, 1-month-old plants were used. For salt stress, plants were treated with 300 mM NaCl. For drought stress, water in vermiculite was withheld for 3 days and then resupplied. Low-temperature treatment was carried out by incubation at 5°C under continuous irradiation. The quantum yield of photosystem II was measured with a MiniPAM chlorophyll fluorescence system (Walz, Effeltrich, Germany), as described (13).
Amino Acid Analysis. The cyanobacterial cells or plant tissues were homogenized in absolute methanol. The supernatant was collected and the pellet was reextracted with 90% methanol. The combined methanol extract was dried in a vacuum rotary evaporator at 45°C. The dry residues were reextracted with a mixture of water and chloroform (1:1, vol/vol). The upper aqueous phase was filtered through a 0.22-μm membrane filter. The filtrate was dried in a vacuum and stored at -20°C until use. At the time of analysis, samples were dissolved in mobile-phase solution (pH 2.6) containing 14.1 g of trilithium citrate tetrahydrate, 70 ml of 2-methoxyethanol, and 13.3 ml of 60% HClO4 per liter and injected into an amino acid analyzer with a shim-pack Li column (Shimadzu).
Other Methods. SDS/PAGE and Western blotting analysis were carried out according to the standard protocol, as described (20). Antibodies raised against the ApGSMT and ApDMT proteins were prepared as described (20). Protein contents were determined by the modified Lowry method (25). Betaine was extracted and analyzed by time-of-flight mass spectroscopy, as described (13). Methyltransferase activities were carried out as described (20).
Results
Coexpression of N-Methyltransferases ApGSMT and ApDMT in a Freshwater Cyanobacterium Synechococcus sp. PCC7942 Conferred the Salt Tolerance of Cells Capable of Growth in Seawater. It has been demonstrated that Synechococcus sp. PCC 7942 cells could grow in BG11 medium containing 0.35 M NaCl but not at 0.375 M NaCl, whereas cells expressing choline-oxidizing genes, such as bet-cluster genes (21) or a choline-oxidase gene (22), could grow in BG11 medium containing 0.375 M NaCl. It was also shown that Synechococcus cells expressing ApNhaP1, but not bet-cluster genes, could grow in BG11 medium containing 0.5 M NaCl and also in seawater (23). These results suggested that the improvement of salt tolerance by genetic engineering of betaine synthesis via choline-oxidizing gene is rather small.
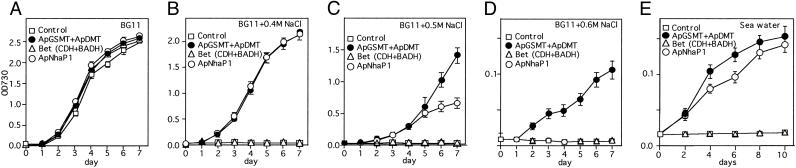
Due to the finding of a pathway for betaine synthesis (20), we tested the potential role of N-methyltransferases for betaine accumulation in a freshwater cyanobacterium. The plasmids for control (pUC303-Bm) and for expressions of E. coli bet-cluster (CDH and betaine aldehyde dehydrogenase), ApNhaP1, and ApGSMT/ApDMT were introduced into Synechococcus cells. Under normal conditions, Synechococcus cells transformed with these vectors could grow at almost the same rate (Fig. 1A). When the growth medium contained 0.4 M NaCl, cells expressing vector alone and bet-cluster genes could not grow, whereas cells expressing ApGSMT/ApDMT could grow (Fig. 1B). Thus betaine synthesis by three-step methylation of glycine conferred salt tolerance more than that by choline oxidation. Interestingly, cells expressing ApGSMT/ApDMT could grow in BG11 medium containing 0.5 M and 0.6 M NaCl, whereas cells expressing ApNhaP1 could not grow in medium containing 0.6 M NaCl (Fig. 1 C and D). Neither control nor bet-cluster-expressing cells could grow in seawater from Mikawa Bay of the Aichi Prefecture in Japan, with practical salinity units 30.4 (Fig. 1E). However, cells expressing ApGSMT/ApDMT could grow at a rate slightly faster than those expressing ApNhaP1 (Fig. 1E). These data indicate that betaine synthesis by three-step methylation of glycine could improve the salt tolerance of Synechococcus cells to a slightly greater extent than that by overexpression of ApNhaP1.
