Abstract
Complement-mediated opsonization, phagocytosis, and immune stimulation are critical processes in host defense and homeostasis, with the complement activation fragment iC3b playing a key effector role. To date, however, there is no high-resolution structure of iC3b, and some aspects of its structure-activity profile remain controversial. Here, we employed hydrogen-deuterium exchange mass spectrometry (HX-MS) to describe the structure and dynamics of iC3b at a peptide resolution level in direct comparison with its parent protein C3b. In our HX-MS study, 264 peptides were analyzed for their deuterium content, providing almost complete sequence coverage for this 175-kDa protein. Several peptides in iC3b showed significantly higher deuterium uptake when compared to C3b, revealing more dynamic, solvent-exposed regions. Most of them resided in the CUB domain, which contains the heptadecapeptide C3f that is liberated during the conversion of C3b to iC3b. Our data suggest a highly disordered CUB, which has acquired a state similar to that of intrinsically disordered proteins, resulting in a predominant form of iC3b that features high structural flexibility. The structure was further validated using an anti-iC3b monoclonal antibody that was shown to target an epitope in the CUB region. The information obtained in this work allows us to elucidate determinants of iC3b specificity and activity and provide functional insights into the protein’s recognition pattern with respect to regulators and receptors of the complement system.
Keywords: iC3b, HX-MS, alternative pathway, CUB, epitope mapping
Introduction
The complement system is comprised of a network of plasma and membrane-associated proteins that recognize and clear invading pathogens and apoptotic cells and also modulate immune responses (1). Central to its function is C3, an abundant multi-domain plasma protein of ~185 kDa that, once activated by distinct initiation pathways, forms the opsonin C3b, which binds covalently to target surfaces (Fig. 1) (2, 3). C3b formation induces marked conformational changes that enable the assembly of C3 convertases that activate more C3 to amplify the complement response (4). Under most circumstances, C3b is readily degraded to its iC3b fragment (Fig. 1), which remains bound to surfaces but can no longer participate in convertase formation. However, iC3b exerts important bridging and signaling functions by binding to complement receptors (CR) on immune cells, making it a versatile effector molecule in host defense and homeostasis (1). Whereas binding of iC3b to CR2 (CD21) on B- and follicular dendritic cells elicits adaptive immune responses (5), interaction with integrin receptors CR3 (CD11b/CD18) and CR4 (CD11c/CD18) enables phagocytosis of opsonized particles (6). This phagocytic function may be enhanced through low-affinity binding to the adhesion receptor CR1 (CD35) and to CRIg, primarily expressed in Kupffer cells (1). The interaction with CR1 also enables further degradation of iC3b to C3dg (Fig. 1), which retains activity for CR2 and potentially CR3 (7).
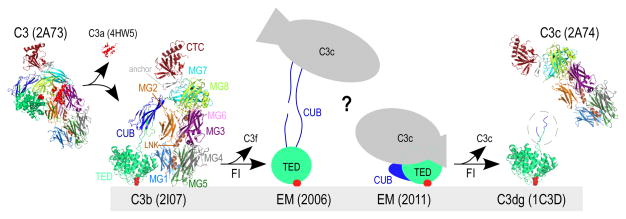
Figure 1. Structures of complement component C3 and activated products.
A simplified schematic of C3-mediated opsonization is shown. Transformation of inert C3 [PDB: 2A73 (2)] through cleavage by C3 convertases leads to the release of C3a [anaphylatoxin domain, PDB: 4HW5 (44)] and hydrolysis of the thioester bond (indicated with red circles). The product of this transformation is termed C3b [PDB: 2I07 (4)] and possesses marked conformational differences from its precursor molecule. C3b molecules close to cell surfaces attach via the activated thioester bond and tag them for opsonization. Sequential cleavages of C3b by FI yield iC3b, for which two controversial EM structures have been reported to date (depicted with cartoons drawn based on (16, 17)). Further cleavage of iC3b by FI yields two end products, surface-bound C3dg (the C3g sequence not crystallized in C3d is encircled, PDB: 1C3D (8)) and fluid-phase C3c (PDB: 2A74 (2)). Domain color annotation follows that in Janssen et al (2); PDB entries are shown in brackets.
