Abstract
Docosahexaenoic acid (DHA) plays important physiological roles in vertebrates. Studies in rats and rainbow trout confirmed that DHA biosynthesis proceeds through the so-called “Sprecher pathway”, a biosynthetic process requiring a Δ6 desaturation of 24:5n−3 to 24:6n−3. Alternatively, some teleosts possess fatty acyl desaturases 2 (Fads2) that enable them to biosynthesis DHA through a more direct route termed the “Δ4 pathway”. In order to elucidate the prevalence of both pathways among teleosts, we investigated the Δ6 ability towards C24 substrates of Fads2 from fish with different evolutionary and ecological backgrounds. Subsequently, we retrieved public databases to identify Fads2 containing the YXXN domain responsible for the Δ4 desaturase function, and consequently enabling these species to operate the Δ4 pathway. We demonstrated that, with the exception of Δ4 desaturases, fish Fads2 have the ability to operate as Δ6 desaturases towards C24 PUFA enabling them to synthesise DHA through the Sprecher pathway. Nevertheless, the Δ4 pathway represents an alternative route in some teleosts and we identified the presence of putative Δ4 Fads2 in a further 11 species and confirmed the function as Δ4 desaturases of Fads2 from medaka and Nile tilapia. Our results demonstrated that two alternative pathways for DHA biosynthesis exist in teleosts.
Introduction
Long chain (≥C20) polyunsaturated fatty acids (LC-PUFA) including arachidonic acid (ARA, 20:4n−6), eicosapentaenoic acid (EPA, 20:5n−3) and docosahexaenoic acid (DHA, 22:6n−3) play numerous physiologically important roles essential to health in humans1, 2. Although humans have some ability to synthesise LC-PUFA from the C18 precursors linoleic acid (LOA, 18:2n−6) and α-linolenic acid (ALA, 18:3n−3), dietary supply of these LC-PUFA is still required to meet physiological demands1. Fish are the primary source of n−3 LC-PUFA for humans3 and this has prompted increasing interest in LC-PUFA metabolism in fish4, with biosynthesis being one of the most targeted pathways under investigation5, 6. The biosynthesis of C20–22 LC-PUFA in vertebrates including fish involves alternating steps of desaturation and elongation of the dietary essential C18 fatty acids (FA), LOA and ALA. Fatty acyl desaturases (Fads) catalyse the introduction of a double bond at a specific position of the acyl chain and have been named accordingly as ∆6, ∆5, ∆4 and ∆8 desaturases7. Elongation of very long-chain fatty acid (Elovl) proteins catalyse the condensation and rate-limiting reaction of the FA elongation pathway8, 9. Biosynthesis of ARA and EPA from the C18 precursors LOA and ALA, respectively, follows the same pathways and involves the same enzymes (Fig. 1). The pathways revealed from studies in vertebrates are the so-called “∆6 pathway” (∆6 desaturation − elongation − ∆5 desaturation) and the “∆8 pathway” (elongation − ∆8 desaturation − ∆5 desaturation) (Fig. 1)6, 10–13.
Figure 1.
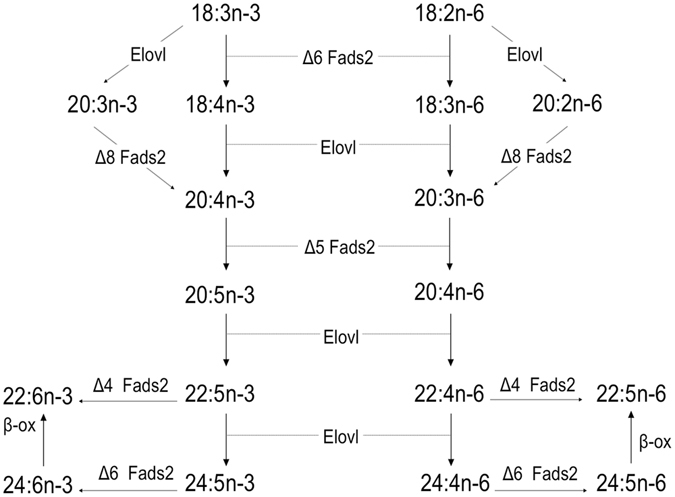
The biosynthetic pathways of long-chain (≥C20) polyunsaturated fatty acids from α-linolenic (18:3n−3) and linoleic (18:2n−6) acids accepted for teleosts6. Enzymatic activities shown in the diagram are predicted from heterologous expression in yeast (Saccharomyces cerevisiae) of fish fatty acyl desaturase 2 (Fads2) and Elongase of very long-chain fatty acid (Elovl) proteins. β-ox, partial β-oxidation
Since the studies of Sprecher and co-workers in rats14–16, it had been generally accepted that the biosynthesis of DHA in vertebrates was achieved by two consecutive elongations from EPA to produce tetracosapentaenoic acid (TPA, 24:5n−3), which then undergoes a ∆6 desaturation to tetracosahexaenoic acid (THA, 24:6n−3), the latter being β-oxidised to DHA in peroxisomes17. This pathway, known as the “Sprecher pathway”, was subsequently confirmed to be operative in rainbow trout Oncorhynchus mykiss 18, 19. The first question that arose after the demonstration of this pathway was whether the same or different ∆6 Fads catalysed the reactions with C18 and C24 substrates15. It was demonstrated that the same ∆6 Fads carried out the conversions of 18:3n−3 to 18:4n−3 and 24:5n−3 to 24:6n−3 in humans20 and rat21, 22. In fish species, it is still unclear whether the same Fads catalyses the two ∆6 desaturation reactions or if two ∆6 Fads (isoenzymes) are involved12, 23, 24. Studies using yeast as a heterologous expression system confirmed that the bifunctional ∆6∆5 Fads from zebrafish (Danio rerio) had ability to desaturate both C18 and C24 substrates at the ∆6 position24. However, the Nibe croaker (Nibea mitsukurii) ∆6 Fads catalysed the desaturation of C18 but not C24 substrates25. These findings suggested that the DHA biosynthetic capability varied among teleost fish and, interestingly, recent findings have demonstrated that, unlike other vertebrates, teleost fish have acquired alternative pathways for DHA biosynthesis during evolution6.
The “∆4 pathway”, first described in the marine protist Thraustochytrium sp.26, is a more direct pathway involving one single elongation of EPA to docosapentaenoic acid (DPA, 22:5n−3), which is subsequently desaturated at the ∆4 position to produce DHA. Although for many years Δ4 desaturases had not been found in any vertebrate species, a Fads2 with Δ4 desaturase activity was first discovered in rabbitfish (Siganus canaliculatus)27. Since then, Fads with ∆4 desaturases have been found in several teleost species such as Senegalese sole (Solea senegalensis)28, pike silverside (Chirostoma estor)29 and striped snakehead (Channa striata)30. Recently, human cells expressing the baboon FADS2 had the ability for direct ∆4 desaturation of 22:5n−3 to 22:6n−331. Thus, the existence of the ∆4 pathway among teleosts appeared to be more widespread than initially believed.
It is interesting to note that, unlike other vertebrates, current evidence suggests that all fads-like genes found in teleost fish are Fads2 orthologues32. Thus the functional diversity among fish Fads2 described above has been hypothesised to be dependent upon various factors including the phylogenetic position of species, in combination with environmental and ecological factors6. In the present study, we aimed to elucidate the pathways for DHA biosynthesis existing in species representing major lineages along the tree of life of teleost fish33. In particular, we have investigated the prevalence of the Sprecher pathway among teleost fish by determining the Δ6 activity towards C24 substrates (24:5n − 3 and 24:4n−6) of desaturases with different substrate specificities (Δ6, Δ5 and Δ4), and derived from fish species with different evolutionary and ecological backgrounds. Furthermore, we have taken advantage of the now known key amino acid (aa) residues determining Δ4 desaturase ability of Fads34 to identify teleost taxa, with publically available genomic or transcriptomic databases, in which their desaturase repertoire enables them to biosynthesise DHA through the more direct Δ4 pathway.