Fig. 1.
Effects of NaCl on the growth of wild-type, ApGSMT/ApDMT, ApNhaP1, and bet-cluster gene-expressing cells. (A-D) Wild-type, ApGSMT/ApDMT, ApNhaP1, and bet-cluster gene-expressing cells at logarithmic phase were transferred to BG11 medium containing 0 M (A), 0.4 M (B), 0.5 M (C), and 0.6 M (D) NaCl. (E) Wild-type, ApGSMT/ApDMT, ApNhaP1, and bet-cluster gene-expressing cells were transferred to seawater. Growth rate was monitored by absorbance change at 730 nm. Data are the means ± SD of three independent experiments with three replicates per experiment.
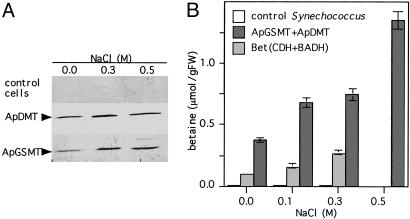
Coexpression of ApGSMT and ApDMT Caused Accumulation of Significant Amounts of Betaine in the Freshwater Cyanobacterium Synechococcus sp. PCC7942. Western blot analysis was carried out to examine the expression level of ApGSMT and ApDMT in Synechococcus cells. The single crossreacting band with antibodies raised against ApGSMT and ApDMT was detected in cells expressing ApGSMT/ApDMT (Fig. 2A) but not in control cells. The accumulation levels of ApGSMT and ApDMT increased at high salinity as observed in A. halophytica cells (20), suggesting the promoters of ApGSMT and ApDMT were recognized in Synechococcus cells.
Fig. 2.
Western blotting of ApGSMT and ApDMT and the accumulation of betaine in Synechococcus sp. PCC 7942 cells expressing ApGSMT/ApDMT. (A) Synechococcus sp. PCC 7942 cells were grown in BG11 medium containing the indicated concentrations of NaCl. The soluble fractions of Synechococcus cells expressing ApGSMT/ApDMT were prepared by sonication. Equal amounts of protein (50 μg) were subjected to 12.5% SDS/PAGE and visualized by using antibodies raised against ApGSMT and ApDMT. (B) Betaine content in wild-type, ApGSMT/ApDMT, and bet-cluster gene-expressing cells. Wild-type, ApGSMT/ApDMT, and bet-cluster gene-expressing cells were harvested at the logarithmic phase. Betaine was extracted as described in Materials and Methods. Betaine content was analyzed by time-of-flight mass spectroscopy. Data are the means ± SD of three independent experiments, with three replicates per experiment.
Next, accumulation levels of betaine in cells expressing ApGSMT/ApDMT and bet-cluster genes were compared. The levels of betaine in both cells increased with increasing the salinity (Fig. 2B). However, betaine levels in cells expressing ApGSMT/ApDMT were always higher than those in cells expressing bet-cluster genes. Especially at 0.5 M NaCl, the betaine level was significantly higher in cells expressing ApGSMT/ApDMT, whereas betaine could not be measured in cells expressing bet-cluster genes due to no growth at this salinity. The betaine level of cells expressing ApGSMT/ApDMT at 0.5 M NaCl was ≈5-fold higher than in cells expressing bet-cluster genes at 0.3 M NaCl and was estimated as ≈200 mM.
In plants transformed with choline-oxidizing enzymes, it was shown that the exogenous supply of choline enhanced accumulation levels of betaine (9, 11-13). Therefore, we tested the effects of substrate supply for betaine accumulation. It was found that the exogenous addition of glycine, sarcosine, dimethylglycine, and SAM to cells with ApGSMT/ApDMT did not enhance accumulation levels of betaine (data not shown). These results suggest that levels of substrates were sufficient and did not limit rates of betaine synthesis in transformed Synechococcus cells.