Despite a wealth of structural information on C3, its fragments and resulting complexes [i.e. (4, 8–14)], detailed molecular characterization of iC3b has remained elusive. The 173-kDa protein iC3b is generated upon proteolytic release of a heptadecapeptide (C3f) from the C1r/C1s, UEGF, BMP1 (CUB) domain of C3b by factor I (FI) and cofactors, typically FH (15). To date, no crystal structure of iC3b has been determined, and medium-resolution EM structures indicate distinct conformations that appear inconsistent with each other (16, 17). The CUB domain, connecting the macroglobulin (MG) core and surface-active thioester-containing domain (TED) of iC3b, is of particular significance; it is viewed either as a flexible linker resulting in a dynamic, extended molecule (16, 18)] or having a denser conformation (17), corroborating early spectroscopic observations in which iC3b was shown to fold back into a C3-like structure (19). The CUB in C3b is directly implicated in binding the convertase component FB (11); the regulators/cofactors FH (10), CD35, CD46, and CD55 (14); and the protease FI (20). Structural rearrangements in the CUB lead to concomitant changes in reactivity toward these interactors. Conformational changes in the region are also thought to relieve steric constraints on TED and make it accessible to CR2 (12) and CR3 (13).
Therefore, we have now employed hydrogen-deuterium exchange mass spectrometry (HX-MS) to provide structural information for surface opsonins C3b and iC3b at peptide resolution. In the absence of an iC3b crystal structure, D-uptake profiles of individual peptides were compared to the respective theoretical HX profiles calculated for random-coil polypeptides (21). Combining this analysis with biophysical and biochemical assays and interpreting our results in the context of available EM structures, we provide essential insights into the structure and dynamics of iC3b that underlie its activity and specificity.
Materials & Methods
Proteins and reagents
Human purified proteins C3b (1 mg/mL; order no. A114), iC3b (1 mg/mL; A115), factor H (1 mg/ml; A137) and factor I (1 mg/ml; A138) were purchased from Complement Tech (Tyler, TX). The N138A mutant of Efb-C was expressed and purified as described before (22). Monoclonal antibody to human iC3b (anti-iC3b mAb, IgG2bκ, clone 013III-1.1.6; A209) was obtained from Quidel (San Diego, CA). Deuterium oxide (99.9 atom % D; 151882) was obtained from Aldrich (St. Louis, MO). Tris(2-carboxyethyl)-phosphine hydrochloride (TCEP·HCl; 20491) and immobilized pepsin on cross-linked agarose beads (6%; 20343) were from Thermo Scientific (Rockford, IL). Guanidine hydrochloride (>99.5% purity; BP178–500), acetonitrile (99.9%; A998) and formic acid (>99.5% purity; A117) were purchased from Fisher (Fair Lawn, NJ). Leucine enkephalin (MS Leucine Enkephalin Kit; 700002456) used for calibration of the MS instrument was from Waters (Milford, MA).
Hydrogen exchange experiments
For the HX experiments, 4 μL of the C3b or iC3b protein stock (198 ng/mL in phosphate-buffered saline (PBS; 10 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl and 137 mM NaCl, pH 7.3)) was mixed with 40 μL of deuterated PBS (final D2O content during reaction 90.9% v/v), prepared by two cycles of lyophilization and reconstitution in D2O. Isotopic exchange was performed at 0±1 °C for 10, 30, 100, 300, 1000, 3000, 10000 and 25200 sec. Reaction mixtures were quenched with an equal volume (44 μL) of pre-chilled guanidinium hydrochloride-TCEP (3.2 M and 0.8 M, respectively) that was adjusted using 2 N NaOH (~11% v/v) so that the pH of quenched samples was 2.4. Samples were incubated on ice for 2 min and snap-frozen in liquid nitrogen prior to LC-MS analysis. Fully deuterated samples were prepared by incubating proteins as described above for 48 h at 37±1 °C, and non-deuterated samples were prepared similarly in protiated PBS. Samples were prepared and analyzed in duplicate.