Results
Determination of Δ6 desaturase activity of fish Fads towards C24 PUFA
The capabilities of fish Fads to desaturate C24 PUFA (24:4n−6 and 24:5n−3) at Δ6 position were determined by co-transforming yeast with D. rerio elovl2 and the individual fish fads to be assayed. Control yeast co-transformed with empty p415TEF and pYES2 vectors did not show any activity towards any of the PUFA substrates assayed (data not shown) and the yeast showed typical FA profiles consisting primarily of 16:0, 16:1 isomers, 18:0 and 18:1n-9 (Fig. 2). Independent of the desaturase cloned into the inducible expression vector pYES2, all the co-transformant yeast were able to elongate the exogenously added 22:4n−6 and 22:5n−3 to 24:4n−6 and 24:5n−3, respectively, confirming the activity of the D. rerio Elovl2 cloned into the constitutive expression vector p415TEF. Importantly, the incubation of all the co-transformant yeast in the presence of the corresponding FA substrate as controls (i.e. 18:3n−3 for Δ6 and Δ6Δ5 desaturases, 20:4n−3 for Δ5 desaturases, and 22:5n−3 for Δ4 desaturases) confirmed that the desaturases were functional, with activities as previously reported (Table 1).
Figure 2.
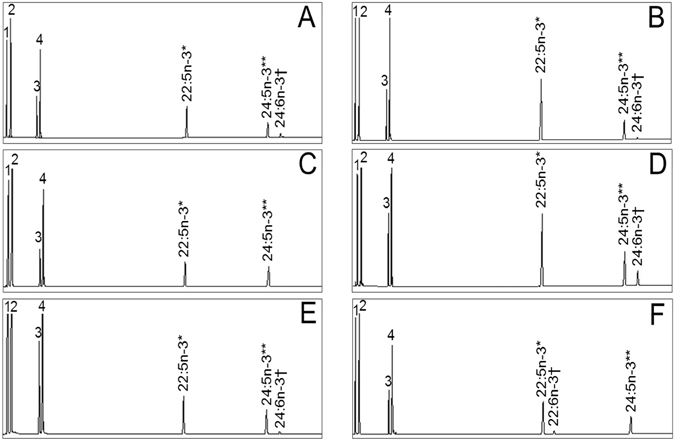
Characterisation of fish fatty acyl desaturases 2 (Fads2) ability to desaturate 24:5n−3. Fatty acid (FA) profiles of yeast (Saccharomyces cerevisiae) co-transformed with the Danio rerio elovl2, and the Arapaima gigas ∆6 fads2 (A), Sparus aurata ∆6 fads2 (B), Nibea mitsukurii ∆6 fads2 (C), Clarias gariepinus ∆6∆5 fads2 (D), Salmo salar ∆5 fads2 (E) and Chirostoma estor ∆4 fads2 (F) and grown in the presence of an exogenously added FA substrates (indicated as “*” in all panels). Peaks 1–4 represent the S. cerevisiae endogenous FA, namely 16:0 (1), 16:1 isomers (2), 18:0 (3) and 18:1n-9 (4). Elongation (**) and desaturation (†) products from exogenously added or endogenously produced FA are indicated accordingly.
Table 1.
Capability of fish Fads2 for Δ6 desaturation of C24 substrates 24:4n−6 and 24:5n−3 using a yeast Saccharomyces cerevisiae heterologous system as described in Materials and Methods. Fatty acid (FA) conversions were calculated as the percentage of 24:4n−6 and 24:5n−3 desaturated to 24:5n−6 and 24:6n−3, respectively, as [product area/(product area + substrate area)] × 100. Conversions towards the control FA substrate (18:3n−3 as controls for Δ6 and Δ6Δ5 desaturases, 20:4n−3 for Δ5 desaturases and 22:5n−3 for Δ4 desaturases) are also indicated. In order to normalise the % conversions obtained throughout the Fads2 dataset, ratios between the activities on 24:5n−3 and those on the control FA (“Δ24:5n−3/Δcontrol”) are also presented.
| Desaturasea | % Conversion | |||
|---|---|---|---|---|
| 24:4n−6 → 24:5n−6 | 24:5n−3 → 24:6n−3 | Control → Product | Δ24:5n−3/Δcontrol | |
| ScyΔ6Fads2 | 29.3 | 34.3 | 41.9 | 0.82 |
| AgΔ6Fads2 | 25.4 | 19.0 | 15.3 | 1.24 |
| AjΔ6Fads2 | 14.0 | 15.8 | 17.8 | 0.89 |
| DrΔ6Δ5Fads2 | 10.4 | 15.8 | 11.9 | 1.33 |
| CgΔ6Δ5Fads2 | 29.9 | 28.1 | 31.5 | 0.89 |
| SsΔ6Fads2 | 18.5 | 26.0 | 23.9 | 1.09 |
| SsΔ5Fads2 | 1.4 | 6.4 | 3.4 | 1.88 |
| OmΔ6Fads2 | 7.5 | 19.7 | 20.4 | 0.97 |
| CeΔ6Δ5Fads2 | 4.2 | 9.0 | 22.9 | 0.39 |
| CeΔ4Fads2 | ND | ND | 9.9 | 0.00 |
| ScΔ6Δ5Fads2 | 6.0 | 7.4 | 36.4 | 0.20 |
| ScΔ4Fads2 | ND | ND | 6.9 | 0.00 |
| SaΔ6Fads2 | 4.8 | 6.5 | 15.0 | 0.43 |
| NmΔ6Fads2 | ND | ND | 10.5 | 0.00 |
| OnΔ4Fads2 | ND | ND | 4.5 | 0.00 |
ND, Not detected
aScy, Scyliorhinus canicula; Ag, Arapaima gigas; Aj, Anguilla japonica; Dr, Danio rerio; Cg, Clarias gariepinus; Ss, Salmo salar; Om, Oncorhychus mykiss; Ce, Chirostoma estor; Sc, Siganus canaliculatus; Sa, Sparus aurata; Nm, Nibea mitsukurii; On, Oreochromis niloticus.
The ability for Δ6 desaturation of C24 PUFA such as 24:4n−6 and 24:5n−3 varied among fish Fads (Fig. 2; Table 1). Interestingly, none of the three Δ4 Fads2 assayed (C. estor, S. canaliculatus and Oreochromis niloticus) showed any ability to desaturate either 24:4n−6 or 24:5n−3 (Table 1). However, most of the fish Fads2 with Δ6 and/or Δ5 specificities were capable of desaturating both 24:4n−6 and 24:5n−3 to their corresponding Δ6 desaturated products, namely 24:5n−6 and 24:6n−3, respectively (Fig. 2; Table 1). Due to the intrinsic variability of desaturation activities in the yeast system, we normalised the Δ6 desaturase activities measured on C24 substrates with those obtained on the corresponding control FA substrate. For that purpose, we calculated the ratio “Δ24:5n−3/Δcontrol” (Table 1) that allowed comparisons among the fish Fads investigated herein. Generally, desaturases from species within relatively ancient fish lineages including Scyliorhinus canicula, Arapaima gigas, Anguilla japonica, Clarias gariepinus, Salmo salar and O. mykiss showed high capacity for Δ6 desaturation towards 24:5n−3, with Δ24:5n−3/Δcontrol ratios ≥ 0.82 (Table 1). On the other hand, more modern species such as S. canaliculatus, Sparus aurata and N. mitsukurii had Fads2 with Δ24:5n−3/Δcontrol ratios ≤ 0.43 (Table 1). It is interesting to note that the S. salar Δ5 (SsΔ5Fads2) showed the ability to desaturate 24:5n−3 to 24:6n−3 (Fig. 2E; Table 1), denoting Δ6 desaturase activity. In order to confirm these results, we incubated the SsΔ5Fads2 co-transformant yeast in the presence of 18:3n−3 and confirmed the presence of Δ6 desaturated product 18:4n−3 (3.5% conversion). Among all the non-Δ4 Fads2, the N. mitsukurii NmΔ6Fads2 was the only tested desaturase with no activity on either 24:4n−6 nor 24:5n−3 (Fig. 2C).