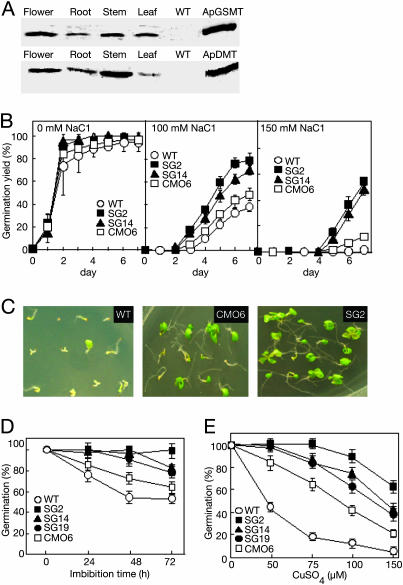
ApGSMT and ApDMT Were Expressed in All Organs of Arabidopsis Plants. Previously, genetic engineering of higher plants with betaine via choline oxidase (11, 26), CDH (27), and CMO (12, 13) produced betaine and conferred some abiotic-stress tolerance. However, the accumulation levels of betaine were small even after supplying exogenous choline (9, 12, 13, 28, 29). Here, we tested the potential role of N-methyltransferases for the accumulation of betaine in plants. Genes encoding ApGSMT and ApDMT with the 35S promoter were transferred into Arabidopsis plants. Among 20 transgenic lines, two homogenous lines accumulating relatively high levels of betaine, SG2 and SG14, were used to analyze stress tolerance. Western blot analysis showed that ApGSMT and Ap-DMT were detected in all organs of transgenic plants but not in wild-type plants (Fig. 3A). Their accumulation levels were not altered or slightly increased at high salinity (data not shown). The methyltransferase activities of ApGSMT and ApDMT were detected in transgenic but not in wild-type plants (data not shown).
Fig. 3.
Expression of ApGSMT and ApDMT and its effect on germination. Western blotting (A), salt stress (B and C), low temperature (D), and oxidative stress by CuSO4 (E). (A) Arabidopsis plants were grown under control conditions. Soluble fractions from different organs were extracted and electrophoresed. Equal amounts of protein (50 μg) were subjected to 12.5% SDS/PAGE and visualized by using antibodies raised against ApGSMT and ApDMT. (B) Seeds were plated on MS agar medium in the presence of different concentrations of NaCl. (C) Photograph taken after 14 days of germination. (D) Seeds were treated with low (0°C) temperature for the indicated time, then returned to normal growth conditions. (E) Seeds were plated on MS agar medium in the presence of different concentrations of CuSO4. The germination of seeds was scored as positive when the tip of the radical had fully penetrated the seed coat. (B, D, and E) Data are the means ± SD of three independent experiments of 19 seeds.
Transgenic Arabidopsis Plants Expressing ApGSMT/ApDMT Increased Tolerance for Various Abiotic Stresses During Germination and Developmental Stages. A potential role of N-methyltransferases for stress tolerance during germination was examined. Three kinds of plants, wild-type, CMO transformant, and ApGSMT/ApDMT transformant, were used. Almost all seeds of wild-type and transgenic plants could germinate in MS agar medium (Fig. 3B). Upon increase of NaCl concentrations, germination was delayed and germination yields decreased. The germination of wild-type seeds was more severely inhibited than in transformants (Fig. 3B). Among the transformants, the CMO transformant was more severely inhibited than the ApGSMT/ApDMT transformant. When 150 mM NaCl was included in the growth medium, all wild-type seeds could not germinate, whereas seeds transformed with CMO and ApGSMT/ApDMT genes germinated ≈20% and 60%, respectively (Fig. 3B). Photographs taken after 14 days of germination in medium containing 100 mM NaCl are shown in Fig. 3C, which clearly displays the survival capability of three different types of seedlings under high salinity. Almost all wild-type and 40% of CMO seedlings showed yellowish cotyledons and less growth, but ≈90% of the SG2 seedlings had green cotyledons. Fig. 3D shows the effects of low temperature (0°C) during imbibition. Upon the increase of the imbibition time at 0°C, the germination yield of wild-type seeds drastically decreased, whereas the transformants retained relatively high values. In this case also, the transformant expressing CMO was more inhibited than that expressing ApGSMT/ApDMT. Similar results were also observed for oxidative stress by copper ion (Fig. 3E) and high-temperature stress (data not shown).