For the anti-iC3b mAb neo-epitope mapping, iC3b (1.1 μM) was incubated with slight molar excess of anti-iC3b mAb (1.4 μM) at 4±1 °C for 10 min prior to labeling reactions. The pre-incubated complex (4 μL) was mixed with 40 μL of deuterated PBS on ice and quenched as described above. The final concentrations of iC3b and anti-iC3b mAb during labeling were 0.1 μM and 0.13 μM, respectively. Labeling reactions were performed in duplicate for 10, 100, 1000 and 10000 sec. As a control, C3b that was described not to bind to anti-iC3b mAb was incubated with the mAb, and samples were prepared as for iC3b:anti-iC3b mAb.
Analysis of HX samples by liquid chromatography-mass spectrometry
HX exchange samples were analyzed on a SYNAPT G2S ESI-Q-TOF spectrometer coupled to a nanoACQUITY UPLC system with HDX technology (Waters), using MassLynxTM 4.1 (SCN 916, Waters) for data acquisition. Spectra were acquired in ES+ sensitivity mode (m/z 300–1500) using the following MS parameters: capillary voltage 3 kV, cone voltage 40 V, ion source block temperature 80 °C, desolvation gas flow 600 L/h at 150 °C, and nebulizer gas flow 6 bar. During data acquisition, leucine enkephalin (2 ng/μL in 50% [v/v] acetonitrile and 0.23% [v/v] formic acid) was infused as a lock spray standard through the lock mass spray line at 4 μL/min; three spectra of 0.5 sec were acquired every 20 sec. Samples were loaded on an ACQUITY UPLC R BEH C18 VanGuard Pre-column (130 Å, 1.7 μm, 2.1 x 5 mm, Waters P/N 186003975) through a 50-μL loop and desalted at 300 μL/min for 3 min using 0.23% (v/v) formic acid. Proteins were digested online with immobilized pepsin packed in a guard column (2 mm x 2 cm; C-135B, IDEX, Middleboro, MA). Peptide separation was carried out on an ACQUITY UPLC R BEH C18 analytical column (130 Å, 1.7 μm, 1 x 100 mm; 186002346, Waters) at 40 μL/min using solvents A (0.23% v/v formic acid) and B (0.23% v/v formic acid in acetonitrile). The following gradient was applied: 3% B to 10% B in 0.2 min, to 38.5% B in 19.8 min, to 90% B in 2 min; kept at 90% B for 2 min and returned to initial conditions in 0.2 min.
Identification of C3b- and iC3b-derived peptides
Peptide identification runs of C3b and iC3b (4 pmol per protein) were performed in the ES+ resolution mode using data-dependent acquisition (DDA). MS survey scans were recorded in the m/z 300–1500 Da region for 0.8 s; the three most intense multiply charged ions with >2,000 intensity/sec were chosen for fragmentation. MS/MS scans of the selected ions were acquired in the m/z 100–1200 Da range for 1 sec using a ramped collision energy profile of 18–30 eV. Fragmented precursor ions were excluded from further MS/MS for 20 sec. To increase the sampling depth of co-eluting peptides, a second identification run for each protein was performed by enabling the fixed peak exclusion feature of the software and incorporating an exclusion list of the precursor ions chosen for fragmentation in the first run. The lock mass spray and UPLC parameters were as described above. Peptides were identified using ProteinLynx Global ServerTM (version 3.0.2, Waters) and Proteome Discoverer (version 1.4, Thermo). In both cases, a non-specific enzyme was chosen, and the peptide and fragment tolerances were 10 ppm and 0.8 Da, respectively. For Proteome Discoverer, .raw format data files were converted to .mgf format using MSConvert (23).