Putative Δ4 desaturase collection and phylogenetics
The phylogenetic tree comparing the deduced aa sequence of the fish Fads with those of human and rat is shown in Fig. 3. All Fads1 clustered together and were separate from all Fads2 in the tree. All teleost Fads2 studied in the present study strongly clustered within the teleost group (99% bootstraps), with desaturases from early divergent teleost species (e.g. A. gigas, A. japonica, C. gariepinus, S. salar and O. mykiss) clustering separately from desaturases from species belonging to more recent lineages (95% bootstraps) (Fig. 3). Among the latter, one can find all the sequences with YXXN residues determining Δ4 activity34 including the previously studied Δ4 desaturases from S. canaliculatus, S. senegalensis and C. estor and the herein characterised Fads2 from medaka (Oryzias latipes) and Nile tilapia (O. niloticus). Clearly, all Fads2-like proteins from Nile tilapia and other cichlids formed a monophyletic clade (99% bootstraps), itself comprising a subgroup with Fads2 sequences possessing the abovementioned distinctive YXXN motif for Δ4 desaturases and another group that includes the Δ6Δ5 Fads2 from Nile tilapia (Fig. 3).
Figure 3.
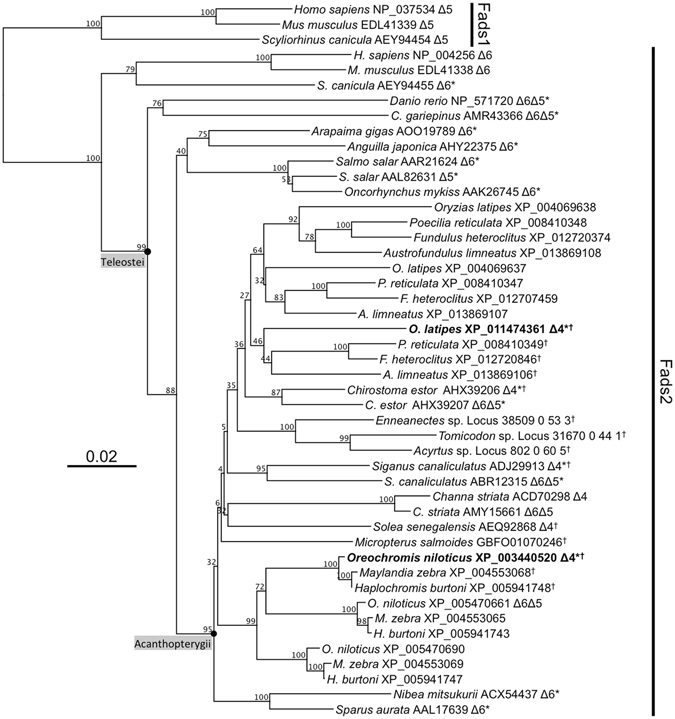
Phylogenetic tree comparing the amino acid sequences of teleost Fads2 with non-teleost vertebrate Fads-like from the cartilaginous fish and mammals (human and mouse). The numbers represent the frequencies (%) with which the tree topology presented was replicated after 1,000 iterations. The functionally characterised Fads were shown with their corresponding regioselectivity (Δ6, Δ5, Δ6Δ5 and Δ4). Asterisks (“*”) indicate Fads2 that have been subjected to further functional analyses in the present study, including the newly cloned Δ4 Fads2 from medaka (Oryzias latipes) and Nile tilapia (Oreochromis niloticus) highlighted in bold. Crosses (“†”) indicate Fads2 that possess the YXXN amino acid residues determining Δ4 desaturase activity32. Branches including Teleostei and Acanthopterygii Fads2 sequences are indicated.
Functional characterisation of Oryzias latipes and Oreochromis niloticus Δ4 desaturase
In order to confirm the function of the putative Δ4 desaturases retrieved by in silico searches, we selected those from medaka O. latipes (XM_011476059) and Nile tilapia O. niloticus (XM_003440472) and characterised their function in yeast. Both desaturases were able to desaturate 22:5n−3 and 22:4n−6 to 22:6n−3 (DHA) and 22:5n−6, respectively (Fig. 4; Table 2), demonstrating that they were both Δ4 desaturases as predicted. Interestingly, both enzymes showed some activity as Δ5 desaturases, since they were able to convert 20:4n−3 and 20:3n−6 into 20:5n−3 (EPA) and 20:4n−6 (ARA), respectively (Table 2). Generally, the Nile tilapia Δ4 Fads2 showed higher conversion activities compared to that of medaka and both proteins were more efficient towards n−3 substrates compared to n−6 substrates.
Figure 4.
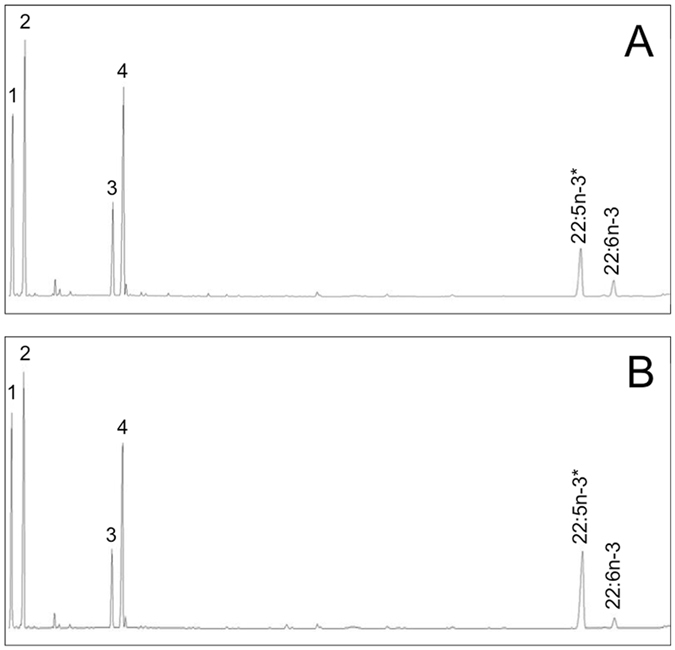
Δ4 desaturase activity towards 22:5n−3 of the newly cloned fads2 from Oryzias latipes (A) and Oreochromis niloticus (B). Peaks 1–4 represent Saccharomyces cerevisiae endogenous FA, namely 16:0 (1), 16:1 isomers (2), 18:0 (3) and 18:1n-9 (4). Peaks derived from exogenously added substrates (*) and the desaturation product 22:6n−3 (DHA) are indicated accordingly.
Table 2.