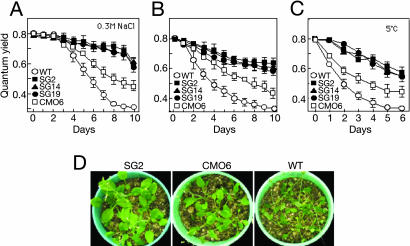
The effects of various stresses on the growth of developmental stage plants were also examined. One-month-old plants grown on vermiculite were subjected to various stresses. Fig. 4 A-C show the effects of salt (0.3 M NaCl), drought, and low temperature (5°C) on the quantum yield of photosystem II. The quantum yield of wild-type plants decreased most rapidly, then CMO transformants and ApGSMT/ApDMT transformants followed. Photographs taken at day 14 after drought stress (Fig. 4D) are consistent with the above results. Taken together with the results of germination, these data clearly indicate that Arabidopsis plants expressing ApGSMT/ApDMT increased tolerance for various abiotic stresses during germination and developmental stages more than did wild-type and CMO transformants.
Fig. 4.
Stress tolerance of wild-type, ApGSMT/ApDMT-expressing, and CMO-expressing Arabidopsis plants during the developmental stage. Salt stress (A), drought stress (B), low temperature (C), and photograph taken at the 14th day after drought stress (D). One-month-old plants grown in vermiculite were subjected to salt, drought, and low-temperature stresses. The quantum yield of photosystem II was measured. Data are the means ± SD of three independent experiments of 10 plants.
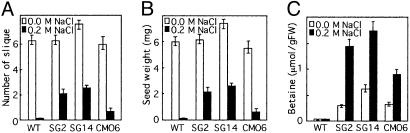
ApGSMT/ApDMT-Expressing Arabidopsis Plants Increased Tolerance for Salt Stress During the Reproductive Stage and Produced Higher Seed Yields. Next, the stress tolerance of reproductive stage plants was examined. Just before the appearance of the primary inflorescence, plants on vermiculite were irrigated with 1/10 MS medium containing 0.2 M NaCl until the harvesting of seeds. Fig. 5 A and B show that the numbers of siliques and seed weight in wild-type and transgenic plants were similar under normal conditions. However, when salt stress was applied during the reproductive stage, transgenic plants expressing ApGSMT/ApDMT could produce siliques 15-fold higher than those in wild-type plants. The ApGSMT/ApDMT-expressing plants produced ≈3- to 4-fold more siliques than those in CMO-expressing plants (Fig. 5A). Similar results were observed for seed weight (Fig. 5B). Transgenic plants accumulated a significant amount of betaine in seeds, and the content in ApGSMT/ApDMT seeds was ≈2-fold higher than that in CMO-expressing seeds (Fig. 5C).
Fig. 5.
Seed yields of wild-type and transformed Arabidopsis plants upon abiotic stresses. Two-week-old seedlings of wild type, ApGSMT/ApDMT-expressing, and CMO-expressing Arabidopsis plants were transferred to vermiculite and allowed to adapt for 5 days, then plants were salinized with 1/10 MS solution containing 200 mM NaCl. Silique formation was counted after 15 days of salt stress. Seed weight and betaine content were measured from dried mature siliques. (A) Number of siliques per plant. (B) Seed weight per plant. (C) Betaine content in seeds. Data are the means ± SD of three independent experiments of 10 plants.
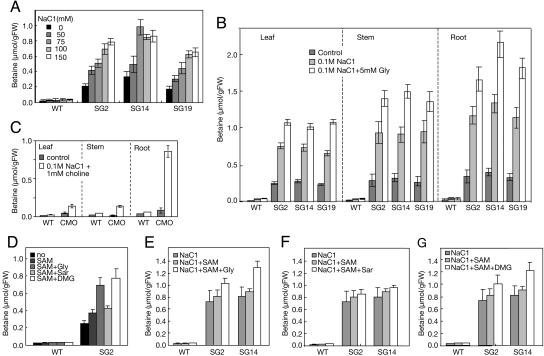
Betaine Accumulated in All Organs in Transgenic Plants, and Their Levels Increased upon Salt Stress. Next, betaine levels in plants were examined. The levels of betaine in wild-type plants were very low or negligible in all conditions tested (Fig. 6). By contrast, transgenic plants expressing ApGSMT/ApDMT accumulated betaine, and the amounts rose as the NaCl level was increased. Betaine levels at 150 mM NaCl were ≈3-fold higher than those without the addition of NaCl (Fig. 6A). Because the expression levels of ApGSMT and ApDMT did not change so much upon salt stress in transgenic plants, the increase of betaine would be caused by the activation of enzymes or by an increased supply of the precursors, glycine and SAM, upon salt stress. Large amounts of betaine were detected in leaves, stems, roots (Fig. 6B), and also flowers (data not shown). By contrast, CMO plants accumulated betaine specifically in roots, as shown in Fig. 6C.