Data analysis of labeled HX samples
Mass spectra of the exchange experiments were loaded in DynamX (version 2.0, Waters). The deuterium uptake of individual peptides over time was calculated based on the assigned centroid m/z value of the isotopic cluster envelopes. Deuterium uptake values of individual peptides were normalized to respective experimental values obtained from the analysis of full deuteration controls divided by the deuterium fraction (0.909) in the solvent during the exchange and the theoretical maximum number of exchangeable amides (24). HX data were fitted in stretched exponential curves using the same stretching factor (β, 0.8 ± 0.03) as calculated for predicted reference curves for the case of no protection (21, 25). Structural protection of peptides was quantified through the calculation of protection factors (21).
Synthesis of anti-iC3b mAb epitope peptides
Peptides were synthesized on an automatic Microwave Peptide Synthesizer (Liberty 1; CEM, Matthews, NC) using Rink Amide MBHA resin (Novabiochem; 0.38 mmol/g) and Fmoc/tBu methodology. Fmoc-protected amino acids and coupling reagents were pre-dissolved in DMF (0.2 M). Deprotection of the peptides was performed with piperidine (20%) in DMF (0.1 M HOBt). Each coupling reaction and deprotection step was performed with microwave energy and nitrogen bubbling. Peptide cleavage from the resin and removal of the protective groups of the side chain of the amino acids were carried out with TFA/EDT/TIS/H2O (94:2:2:2 v/v/v/v) for 2.5 h at ambient temperature. The resins were washed with TFA and the filtrates were partially evaporated. Crude products were precipitated with diethyl ether, collected by centrifugation at 14,000 rpm for 15 min, dissolved in H2O and lyophilized. Crude products were purified by HPLC using an XBridge BEH130 C18 column (150 x 4.6 mm, 5 μm; Waters) with an acetonitrile gradient of 5–100% (0.1% formic acid) in 25 min at a flow rate of 1 mL/min. Mass spectra were collected on a MALDI Micro MX (Waters) mass spectrometer using α-cyano-4-hydroxycinammic acid as matrix.
Detection of epitope peptide binding by ELISA
A competitive enzyme-linked immunosorbent assay (ELISA) was developed for detecting the binding activity of synthesized peptides for the anti-human iC3b mAb. iC3b was immobilized on a 96-well polystyrene plate (50 μg/ml, 50 μL). Following blocking of the plate with BSA (1% in PBS, 200 μL), serial dilutions of the peptides or iC3b (0.0056–5.7 μM, 50 μL) were pre-incubated with anti-human iC3b mAb (1:2000, 50 μL) and added to the plate for 1 h at room temperature (RT) to compete with the immobilized iC3b for binding to the mAb. A peroxidase-conjugated polyclonal goat anti-mouse IgG (BioRad) was then added (1:1000, 100 μL) and incubated for 1 h at RT. The plate was developed by adding hydrogen peroxide with 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS, 50 μL) according to manufacturer’s instructions (Roche). Plates were analyzed at 405 nm.
A direct ELISA was developed for detecting the binding activity of each peptide or C3 fragment for the anti-iC3b mAb: The peptides and C3 fragments (blue circle) were immobilized on a 96-well polystyrene plate at 50 μg/ml. The plate was blocked with 1% BSA in PBS and a dilution series of anti-iC3b mAb was added, starting from 1:250 (1 h, room temperature), followed by the addition of a peroxidase-conjugated polyclonal rabbit anti-mouse IgG (1:1000, 1 h, room temperature). The plate was developed as described above.
Western blot analysis of anti-iC3b mAb specificity
Purified C3 fragments (C3b, iC3b, C3dg; 0.25 μg in PBS each) were subjected to electrophoresis (4–15% SDS-PAGE) under reducing and denaturing conditions. Proteins were transferred onto a polyvinylidene difluoride membrane and blocked by incubation for 1 h in PBS containing 5% nonfat dry milk). The membrane was incubated for 1 h at RT with mouse anti-iC3b mAb (1:5000). Following washes, the membrane was incubated with anti-mouse HRP-conjugated secondary antibody (1:2000, #170-6516, BioRad). Bands were developed with HRP substrate (Immobilon, Millipore) and images were acquired using a ChemiDoc system with ImageLabTM software (v5.1, BioRad).