Substrate conversions of Saccharomyces cerevisiae transformed with Oryzias latipes and Oreochromis niloticus fads2 coding region and grown in the presence of one exogenously added fatty acid (FA) substrate (18:3n−3, 18:2n−6, 20:4n−3, 20:3n−6, 22:5n−3 or 22:4n−6). Conversions were calculated according to the formula [product area/(product area + substrate area)] × 100.
| Species | FA substrate | FA product | Conversion (%) | Activity |
|---|---|---|---|---|
| O. latipes | 18:3n−3 | 18:4n−3 | ND | Δ6 |
| 18:2n−6 | 18:3n−6 | ND | Δ6 | |
| 20:4n−3 | 20:5n−3 | 11.8 | Δ5 | |
| 20:3n−6 | 20:4n−6 | 2.3 | Δ5 | |
| 22:5n−3 | 22:6n−3 | 24.1 | Δ4 | |
| 22:4n−6 | 22:5n−6 | 13.7 | Δ4 | |
| O. niloticus | 18:3n−3 | 18:4n−3 | ND | Δ6 |
| 18:2n−6 | 18:3n−6 | ND | Δ6 | |
| 20:4n−3 | 20:5n−3 | 1.6 | Δ5 | |
| 20:3n−6 | 20:4n−6 | 0.3 | Δ5 | |
| 22:5n−3 | 22:6n−3 | 10.8 | Δ4 | |
| 22:4n−6 | 22:5n−6 | 8.1 | Δ4 |
ND, Not detected
Discussion
It has been largely accepted that DHA biosynthesis in vertebrates proceeds through the Sprecher pathway6, 9. While most earlier investigations focussed on mammals, studies in O. mykiss confirmed that the Sprecher pathway also operated in fish18, 19. It was subsequently demonstrated that the same ∆6 Fads-like enzyme that acts on C18 PUFA precursors at the initiation of the LC-PUFA biosynthesis (Fig. 1) was also responsible for the desaturation of 24:5n−3 required in the Sprecher pathway20, 22. Despite the plethora of studies reporting on the functions of fish Fads6, the capability of fish Fads to operate towards 24:5n−3, and therefore to contribute to DHA biosynthesis through the Sprecher pathway, had not been fully established. For that purpose, we herein conducted a retrospective study investigating the ability to operate as ∆6 desaturases towards 24:5n−3 and 24:4n−6 of a range of previously characterised Fads2 from fish belonging to lineages distributed along the phylogenetic tree of teleosts33.
Using a newly developed method involving yeast, we were able to establish that, with the exception of the Nibe croaker Fads2, all teleost non-∆4 desaturases tested in this study had the ability to efficiently convert 24:5n−3 and 24:4n−6 into 24:6n−3 and 24:5n−6, respectively, confirming their ability for ∆6 desaturation of C24 PUFA substrates. Such ability was observed in Fads2 from species spread across the evolutionary history of teleosts from basal (e.g. A. gigas and A. japonica) and recent (e.g. S. canaliculatus and S. aurata) lineages, and with different regioselectivities including ∆6 desaturases (e.g. O. mykiss and S. aurata) and bifunctional ∆6∆5 desaturases (e.g. C. estor and S. canaliculatus). Since all these Fads2 also showed ∆6 desaturase activity towards C18 PUFA (18:3n−3 and 18:2n−6), the present results confirmed that the same Fads2 can function as ∆6 desaturases at both steps of the LC-PUFA biosynthetic pathway as described above for mammals20, 22. This is in agreement with studies on zebrafish ∆6∆5 Fads2, which showed the ability to operate as ∆6 desaturase on C18 35 and C24 PUFA substrates, using for the latter a yeast supplemented with 24:5n−324. Interestingly, we could also confirm that the previously characterised ∆5 Fads2 from Atlantic salmon S. salar 36, also showed ∆6 activity on C24 PUFA (24:4n−6 and 24:5n−3) and 18:3n−3, suggesting that this enzyme is indeed a bifunctional ∆6∆5 desaturase. Bifunctionality appears a relatively widespread feature among fish Fads2 as a consequence of sub- (acquisition of additional substrate specificities) and neo-functionalisation (substitution and/or acquisition of new substrate specificities) events that have occurred in teleost Fads26, 29. More specifically, bifunctional ∆6∆5 Fads2 have been found in D. rerio 35, S. canaliculatus 27, O. niloticus 37, C. estor 29, C. gariepinus 38 and C. striata 39. Moreover, all the ∆4 Fads2 found so far in fish also exhibited some ∆5 desaturase activity27–30, although none of them had ∆6 activity, which is consistent with the lack of ∆6 desaturase activity towards C24 PUFA substrates observed in all the ∆4 Fads2 assayed in the present study. Interestingly, our results show that the two pathways of DHA biosynthesis, namely the Sprecher and ∆4 pathways, co-exist within some species such as S. canaliculatus and C. estor since, in addition to the role of their ∆6∆5 Fads2 in the Sprecher pathway uncovered in the present study, the existence of ∆4 desaturases in their genomes potentially enables them to further operate via the ∆4 pathway27, 29.
The Δ4 pathway was first reported in the rabbitfish S. canaliculatus 27, with further Δ4 desaturases subsequently described in S. senegalensis, C. estor and C. striata 28–30. In the present study, we have expanded the number of fish lineages and species in which putative Δ4 desaturases exist. In particular, putative Δ4 desaturases were identified in 11 species belonging to Cichliformes (O. niloticus, Maylandia zebra and Haplochromis burtoni), Beloniformes (O. latipes), Blenniiformes (Tomicodon sp., Acyrtus sp. and Enneanectes sp.), Cyprinodontiformes (Poecilia reticulata, Fundulus heteroclitus and Austrofundulus limnaeus) and Centrarchiformes (Micropterus salmoides). It is very likely that the number of species with Δ4 Fads2 will expand when further genomic and/or transcriptomic data become available. This is particularly true for species within groups such as Cichliformes and Beloniformes, in which we found putative Δ4 Fads2 in all species studied in each group. While the actual functional activity of the retrieved desaturases should be assessed in each individual species, the characterisation of two of the newly identified putative Δ4 desaturases, namely the O. latipes and O. niloticus Fads2, as functional Δ4 enzymes supported the effectiveness of our in silico search strategy using a conserved aa region containing the YXXN motif responsible for Δ4 activity to identify functional Δ4 desaturases34. Overall, these results clearly showed that the presence of Δ4 Fads2 among teleosts was far more common than initially believed when the first vertebrate Δ4 desaturase was discovered in S. canaliculatus 27. However, the presence of Δ4 Fads2 appears to be restricted to teleost species within groups regarded herein as “recent lineages”, indicating that the acquisition of the Δ4 pathway occurred later during the evolution of teleosts6, 29.
In more basal teleost lineages, namely Osteoglossiformes (e.g. A. gigas), Anguilliformes (e.g. A. japonica), Cypriniformes (e.g. D. rerio), Siluriformes (e.g. C. gariepinus) and Salmoniformes (e.g. S. salar and O. mykiss), the Sprecher pathway appears to be the only possible route available for DHA biosynthesis. This is supported by, not only the apparent absence of Δ4 Fads2 in their genomes, but also the relatively higher capacity for Δ6 desaturase towards 24:5n−3 of their Fads2, as denoted by normalising the Δ6 conversions of 24:5n−3 (Δ24:5n−3) with that towards a control substrate (Δcontrol). Thus, Fads2 from early divergent teleosts, along with the cartilaginous fish S. canicula, had relatively high capacity for ∆6 desaturation towards 24:5n−3, with Δ24:5n−3/Δcontrol ≥ 0.82. In contrast, Fads2 from other species (S. aurata, C. estor and S. canaliculatus) had lower Δ24:5n−3/Δcontrol ≤ 0.43, indicating lower activity of the Sprecher pathway. While exceptions to this pattern are likely to exist given the functional diversity among teleost Fads26, 29, the apparent lower contribution of the Sprecher pathway to DHA biosynthesis in late-diverging teleosts coincided with the occurrence of Δ4 Fads2 enabling certain species an alternative route for DHA biosynthesis. The limited activity of the Sprecher pathway among these teleost species might be not only restricted to their lower desaturation capability on 24:5n−3 stated above, but also to the absence of key elongase enzymes such as Elovl2, responsible for the production of the Δ6 desaturase substrate 24:5n−340–42. Although Elovl4 can partly compensate such an absence in certain tissues42–45, loss of Elovl2 in the genomes of Acanthopterygii, a group that includes all the late-diverging species considered in this study46, can notably compromise the efficient production of 24:5n−3 as precursor for DHA biosynthesis via the Sprecher pathway. Lack of key enzymatic capabilities in LC-PUFA biosynthetic pathways has been speculated to be a consequence of species having readily available essential LC-PUFA in their diets24, 41. This is the case of marine teleosts, particularly higher trophic species, in which no selection pressure to retain complete and active LC-PUFA biosynthetic pathways has been exerted. For example, extreme cases of marine teleosts with loss of enzymatic activities include the pufferfish (e.g. Tetraodon nigroviridis and Takifugu rubripes), which lack Fads2 in their genomes46. In the present study, we observed that the marine carnivore Nibe croaker N. mitsukurii possess a Fads2 that was the only non-Δ4 Fads2 studied that showed no detectable activity towards 24:5n−3. These results were consistent with the inability of N. mitsukurii Fads2 to desaturate 24:5n−3 to 24:6n−3 in yeast25 and the accumulation of 24:5n−3, but not DHA, in transgenic N. mitsukurii carrying an elovl2 47.