Fig. 6.
Effects of exogenous supply of substrates for the accumulation of betaine. Three- to four-week-old wild-type and transformed Arabidopsis plants grown in different conditions were used to extract betaine, as described in Materials and Methods. (A) Levels of betaine in ApGSMT/ApDMT-expressing Arabidopsis plants grown in MS medium containing different salt concentrations. (B) Effects of NaCl and glycine for the accumulation of betaine in different organs of ApGSMT/ApDMT-expressing Arabidopsis plants. (C) Levels of betaine in Arabidopsis plants expressing CMO. (D) Effects of SAM, glycine, sarcosine, and dimethylglycine. (E) Effects of NaCl, SAM, and glycine. (F) Effects of NaCl, SAM, and sarcosine. (G) Effects of NaCl, SAM, and dimethylglycine. Data are the means ± SD of three independent experiments of five plants.
Next, we examined the effects of substrates on the accumulation of betaine. Adding an exogenous supply of glycine or dimethylglycine to the control growth medium enhanced the accumulation levels of betaine by ApGSMT/ApDMT plants >2-fold, whereas sarcosine did not enhance the levels (data not shown). Fig. 6B shows that when NaCl and glycine were included in the growth medium, the levels of betaine in leaves, stems, and roots of Arabidopsis plants expressing ApGSMT/ApDMT were ≈1-2 μmol per gram of fresh weight (gFW). In contrast, Arabidopsis plants expressing CMO accumulated betaine ≈0.8 μmol per gFW in roots but very low levels in leaves and stems (Fig. 6C). The accumulation of betaine at high levels in various organs of Arabidopsis plants expressing ApGSMT/ApDMT would be the reason for the significant improvement against various abiotic stresses.
Effects of Adding Substrates to the Medium on Betaine Content and Tolerance to Salt Stress of Plants Expressing ApGSMT/ApDMT Genes. The effects of the methyl donor SAM on the accumulation of betaine were examined. The addition of SAM to 2-week-old plants grown in MS medium enhanced betaine levels slightly, ≈1.3-fold (Fig. 6D). The addition of SAM to the growth medium containing 100 mM NaCl slightly increased betaine levels (Fig. 6E). On the other hand, the addition of glycine and dimethylglycine, but not sarcosine, brought about much higher levels of betaine (Fig. 6 D-G). These data suggest that SAM is not a limiting factor for the accumulation of betaine in transgenic plants, but glycine levels are somewhat limiting.
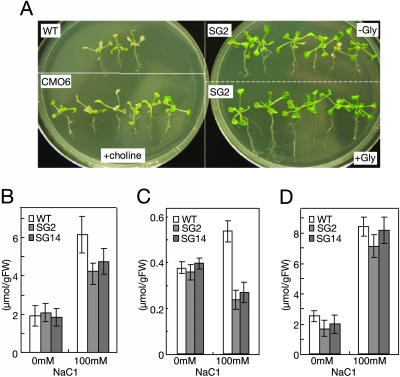
Next, we examined whether the addition of glycine enhances the stress tolerance of transgenic plants expressing ApGSMT/ApDMT. Fig. 7A shows the photographs of wild-type-, CMO-, and ApGSMT/ApDMT-expressing plants after 6 days of salt stress (0.2 M NaCl). Whereas wild-type plants were almost all bleached and/or dead, the CMO-expressing plants supplied with choline survived. Almost all ApGSMT/ApDMT-expressing plants without supply of glycine (SG2 -Gly) still retained green leaves (Fig. 7A). However, if glycine was supplied to the ApGSMT/ApDMT-expressing plants (SG2 + Gly), the plant size was greater, and especially root length was longer than those plants to which glycine was not supplied. These results indicate that glycine was limiting for maximal salt-stress tolerance in plants expressing ApGSMT/ApDMT. Consistent with the above conclusion, the levels of serine (Fig. 7B) and glycine (Fig. 7C) in plants expressing ApGSMT/ApDMT were significantly lower than those in wild-type plants under salt-stress conditions, whereas proline levels were similar (Fig. 7D). Levels of these amino acids were similar between the wild-type and transformants under nonstress conditions (Figs. 7 B-D).