Surface plasmon resonance analysis
All surface plasmon resonance (SPR)-based interaction studies were performed on a Biacore 3000 instrument (GE Healthcare, Piscataway, NJ) at 25 °C using PBS-T (10 mM NaHPO4, 150 mM NaCl, 0.005% Tween 20, pH 7.4) as running buffer.
Two distinct assay formats were employed for analyzing the interaction between the anti-iC3b mAb and C3-derived opsonins. In the first assay setup, opsonins were deposited on a sensor chip surface in physiological orientation. For this purpose, C3b that was site-specifically biotinylated at its thioester moiety (as described before (26)) was captured on three flow cells of a streptavidin-coated chip (Biotin CAPture kit, GE Healthcare) at a concentration of 15 μg/ml to reach densities of 1,400–1,600 resonance units (RU). One C3b surface was converted to iC3b by injection of 0.1 μM factor I and 6 μM factor H for 4 min, whereas another C3b surface was converted to C3dg by 0. 1 μM factor I and 7 μM soluble complement receptor 1 (sCR1; kind gift from Celldex); the surfaces were washed by injecting 2M NaCl for 1 min. A ten-fold serial dilution of the anti-iC3b mAb (0.01–10 μg/mL in PBS-T) was injected over all three surfaces (C3b, iC3b, C3dg) using consecutive injections of 2 min each at a flow rate of 10 μl/min. An unmodified streptavidin surface was used as a reference and the signals were processed using Scrubber (v2.0c; BioLogic, Campbell, Australia). In a reversed assay format, the anti-iC3b mAb was captured on a sensor chip via its Fc part. The surface on a CM5 chip (GE Healthcare) was activated for 7 min according to the manufacturer’s instructions, an anti-mouse IgG Fc antibody (Jackson ImmunoResearch, No. 315-005-046; 20 μg/ml in 10 mM sodium acetate pH 5.0) was injected to reach a density of 8,500 RU before surface deactivation for 7 min. In each injection cycle, anti-iC3b mAb was injected for 5 min to reach ~750 RU, followed by a 2 min injection of C3 fragments (C3b, iC3b or C3dg) at concentrations of 200 nM with a 4-min dissociation phase, and regeneration using 10 mM glycine pH 1.7. Referencing and data processing was performed as described above and SPR signals were normalized by dividing the signals by the molecular weight of each opsonin.
The differential binding of the bacterial inhibitor Efb-C to C3b and iC3b was probed by site-specifically capturing C3b-biotin on two sensor chip surfaces (1,900–2,000 RU) as described above, and injecting Efb-C (N138A mutant reported previously (27)) as six consecutive 1-min injections over a concentration range of 3–100 nM at a flow rate of 10 μl/min. One of the C3b surfaces was subsequently converted to iC3b using factor H and factor I as described above, and the Efb-C injection series was repeated. Since kinetic titration analyses were performed for all analytes, no regeneration step was applied between injections (28). The data sets were processed in Scrubber and superimposed to reveal the effect of iC3b conversion. For kinetic analysis, a two-fold dilution series of Efb-C N138A (6–100 nM) was injected as five consecutive injection of 1 min each at a flow rate of 30 μl/min over the C3b and iC3b surfaces. The processed data sets were analyzed in BIAevaluation (v4.1; GE Healthcare) using a single cycle kinetic analysis model (kindly provided by GE Healthcare (28) assuming Langmuir 1:1 binding to extract association (ka) and dissociation rates (kd) and calculate the binding affinity (KD = kd/ka).