Conclusions
The present study demonstrated that, with the notable exception of Δ4 desaturases, fish Fads2 have the ability to operate as Δ6 desaturases towards C24 PUFA enabling them to synthesise DHA through the Sprecher pathway. However, the so-called “Δ4 pathway” represents an alternative route in some species. Through in silico searches, the present study revealed that the presence of Δ4 Fads was more common than initially believed, and reported three new orders and 11 species in which putative Δ4 desaturases were identified. Importantly, two putative Δ4 Fads2 desaturases retrieved from medaka and Nile tilapia in silico were confirmed as functional Δ4 enzymes. Interestingly, functional characterisation of the S. salar Fads2 previously characterised as a Δ5 desaturase confirmed this enzyme has also Δ6 desaturase activity and should be therefore regarded as a bifunctional Δ6Δ5 desaturase. Overall our results demonstrate that two alternative routes for DHA biosynthesis can exist in teleost fish. Whereas the Sprecher pathway appeared to be widely spread across the entire clade, a more scattered distribution was observed for the Δ4 pathway.
Methods
Fish lineages
A comprehensive set of Fads2-like sequences was collected by screening genomic and transcriptomic databases from fish species representing a sample group of lineages such as the basal gnathostome S. canicula; early diverging post-3R teleosts Osteoglossiformes (A. gigas) and Anguilliformes (A. japonica); and various other teleostei such as Cypriniformes (D. rerio), Siluriformes (C. gariepinus) and Salmoniformes (S. salar and O. mykiss), to relatively modern groups like Anabantiformes (C. striata), Atheriniformes (C. estor), Cichliformes (O. niloticus, M. zebra and H. burtoni), Blenniiformes (Tomicodon sp., Acyrtus sp. and Enneanectes sp.), Beloniformes (O. latipes), Cyprinodontiformes (P. reticulata, F. heteroclitus and A. limnaeus), Pleuronectiformes (S. senegalensis), Spariformes (S. aurata), Centrarchiformes (M. salmoides) and Eupercaria (S. canaliculatus and N. mitsukurii). The desaturase sequences from fish species listed above were used for phylogenetic analysis and selected sequences were subjected to functional characterisation as described.
Determination of Δ6 desaturase activity of fish Fads2 towards C24 PUFA in co-transformant Saccharomyces cerevisiae
We first investigated the ability for Δ6 desaturase activity towards C24 PUFA substrates, i.e. 24:4n−6 and 24:5n−3, the latter being an intermediate in the Sprecher pathway for DHA biosynthesis. Such activities were tested in a total of 15 Fads sequences belonging to 12 species of fish (Table 3), through a newly developed yeast-based assayed as follows. Yeast competent cells InvSc1 (Invitrogen) were co-transformed with two different plasmid constructs prepared as described below. First, the D. rerio elovl2 open reading frame (ORF)48 was ligated into the yeast expression vector p415TEF (a centromeric plasmid with a LEU2 selectable marker) to produce the construct p415TEF-elovl2, in which the expression of the D. rerio elovl2 was controlled under the yeast TEF1 promoter (constitutive expression). Second, the ORF of the corresponding fish Fads (Table 3) was cloned into the episomal yeast vector pYES2, in which the Fads expression was under the control of the GAL1 promoter (inducible expression). Selection of transformant yeast containing both constructs was performed by growing the co-transformed yeast on S. cerevisiae minimal medium minus uracil minus leucine (SCMM−ura−leu) plates. One single colony was grown in SCMM−ura−leu broth for 24 h at 30 °C, and subsequently subcultured in individual Erlenmeyer flasks at 0.1 OD600 (t0) and supplemented with either 0.75 mM Na salts of 22:4n−6 (docosatetraenoic acid, DTA) or 22:5n−3 (DPA) (0.75 mM). Co-transformed yeast were then grown for 24 h (t0 + 24 h) allowing the D. rerio Elovl2 to convert the exogenously added C22 substrates (DTA or DPA) into their corresponding C24 elongation products 24:4n−6 and 24:5n−3, respectively. In order to test the ability of the fish desaturases to introduce Δ6 double bonds into the newly synthesised 24:4n−6 and 24:5n−3 in yeast, the fads expression was then induced (t0 + 24 h) by addition of 2% galactose, after which the recombinant yeast were further grown for 48 h (t0 + 72 h) before collection. As positive controls, a subculture aliquot of the same colony used for the above described assay was supplemented with an n−3 PUFA substrate for which the corresponding assayed Fads had previously shown activity (Table 3) and galactose (2%) at t0. More specifically, co-transformant yeasts were grown in the presence of 18:3n−3 as controls for Δ6 (e.g. AgΔ6Fads2) or Δ6Δ5 (e.g. DrΔ6Δ5Fads2) desaturases, 20:4n−3 for Δ5 desaturases (e.g. SsΔ5Fads2) and 22:5n−3 for Δ4 desaturases (e.g. CeΔ4Fads2). The yeast co-transformed with empty p415TEF and pYES2 vectors were also prepared as negative controls.
Table 3.
Fish fatty acyl desaturases (Fads) investigated for the ability to desaturate tetracosapentaenoic acid (24:5n−3) to tetracosahexaenoic acid (24:6n−3). Their known desaturation activities and the studies in which they were published are indicated accordingly.