Fig. 7.
Effects of exogenous glycine on salt tolerance and levels of amino acids in wild-type and transformed Arabidopsis plants. Ten-day-old plants were transferred to plates containing 200 mM NaCl supplemented with choline or glycine or without supplementation. (A) WT, wild-type plants; CMO, CMO-expressing Arabidopsis plants supplemented with 1 mM choline; SG2 (-Gly), ApGSMT/ApDMT-expressing Arabidopsis plants without supplementation of glycine; SG2 (+Gly), ApGSMT/ApDMT-expressing Arabidopsis plants supplemented with 2.5 mM glycine. Photographs were taken 6 days after transplantation. (B-D) Amino acid content of wild-type and ApGSMT/ApDMT-expressing Arabidopsis plants under non- and salt-stress conditions. (B) Serine. (C) Glycine. (D) Proline. Data are the means ± SD of three independent experiments of five plants.
Discussion
The present data clearly indicate that the coexpression of N-methyltransferases ApGSMT and ApDMT in freshwater cyanobacterium Synechococcus drastically improves the salt tolerance of these cells, enabling them to thrive under high salinity, up to 0.6 M NaCl (Fig. 1D), and seawater (Fig. 1E). The improvement of salt tolerance by ApGSMT and ApDMT was significantly larger than that by choline-oxidizing enzymes. The expression of choline-oxidizing enzymes improved salt tolerance of cyanobacterium by ≈0.03 M NaCl, from 0.35 to 0.375 M NaCl, whereas the coexpression of ApGSMT and ApDMT improved by ≈0.2 M NaCl, from 0.35 to 0.5-0.6 M NaCl.
Fig. 2B showed that different stress tolerance was due to different levels of betaine. The betaine level in the cells expressing bet-cluster genes was ≈24 mM (21), whereas that in cells expressing ApGSMT/ApDMT at 0.5 M NaCl was ≈200 mM. One of the reasons causing this difference might be the use of different promoters. Their own promoters from A. halophytica were used for the ApGSMT and ApDMT genes, whereas the E. coli promoters were used for the expression of bet-cluster genes. Another possibility is the difference of substrate availability. Glycine was used for N-methyltransferase, whereas choline was used for choline-oxidizing enzymes. Glycine is the major amino acid in all living organisms and can be synthesized in vivo. On the other hand, cyanobacteria do not synthesize choline; it must be taken from the environment. However, it is not clear to what extent these factors are really involved in the stress tolerance of Synechococcus cells.
Levels of betaine increased with increasing concentrations of NaCl (Fig. 2B). If levels of betaine still increase at even higher salinity where growth is inhibited, it would be interesting to clarify the limiting factor for growth at very high salinity. We thought that extrusion of Na+ would limit the growth at high salinity and tested whether overexpression of both ApNhaP1 and ApGSMT/ApDMT improves the salt tolerance of Synechococcus cells further. The results so far have been negative. Identification of limiting factor(s) for salt tolerance is an interesting subject for future work. Previously, it was implicated that stress tolerance conferred by the accumulation of betaine might be due to the protective function of betaine molecules, not their osmotic function (9, 10). However, considering the concentrations of K+ and Cl- and the osmoprotectant of Synechococcus cells (5), betaine at ≈200 mM might easily function as an osmoprotectant.
Figs. 3 and 6 show that Arabidopsis expressing ApGSMT and ApDMT catalyzed betaine synthesis from glycine. Arabidopsis plants expressing CMO accumulated betaine exclusively in the roots (Fig. 6C) (13). Other transgenic plants expressing choline-oxidizing enzymes have also been shown to accumulate betaine mainly in leaves (11, 26, 27) or flowers (24). By contrast, plants expressing ApGSMT/ApDMT accumulated high betaine levels in all organs (Fig. 6B). Betaine levels in the roots of transgenic plants expressing ApGSMT/ApDMT were ≈2-fold higher than that of CMO plants and significantly higher in stems and leaves. Accumulation of betaine in various tissues would contribute to confer stress tolerance in plants. Indeed, Figs. 3, 4, 5 clearly showed that ApGSMT/ApDMT Arabidopsis plants improved tolerances for various abiotic stresses more than CMO-expressing plants. The reason for the accumulation of betaine in various tissues of ApGSMT/ApDMT-expressing plants might be the availability of substrates. Glycine and serine are both potential sources of C1 units in plants. They are readily interconvertible via the action of serine hydroxymethyltransferase and glycine decarboxylase enzymes (30). Therefore, glycine formation is favorable in all plant organs. The high expression of SAM synthetase in root and stem has also been reported (31).