Results
Structural dynamics of C3b and iC3b
Conversion of C3b to iC3b results in dynamic structural changes affecting the ligand accessibility of individual domains and, consequently, the effector profile. We therefore used local HX-MS, a method ideally suited for assessing such rearrangements of large proteins in solution by monitoring the differential exchange of free backbone hydrogens with deuterons (D) over time. This approach has already been successfully used to characterize C3 and its derivatives (29, 30).
We found that incubating the proteins for 2 min on ice at optimized quench conditions (TCEP and guanidine) was essential to achieve high sequence coverage. Using this procedure, we generated 401 peptides spanning 96% of the protein’s length. We were able to analyze 264 peptides (sequence coverage 91%) covering all 12 domains of C3b/iC3b (Fig. 1, Fig. 2B), for their D-content; 252 peptides were common to both proteins, whereas 8 and 4 peptides were unique to C3b (e.g., C3f-containing area) and iC3b, respectively (Fig. 2B). Modified sites were taken into consideration during the analysis, such as the hydrolyzed thioester bond (Cys988, Gln991), C3f-containing peptides in C3b, and shortened peptides produced by cleavage of C3f in iC3b. The two glycosylation sites (Asn67, Asn917 (31)) were found to be fully modified, since peptides in their non-glycosylated form were not detected; for Asn67, heterogeneous oligosaccharide populations were detected corresponding to Man5GlcNAc2 and Man6GlcNAc2, with the latter predominating. Finally, to ensure sample uniformity, both proteins were analyzed for their major polymorphic forms, which result from a single amino acid substitution (p.Arg80Gly) and that have been associated with disease risk (32). Two overlapping peptides for C380R (74–83, 74–87) and one for C380G (74–83) were identified. The percentage of C380G within each protein sample, calculated by normalizing the peak areas, was 24.6% in C3b and 24.2% in iC3b, indicating that both proteins possess similar allotype ratios.
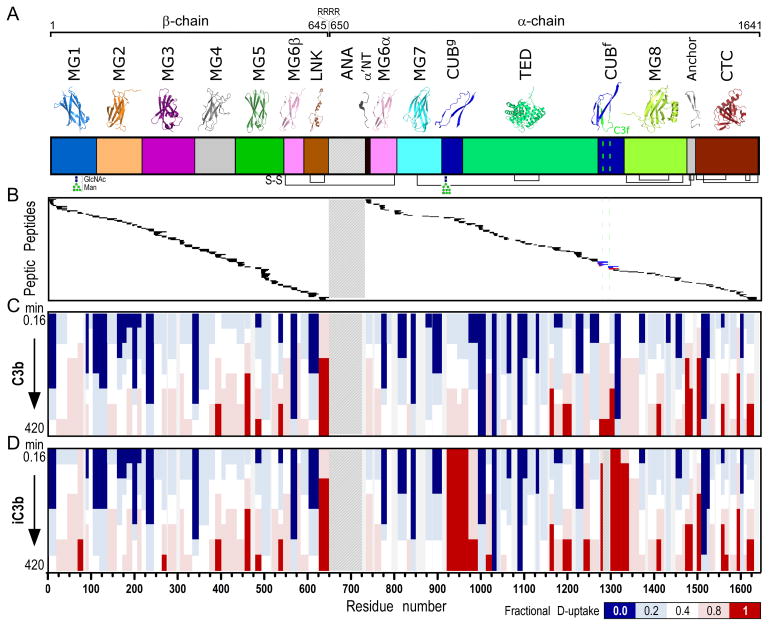
Figure 2. H-to-D exchange protection and heat map of C3b and iC3b.