| Species | Desaturase namea | Reported activityb | GenBank Accession no. | Reference |
|---|---|---|---|---|
| Scyliorhinus canicula | ScyΔ6Fads2 | Δ6 | JN657544 | 32 |
| Arapaima gigas | AgΔ6Fads2 | Δ6 | AOO1978 | 51 |
| Anguilla japonica | AjΔ6Fads2 | Δ6 | AHY22375 | 53 |
| Danio rerio | DrΔ6Δ5Fads2 | Δ6, Δ5 | AAG25710 | 35 |
| Clarias gariepinus | CgΔ6Δ5Fads2 | Δ6, Δ5 | AMR43366 | 38 |
| Salmo salar | SsΔ6Fads2 | Δ6c | AAR21624 | 54 |
| S. salar | SsΔ5Fads2 | Δ5 | AAL82631 | 36 |
| Oncorhynchus mykiss | OmΔ6Fads2 | Δ6 | AAK26745 | 54 |
| Chirostoma estor | CeΔ6Δ5Fads2 | Δ6, Δ5 | AHX39207 | 29 |
| C. estor | CeΔ4Fads2 | Δ4 | AHX39206 | 29 |
| Siganus canaliculatus | ScΔ6Δ5Fads2 | Δ6, Δ5 | ABR12315 | 27 |
| S. canaliculatus | ScΔ4Fads2 | Δ4 | ADJ29913 | 27 |
| Sparus aurata | SaΔ6Fads2 | Δ6 | AAL17639 | 55 |
| Nibea mitsukurii | NmΔ6Fads2 | Δ6 | AJD80650 | 25 |
| Oreochromis niloticus | OnΔ4Fads2 | Δ4d | XP_003440520 | Present study |
aScy, Scyliorhinus canicula; Ag, Arapaima gigas; Aj, Anguilla japonica; Dr, Danio rerio; Cg, Clarias gariepinus; Ss, Salmo salar; Om, Oncorhychus mykiss; Ce, Chirostoma estor; Sc, Siganus canaliculatus; Sa, Sparus aurata; Nm, Nibea mitsukurii; On, Oreochromis niloticus. bΔ8 desaturase activities of some of these desaturases and reported in the corresponding publication are not indicated in the interests of clarity. cRefers to “Fads2_a” as termed by Monroig et al.56. dFunctional characterisation of OnΔ4Fads2 was carried out in the present study.
In silico retrieval of putative Δ4 desaturases
For retrieval of putative Δ4 desaturase sequences from databases, an alignment of the four functionally characterised Δ4 desaturases from rabbitfish (ADJ29913), Senegalese sole (AEQ92868), pike silverside (AHX39206) and striped snakehead (ACD70298) was performed using the Clustal Omega Multiple Sequence Alignment tool (http://www.ebi.ac.uk/Tools/msa/clustalo/). The conserved aa sequence PPLLIPVFYNFNIMXTMISR, which included the four key aa residues (underlined) accounting for Δ4 regioselectivity34, was used as a query for blast searches. The majority of the putative Δ4 desaturase sequences were obtained from the NCBI Non-redundant protein sequences (nr) database using the blastp algorithm. We further explored the Expressed Sequence Tags (EST) and Transcriptome Shotgun Assembly (TSA) databases using the tblastn algorithm. In addition, the Fish-T1K website (http://www.fisht1k.org) was also used for the tblastn search. Among the retrieved sequences, we selected only those that contained “Y” and “N” in positions 1 and +4, respectively, within the four aa domain YXXN, as these have been reported previously to be crucial for Δ4 function34.
Phylogenetic analysis of Fads desaturases
A phylogenetic tree was built to compare the deduced aa sequences of the fish Fads considered in the present study. The neighbour-joining method49, with the CLC Main Workbench 7 (CLC bio, Aarhus, Denmark), was used to construct the phylogenetic tree, with confidence in the resulting tree branch topology measured by bootstrapping through 1,000 iterations. The alignment of Fads aa sequences used for constructing the phylogenetic tree was performed with MAFFT using the L-INS-i method50. Non-teleost fish sequences from S. canicula and mammalian (human and mouse) Fads2 sequences were also included in the analysis.
O. latipes and O. niloticus putative Δ4 desaturases: Molecular cloning and functional characterisation by heterologous expression in Saccharomyces cerevisiae
In order to confirm that the in silico retrieved Fads sequences encoded Δ4 desaturases, we performed the functional analysis of those identified in O. latipes (XM_011476059) and O. niloticus (XM_003440472). The ORF of the putative Δ4 desaturase sequences were amplified using as template cDNA prepared from a mixture of liver, brain and intestine RNA samples and with primers containing BamHI and XhoI restriction sites (underlined). The primers for O. latipes putative Δ4 Fads2 were CCCGGATCCAAGATGGGAGGTGGAGGTCA (forward) and CCGCTCGAGTCATTTATGAAGATATGCATCAAGC (reverse), whereas the primers CCCGGATCCAGGATGGGACGTGGAAGC (forward) and CCGCTCGAGTCATTTATGGAGGTAAGCGT (reverse) were used to amplify O. niloticus putative Δ4 Fads2. For both genes, PCR were performed using the Phusion HF polymerase (Thermo Fisher Scientific, Waltham, MA, USA) with an initial denaturing step at 98 °C for 30 s, followed by 35 cycles of denaturation at 98 °C for 10 s, annealing at 55 °C for 30 s, extension at 72 °C for 1 min followed by a final extension at 72 °C for 10 min. The DNA fragments of the O. latipes and O. niloticus fads2 obtained were purified, digested with the restriction enzymes, and ligated into similarly digested pYES2 yeast expression vector (Invitrogen) producing the constructs pYES2-Olfads2 and pYES2-Onfads2, respectively. Yeast competent cells InvSc1 (Invitrogen) were transformed with the plasmid constructs using the S.c. EasyCompTM Transformation Kit (Invitrogen). Selection of yeast containing the pYES2 constructs was performed on S. cerevisiae minimal medium minus uracil (SCMM−ura) plates. One single yeast colony of each transformation (pYES2-Olfads2 or pYES2-Onfads2) was grown in SCMM−ura broth for 2 days at 30 °C, and subsequently subcultured in individual Erlenmeyer flasks until optical density measured at a wavelength of 600 nm (OD600) reached 1, after which galactose (2%, w/v) and a PUFA substrate were added as sodium (Na) salts. The exogenous supplemented PUFA included Δ6 (18:3n−3 and 18:2n−6), Δ5 (20:4n−3 and 20:3n−6), and Δ4 (22:5n−3 and 22:4n−6) desaturase substrates, at final concentrations of 0.5 mM (C18), 0.75 mM (C20) and 1.0 mM (C22) to compensate for differential uptake related to fatty acyl chain51. After 2 days, transgenic yeast expressing either the O. latipes or O. niloticus fads2 were harvested and processed for fatty acid analysis as below. All FA substrates (>98–99% pure) used for the functional characterisation assays, except for stearidonic acid (18:4n−3) and eicosatetraenoic acid (20:4n−3), were obtained from Nu-Chek Prep, Inc. (Elysian, MN, USA). Stearidonic acid (>99% pure) and yeast culture reagents including galactose, nitrogen base, raffinose, tergitol NP-40 and uracil dropout medium were obtained from Sigma-Aldrich (UK). Eicosatetraenoic acid was purchased from Cayman Chemical Co. (Ann Arbor, USA).
Fatty acid analysis of yeast
Total lipids extracted from yeast samples52 were used to prepare fatty acid methyl esters (FAME). FAME extraction, purification and analysis were performed as described by Li et al.27. For functional characterisation of the desaturases from O. latipes and O. niloticus, substrate FA conversions were calculated as the proportion of exogenously added FA substrate desaturated as [product area/(product area + substrate area)] × 10051. Substrate FA conversions for the Δ6 desaturase activity towards C24 substrates were calculated using the same formula as above considering the areas of 24:5n−3 and 24:4n−6 produced endogenously by the D. rerio Elovl2 as substrates for calculations. When necessary, GC-MS was used to confirm the identity of the products27.
Acknowledgements
A.O. is a Commonwealth scholar funded by the Commonwealth Scholarship Commission (NGCS-2014-438) in the UK. N.K. was funded by the Japan Society for the Promotion of Science through Grant-in-Aid for JSPS Research Fellow. DRT and OM were partly supported by the Major International Joint Research Project from the National Natural Science Foundation of China (NSFC) (No. 31110103913). The authors will also like to thank Dr. Francisco Hontoria and Dr. Juan C. Navarro (Instituto de Acuicultura Torre de la Sal, CSIC, Spain) for assistance in developing the dual transformation yeast assay used in the present study.