Fig. 6 shows that the betaine level in ApGSMT/ApDMT plants increased at high salinity, although their enzyme levels did not change very much. This increase might be due to the increased accumulation of glycine and serine at high salinity, as shown in wild-type plants (Fig. 7B). The high accumulation of serine at high salinity is also reported by Ho and Saito (32). Moreover, the induction of SAM synthetase expression at high salinity (33) may also increase the SAM level.
Glycine and serine levels in transgenic plants at high salinity were lower than those in wild-type plants (Fig. 7). Considerable amounts of glycine may have been used for betaine synthesis under salt-stress conditions in ApGSMT/ApDMT transformants. Levels of proline, which is not involved in betaine synthesis, were also higher at high salinity, but these were the same in wild-type and transgenic plants. Supplying exogenous glycine in Arabidopsis transformants enhances the accumulation levels of betaine and stress tolerance (Figs. 6 and 7). This did not occur in Synechoccus cells expressing ApGSMT/ApDMT, where the levels of glycine were increased 4.5-fold, from 0.15 to 0.70 nmol/mg fresh weight, at high salinity (0.3 M NaCl), which seems to be sufficient for the synthesis of betaine.
Compared with the success of manipulation of Na+/H+ antiporter genes (6-8), the genetic engineering of betaine synthesis was previously hampered because of low accumulation of betaine (9-13). However, present data demonstrate the potential importance of this osmoprotectant for abiotic stress tolerance. It now seems clear that overexpression of both the Na+/H+ antiporter gene (23) and N-methyltransferase genes (20) could increase abiotic stress tolerance to similar extents. Further increase of stress tolerance by the combination of the Na+/H+ antiporter gene and N-methyltransferase genes in an individual plant is an interesting possibility requiring additional effort.
Acknowledgments
We thank Eiko Tsunekawa for expert technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan and the High-Tech Research Center of Meijo University.
Abbreviations: ApNhaP1, Na+/H+ antiporter from A. halophytica; ApDMT, A. halophytica dimethylglycine methyltransferase; ApGSMT, A. halophytica glycine sarcosine methyltransferase; CDH, choline dehydrogenase; CMO, choline monooxygenase; MS, Murashige and Skoog; SAM, S-adenosylmethionine.
References
- 1.Flower, T. J. & Yeo, A. R. (1995) Aust. J. Plant Physiol. 22, 875-884. [Google Scholar]
- 2.Rhodes, D. & Hanson, A. D. (1993) Annu. Rev. Plant Physiol. Mol. Biol. 44, 357-384. [Google Scholar]
- 3.Hanson, A. D., Rathinasabapathi, B., Rivoal, J., Burnet, M., Dillon, M. O. & Gage, D. A. (1994) Proc. Natl. Acad. Sci. USA 91, 306-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kempf, B. & Bremer, E. (1998) Arch. Microbiol. 170, 319-330. [DOI] [PubMed] [Google Scholar]
- 5.Serrano, R. & Rodriguez-Navarro, A. (2001) Curr. Opin. Cell Biol. 13, 399-404. [DOI] [PubMed] [Google Scholar]
- 6.Apse, M. P., Aharon, G. S., Snedden, W. A. & Blumwald, E. (1999) Science 285, 1256-1258. [DOI] [PubMed] [Google Scholar]
- 7.Zhang, H. X. & Blumwald, E. (2001) Nat. Biotechnol. 19, 765-768. [DOI] [PubMed] [Google Scholar]
- 8.Shi, H., Lee, B. H., Wu, S. J. & Zhu, J. K. (2003) Nat. Biotechnol. 21, 81-85. [DOI] [PubMed] [Google Scholar]