A. Domain organization of C3 and its activation products (color annotation follows (2)). Ribbon representations of individual domains are shown at the top. C3b is generated upon proteolytic removal of the ANA domain (shown with a crosshatch fill pattern). β- (residues 1–645) and α-chains (residues 650–1641) are covalently linked through a disulfide bond formed between MG6 domain residues Cys537-Cys794. Further release of the heptadecapeptide C3f in the CUBf domain leads to the formation of iC3b (cleavage sites are indicated with a green dashed line). Inter- and intra-domain disulfide bonds are shown at panel bottom with black connecting lines. B. Peptic peptides of C3b and iC3b identified and analyzed for D-content. These correspond to residue numbers as indicated in the x-axis of Fig. 2D. For a detailed view of individual peptides identified and respective peptide boundaries, please refer to Fig. S1. Common peptides identified for both C3b and iC3b are indicated with black. Unique peptides of C3b (indicated with blue) and iC3b (indicated with red) were identified in the C3f-surrounding region. C, D. Heat maps of the exchange observed on the peptide level for C3b and iC3b, respectively. Color-coding is based on the fractional deuterium uptake calculated by normalizing deuterium uptake values to respective values from fully deuterated samples; each value is the average of two replicate experiments. Values for different time points of exchange are depicted; these are 0.16, 0.5, 1.7, 5, 16.7, 50, 166.7 and 420 min (from top to bottom). For simplicity, only non-overlapping peptides are shown.
Proteins were subjected to HX for different periods of time, and the D-content of the generated peptic peptides was measured. Reference curves depicting the exchange behavior at no protection (random coil) were calculated (21) and fit to stretched exponential curves to account for the breadth of the intrinsic chemical exchange rates of amides within peptides with different neighboring amino acid side chains (33) (Fig. S1). For all peptides, uniform populations were detected, suggesting that exchange proceeded in the EX2 regime (34). A heat map of the fractional deuterium uptake for all time points was generated, showing ordered, slow-exchange regions in blue and fast-exchanging regions in red (Fig. 2C–D). Time-resolved exchange profiles were similar for most of the common peptides in C3b and iC3b (173 peptides, Fig. S1); these spanned mainly the MG1–MG8, α’NT, and CTC [i.e., the C-terminal C345c domain (14)] regions. Average deuteration levels in the MG1–MG6 and MG8 domains reached <80–90%, whereas slower-exchanging peptides were detected in the MG7 and TED domains of the α-chain (average deuteration levels <60%). Dynamic exchange behavior was observed in the C-terminal peptides of the β-chain (623–645), supporting the lack of secondary structure seen in the crystal structure (4).
HX-MS reveals a disordered CUB in iC3b
Whereas large parts of the β-chain and the terminal regions of the α-chain remain unchanged between C3b and iC3b, significant differences in relative D-uptake between the two proteins were detected in the CUB domain and in peptides defining the interface between CUB-TED and the MG core (Fig. 2C–D, Fig. 3A, Table S1). The CUB adopts a conserved β-sandwich fold in C3b, with each β-sheet containing four antiparallel strands connected through flexible loops of varying lengths (Fig. 3B). Analysis of the CUB domain from X-ray coordinates of C3b [PDB: 2I07 (4)] revealed a network of 82 hydrogen bonds, with 54 between backbone carbonyls and amide hydrogens (Fig. 3C). Processing of C3b by FI to produce iC3b takes place in two sequential cleavages: first, between residues R1281–S1282 in loop L6 to form iC3b1 and then between residues R1298–S1299 in strand β8 to form iC3b2 (commonly termed iC3b (15)). This process results in liberating C3f (1282–1298), which contains strand β7 and its side loops. As expected, the seven C3f-containing peptides were detected only in C3b, whereas iC3b produced unique peptides containing amino acids flanking the C3f cleavage sites (Fig. 2B); overall, we obtained 92% sequence coverage for the CUB domain.