Author Contributions
A.O., N.K. and O.M. designed research; A.O., N.K., G.C.-A. and J.R.D. performed research; A.O., N.K., D.R.T. and L.F.C.C. analysed data; and Ó.M., A.O. and D.R.T. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brenna JT. Efficiency of conversion of alpha-linolenic acid to long chain n−3 fatty in man. Curr. Opin. Clin. Nutr. Metab. Care. 2002;5:127–132. doi: 10.1097/00075197-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Cardoso C, Afonso C, Bandarra NM. Seafood lipids and cardiovascular health. Nutrire. 2016;41:7. doi: 10.1186/s41110-016-0008-8. [DOI] [Google Scholar]
- 3.Shepherd CJ, Monroig O, Tocher DR. Future availability of raw materials for salmon feeds and supply chain implications: The case of Scottish farmed salmon. Aquaculture. 2017;467:49–62. doi: 10.1016/j.aquaculture.2016.08.021. [DOI] [Google Scholar]
- 4.Tocher DR. Metabolism and functions of lipids and fatty acids in teleost fish. Rev. Fish. Sci. 2003;11:107–184. doi: 10.1080/713610925. [DOI] [Google Scholar]
- 5.Tocher DR. Omega-3 long-chain polyunsaturated fatty acids and aquaculture in perspective. Aquaculture. 2015;449:94–107. doi: 10.1016/j.aquaculture.2015.01.010. [DOI] [Google Scholar]
- 6.Castro LFC, Tocher DR, Monroig O. Long-chain polyunsaturated fatty acid biosynthesis in chordates: Insights into the evolution of Fads and Elovl gene repertoire. Prog. Lipid Res. 2016;62:25–40. doi: 10.1016/j.plipres.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Meesapyodsuk D, Qiu X. The front-end desaturase: Structure, function, evolution and biotechnological use. Lipids. 2012;47:227–237. doi: 10.1007/s11745-011-3617-2. [DOI] [PubMed] [Google Scholar]
- 8.Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid Res. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Guillou H, Zadravec D, Martin PGP, Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010;49:186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Tocher DR. Fatty acid requirements in ontogeny of marine and freshwater fish. Aquacult. Res. 2010;41:717–732. doi: 10.1111/j.1365-2109.2008.02150.x. [DOI] [Google Scholar]
- 11.Monroig Ó, Li Y, Tocher DR. Delta-8 desaturation activity varies among fatty acyl desaturases of teleost fish: high activity in delta-6 desaturases of marine species. Comp. Biochem. Physiol. B. 2011;159:206–213. doi: 10.1016/j.cbpb.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Vagner M, Santigosa E. Characterization and modulation of gene expression and enzymatic activity of delta-6 desaturase in teleosts: A review. Aquaculture. 2011;315:131–143. doi: 10.1016/j.aquaculture.2010.11.031. [DOI] [Google Scholar]
- 13.Park WJ, Kothapalli KS, Lawrence P, Tyburczy C, Brenna JT. An alternate pathway to long-chain polyunsaturates: the FADS2 gene product Delta8-desaturates 20:2n−6 and 20:3n−3. J. Lipid Res. 2009;50:1195–202. doi: 10.1194/jlr.M800630-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprecher H. A reevaluation of the pathway for the biosynthesis of 4,7,10,13,16,19-docosahexaenoic acid. Omega-3 News. 1992;7:1–3. [Google Scholar]
- 15.Sprecher H, Luthria DL, Mohammed BS, Baykousheva SP. Reevaluation of the pathways for the biosynthesis of polyunsaturated fatty acids. J. Lipid Res. 1995;36:2471–2477. [PubMed] [Google Scholar]
- 16.Sprecher H. Metabolism of highly unsaturated n−3 and n−6 fatty acids. Biochim. Biophys. Acta. 2000;1486:219–231. doi: 10.1016/S1388-1981(00)00077-9. [DOI] [PubMed] [Google Scholar]
- 17.Ferdinandusse S, et al. Identification of the peroxisomal-oxidation enzymes involved in the biosynthesis of docosahexaenoic acid. J. Lipid Res. 2001;42:1987–1995. [PubMed] [Google Scholar]
- 18.Buzzi M, Henderson RJ, Sargent JR. The desaturation and elongation of linolenic acid and eicosapentaenoic acid by hepatocytes and liver microsomes from rainbow trout (Oncorhynchus mykiss) fed diets containing fish oil or olive oil. Biochim. Biophys. Acta. 1996;1299:235–244. doi: 10.1016/0005-2760(95)00211-1. [DOI] [PubMed] [Google Scholar]
- 19.Buzzi M, Henderson RJ, Sargent JR. Biosynthesis of docosahexaenoic acid in trout hepatocytes proceeds via 24-carbon intermediates. Comp. Biochem. Physiol. B. 1997;116:263–267. doi: 10.1016/S0305-0491(96)00210-6. [DOI] [PubMed] [Google Scholar]
- 20.De Antueno RJ, et al. Activity of human Delta5 and Delta6 desaturases on multiple n−3 and n−6 polyunsaturated fatty acids. FEBS Lett. 2001;509:77–80. doi: 10.1016/S0014-5793(01)03135-0. [DOI] [PubMed] [Google Scholar]
- 21.Geiger M, Mohammed BS, Sankarappa S, Sprecher H. Studies to determine if rat liver contains chain-length specific acyl-CoA 6-desaturases. Biochim. Biophys. Acta. 1993;1170:137–142. doi: 10.1016/0005-2760(93)90063-F. [DOI] [PubMed] [Google Scholar]
- 22.D’Andrea S, et al. The same rat D6-desaturase not only acts on 18- but also on 24-carbon fatty acids in very-long-chain polyunsaturated fatty acid biosynthesis. Biochem. J. 2002;364:49–55. doi: 10.1042/bj3640049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sargent, J. R., Tocher, D. R. & Bell, J. G. The lipids in Fish Nutrition Third Ed. (eds Halver, J. E. & Hardy, R. W.) 181–257 (Academic Press, San Diego, 2002).
- 24.Tocher, D. R., Agaba, M., Hastings, N. & Teale, A. J. Biochemical and molecular studies of the polyunsaturated fatty acid desaturation pathway in fish in The big fish bang - Proceedings of the 26th annual larval fish conference (eds Browman, H. I. & Skiftesvik, A. B.) 211–227 (Institute of Marine Research, Bergen, Norway, 2003).