- 9.Hanson, A. D. & Gregory, J. F. (2002) Curr. Opin. Plant Biol. 5, 244-249. [DOI] [PubMed] [Google Scholar]
- 10.Chen, T. H. & Murata, N. (2002) Curr. Opin. Plant Biol. 5, 250-257. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi, H., Alia, Mustardy, L., Deshnium, P., Ida, M. & Murata, N. (1997) Plant J. 12, 133-142. [DOI] [PubMed] [Google Scholar]
- 12.Rontein, D., Basset, G. & Hanson, A. D. (2002) Metab. Eng. 4, 49-56. [DOI] [PubMed] [Google Scholar]
- 13.Hibino, T. Waditee, R., Araki, E., Ishikawa, H., Aoki, K., Tanaka, Y. & Takabe, T. (2002) J. Biol. Chem. 277, 41352-41360. [DOI] [PubMed] [Google Scholar]
- 14.Rathinasabapathi, B., Burnet, M., Russell, B. L., Gage, D. A., Liao, P. C., Nye, G. J., Scott, P., Golbeck, J. H. & Hanson, A. D. (1997) Proc. Natl. Acad. Sci. USA 94, 3454-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamark, T., Kaasen, I., Eshoo, M. W., McDougall, J. & Strom, A. R. (1991) Mol. Microbiol. 5, 1049-1064. [DOI] [PubMed] [Google Scholar]
- 16.Boch, J., Kempf, B., Schmid, R. & Bremer, E. (1996) J. Bacteriol. 178, 5121-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada, H., Mori, N. & Tani, Y. (1979) Agric. Biol. Chem. 43, 2173-2177. [Google Scholar]
- 18.Weretilnyk, E. A. & Hanson, A. D. (1990) Proc. Natl. Acad. Sci. USA 87, 2745-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chern, M.-K. & Pietruszko, T. (1995) Biochem. Biophys. Res. Commun. 213, 561-568. [DOI] [PubMed] [Google Scholar]
- 20.Waditee, R., Tanaka, Y., Aoki, K., Hibino, T., Jikuya, H., Takano, J., Takabe, T. & Takabe, T. (2003) J. Biol. Chem. 278, 4932-4942. [DOI] [PubMed] [Google Scholar]
- 21.Nomura, M., Ishitani, M., Takabe, T., Rai, A. K. & Takabe, T. (1995) Plant Physiol. 107, 703-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deshnium, P., Los, D. A., Hayashi, H., Mustardy, L. & Murata, N. (1995) Plant Mol. Biol. 29, 897-907. [DOI] [PubMed] [Google Scholar]
- 23.Waditee, R., Hibino, T., Nakamura, T., Incharoensakdi, A. & Takabe, T. (2002) Proc. Natl. Acad. Sci. USA 99, 4109-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 25.Larson, E., Howlett, B. & Jagendorf, A. (1986) Anal. Biochem. 155, 243-248. [DOI] [PubMed] [Google Scholar]
- 26.Sulpice, R. Tsukaya, H., Nonaka, H., Mustardy, L., Chen, T. H. & Murata, N. (2003) Plant J. 36, 165-176. [DOI] [PubMed] [Google Scholar]
- 27.Holmstrom, K. O., Somersalo, S., Mandal, A., Palva, T. E. & Welin, B. (2000) J. Exp. Bot. 51, 177-185. [DOI] [PubMed] [Google Scholar]
- 28.Nuccio, M. L., Russell, B. L., Nolte, K. D., Rathinasabapathi, B., Gage, D. A. & Hanson, A. D. (1998) Plant J. 16, 487-496. [DOI] [PubMed] [Google Scholar]
- 29.Huang, J., Hirji, R., Adam, L., Rozwadowski, K. L., Hammerlindl, J. K., Keller, W. A. & Selvaraj, G. (2000) Plant Physiol. 122, 747-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mouillon, J. M., Aubert, S., Bourguignon, J., Gout, E., Douce, R. & Rebeille, F. (1999) Plant J. 20, 197-205. [DOI] [PubMed] [Google Scholar]
- 31.Peleman, J. Saito, K., Cottyn, B., Engler, G., Seurinck, J., Van Montagu, M. & Inze, D. (1989) Gene 84, 359-369. [DOI] [PubMed] [Google Scholar]
- 32.Ho, C. L. & Saito, K. (2001) Amino Acids 20, 243-259. [DOI] [PubMed] [Google Scholar]
- 33.Espartero, J., Pintor-Toro, J. A. & Pardo, J. M. (1994) Plant. Mol. Biol. 25, 217-227. [DOI] [PubMed] [Google Scholar]