- 25.Kabeya N, et al. Polyunsaturated fatty acid metabolism in a marine teleost, Nibe croaker Nibea mitsukurii: Functional characterization of Fads2 desaturase and Elovl5 and Elovl4 elongases. Comp. Biochem. Physiol. B. 2015;188:37–45. doi: 10.1016/j.cbpb.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Qiu X, Hong H, MacKenzie SL. Identification of a Delta 4 fatty acid desaturase from Thraustochytrium sp. involved in the biosynthesis of docosahexanoic acid by heterologous expression in Saccharomyces cerevisiae and Brassica juncea. J. Biol. Chem. 2001;276:31561–31566. doi: 10.1074/jbc.M102971200. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, et al. A vertebrate fatty acyl desaturase with Δ4 activity. Proc. Natl. Acad. Sci. USA. 2010;107:16840–16845. doi: 10.1073/pnas.1008429107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morais S, Castanheira F, Martinez-Rubio L, Conceiçao LEC, Tocher DR. Long chain polyunsaturated fatty acid synthesis in a marine vertebrate: Ontogenetic and nutritional regulation of a fatty acyl desaturase with Δ4 activity. Biochim. Biophys. Acta. 2012;1821:660–671. doi: 10.1016/j.bbalip.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Fonseca-Madrigal J, et al. Diversification of substrate specificities in teleostei Fads2: Characterization of Δ4 and Δ6Δ5 desaturases of Chirostoma estor. J. Lipid Res. 2014;55:1408–1419. doi: 10.1194/jlr.M049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuah M, Jaya-Ram A, Shu-Chien AC. The capacity for long-chain polyunsaturated fatty acid synthesis in a carnivorous vertebrate: Functional characterisation and nutritional regulation of a Fads2 fatty acyl desaturase with Δ4 activity and an Elovl5 elongase in striped snakehead (Channa striata) Biochim. Biophys. Acta. 2015;1851:248–260. doi: 10.1016/j.bbalip.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 31.Park HG, Park WJ, Kothapalli KS, Brenna JT. The fatty acid desaturase 2 (FADS2) gene product catalyzes Δ4 desaturation to yield n−3 docosahexaenoic acid and n−6 docosapentaenoic acid in human cells. FASEB J. 2015;29:3911–3919. doi: 10.1096/fj.15-271783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castro LFC, et al. Functional desaturase Fads1 (Δ5) and Fads2 (Δ6) orthologues evolved before the origin of jawed vertebrates. PLoS ONE. 2012;7:e31950. doi: 10.1371/journal.pone.0031950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betancur-R, R. et al. The tree of life and a new classification of bony fishes. PLOS Currents Tree of Life. Edition 1. doi:10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288 (2013). [DOI] [PMC free article] [PubMed]
- 34.Lim Z, Senger T, Vrinten P. Four amino acid residues influence the substrate chain-length and regioselectivity of Siganus canaliculatus Δ4 and Δ5/6 desaturases. Lipids. 2014;49:357–367. doi: 10.1007/s11745-014-3880-0. [DOI] [PubMed] [Google Scholar]
- 35.Hastings N, et al. A vertebrate fatty acid desaturase with Δ5 and Δ6 activities. Proc. Natl. Acad. Sci. USA. 2001;98:14304–14309. doi: 10.1073/pnas.251516598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hastings N, et al. Molecular cloning and functional characterization of fatty acyl desaturase and elongase cDNAs involved in the production of eicosapentaenoic and docosahexaenoic acids from α-linolenic acid in Atlantic salmon (Salmo salar) Mar. Biotechnol. 2005;6:463–474. doi: 10.1007/s10126-004-3002-8. [DOI] [PubMed] [Google Scholar]
- 37.Tanomman S, Ketudat-Cairns M, Jangprai A, Boonanuntanasarn S. Characterization of fatty acid delta-6 desaturase gene in Nile tilapia and heterogenous expression in Saccharomyces cerevisiae. Comp. Biochem. Physiol. B. 2013;166:148–156. doi: 10.1016/j.cbpb.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 38.Oboh A, Betancor MB, Tocher DR, Monroig Ó. Biosynthesis of long-chain polyunsaturated fatty acids in the African catfish Clarias gariepinus: Molecular cloning and functional characterisation of fatty acyl desaturase (fads2) and elongase (elovl2) cDNAs. Aquaculture. 2016;462:70–79. doi: 10.1016/j.aquaculture.2016.05.018. [DOI] [Google Scholar]
- 39.Kuah M, Jaya-Ram A, Shu-Chien AC. A fatty acyl desaturase (fads2) with dual Δ6 and Δ5 activities from the freshwater carnivorous striped snakehead Channa striata. Comp. Biochem. Physiol. A. 2016;201:146–155. doi: 10.1016/j.cbpa.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Leaver MJ, et al. Towards fish lipid nutrigenomics: Current state and prospects for fin-fish aquaculture. Rev. Fish. Sci. 2008;16:71–92. doi: 10.1080/10641260802325278. [DOI] [Google Scholar]
- 41.Bell, M. V. & Tocher, D. R. Biosynthesis of polyunsaturated fatty acids in aquatic ecosystems: general pathways and new directions in Lipids in AquaticEcosystems (eds Arts, M. T., Brett, M. & Kainz, M.) 211–236 (Springer-Verlag, New York, 2009).
- 42.Monroig Ó, et al. Expression and role of Elovl4 elongases in biosynthesis of very long-chain fatty acids during zebrafish Danio rerio early embryonic development. Biochim. Biophys. Acta. 2010;1801:1145–1154. doi: 10.1016/j.bbalip.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Monroig Ó, Webb K, Ibarra-Castro L, Holt GJ, Tocher DR. Biosynthesis of long-chain polyunsaturated fatty acids in marine fish: Characterization of an Elovl4-like elongase from cobia Rachycentron canadum and activation of the pathway during early life stages. Aquaculture. 2011;312:145–153. doi: 10.1016/j.aquaculture.2010.12.024. [DOI] [Google Scholar]
- 44.Carmona-Antoñanzas G, Monroig Ó, Dick JR, Davie A, Tocher DR. Biosynthesis of very long-chain fatty acids (C ≥ 26) in Atlantic salmon: Cloning, functional characterisation, and tissue distribution of an Elovl4. Comp. Biochem. Physiol. B. 2011;159:122–129. doi: 10.1016/j.cbpb.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Li S, et al. Molecular cloning, functional characterization and nutritional regulation by dietary fatty acid profiles of a putative Elovl4 gene in orange-spotted grouper Epinephelus coioides. Aquacult. Res. 2017;48:537–552. doi: 10.1111/are.12902. [DOI] [Google Scholar]
- 46.Morais S, Monroig Ó, Zheng X, Leaver M, Tocher DR. Highly unsaturated fatty acid synthesis in Atlantic salmon: characterization of ELOVL5- and ELOVL2-like elongases. Mar. Biotechnol. 2009;11:627–639. doi: 10.1007/s10126-009-9179-0. [DOI] [PubMed] [Google Scholar]
- 47.Kabeya N, et al. Modification of the n−3 HUFA biosynthetic pathway by transgenesis in a marine teleost, nibe croaker. J. Biotechnol. 2014;172:46–54. doi: 10.1016/j.jbiotec.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 48.Monroig Ó, Rotlant J, Sanchez E, Cerda-Reverter JM, Tocher DR. Expression of long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis genes during zebrafish Danio rerio early embryogenesis. Biochim. Biophys. Acta. 2009;1791:1093–1101. doi: 10.1016/j.bbalip.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 51.Lopes-Marques M, et al. Molecular and functional characterization of a fads2 orthologue in the Amazonian teleost, Arapaima gigas. Comp. Biochem. Physiol. B. 2017;203:84–91. doi: 10.1016/j.cbpb.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 52.Folch J, Lees M, Sloane-Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 53.Wang S, et al. Investigating the long-chain polyunsaturated fatty acid biosynthesis in teleost fish: functional characterisation of fatty acyl desaturase (Fads2) and Elovl5 elongase in the catadromous species, Japanese eel Anguilla japonica. Aquaculture. 2014;434:57–65. doi: 10.1016/j.aquaculture.2014.07.016. [DOI] [Google Scholar]
- 54.Zheng X, Tocher DR, Dickson CA, Bell JG, Teale AJ. Highly unsaturated fatty acids synthesis in vertebrates: New insights with the cloning and characterization of ∆6 desaturase of Atlantic salmon. Lipids. 2005;40:13–24. doi: 10.1007/s11745-005-1355-7. [DOI] [PubMed] [Google Scholar]
- 55.Zheng X, et al. Characterization and comparison of fatty acyl ∆6 desaturase cDNAs from freshwater and marine teleost fish species. Comp. Biochem. Physiol. B. 2004;139:269–279. doi: 10.1016/j.cbpc.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Monroig Ó, et al. Multiple genes for functional ∆6 fatty acyl desaturases (Fad) in Atlantic salmon (Salmo salar L.): Gene and cDNA characterization, functional expression, tissue distribution and nutritional regulation. Biochim Biophys Acta. 2010;1801:1072–1081. doi: 10.1016/j.bbalip.2010.04.007. [DOI] [PubMed] [Google Scholar]