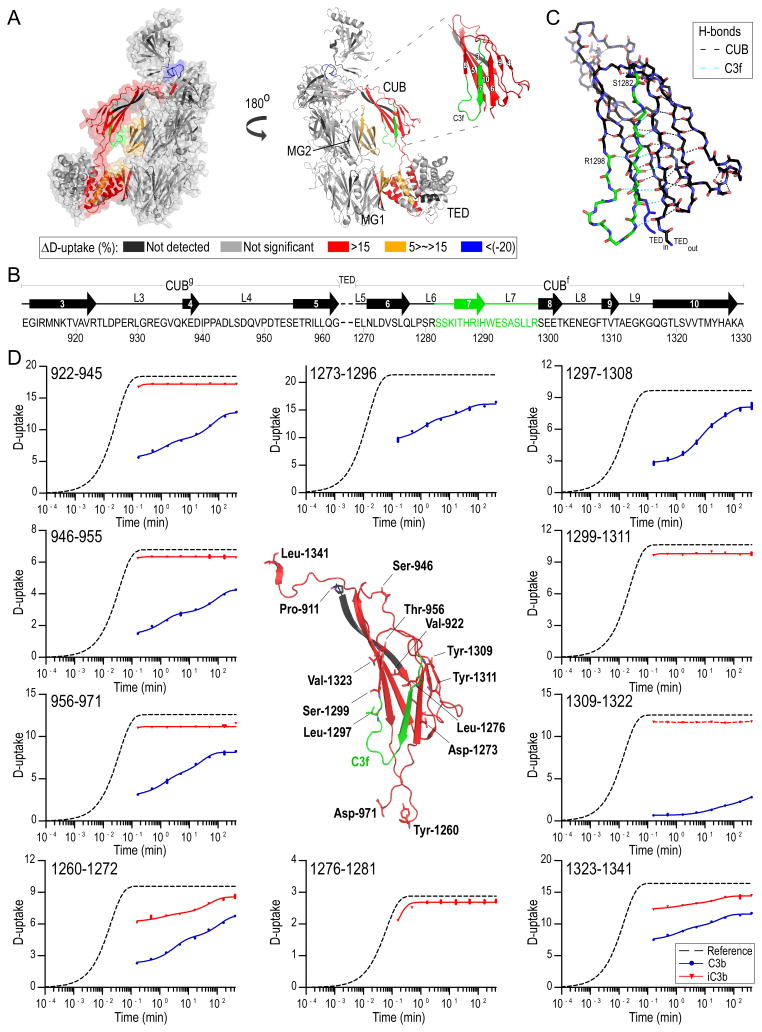
Figure 3. Differential D-uptake between C3b and iC3b.
A. Mapping of HX differences between C3b and iC3b on C3b structure; peptides with ΔD-uptake>5% are mapped on C3b [PDB: 2I07 (4), Table S1]. More exposed regions in iC3b than C3b against D-exchange are colored red, and less exposed are blue. CUB with respective fibronectin type-3 strand numbering is shown in the inset. Prominent differences in the D-exchange are detected for peptides in CUB, whereas marginal but still significant differences are detected on the interface between CUB-TED and the MG core as well as on CUB-TED inter-domain peptides. iC3b is shown to acquire a more dynamic and solvent-exposed conformation. An exception is the 1471–1480 peptide positioned between MG8 and CTC, indicating re-positioning of CTC toward the MG7/MG8 domains. B. Structural organization of the CUB domain; strand numbering follows Fig. 3A. CUBg, TED and CUBf boundaries are depicted. Loops (L) and disordered segments are marked with a solid line. C. Detailed view of the CUB structure. C3f is featured in green, and amino acids denoting the beginning and the end of the peptide are labeled. H-bonds between backbone amide hydrogens and carbonyls are depicted with black dashed lines, and those involving C3f residues with cyan; C3f participates in 9 main-chain hydrogen bonds. D. D-uptake plots of selected CUB domain peptides for C3b (blue) and iC3b (red) (for a complete list of plots, see Fig. S1). For each sequence, a reference curve (for the case of no protection) is calculated and fit to a stretched exponential equation (33). Experimental data points are similarly fit using the same β-factor calculated for the respective reference curves. Amino acids denoting the beginning of measured peptides are labeled, and their main- and side-chains are depicted with sticks. Unique peptides for C3b entailing a part (1297–1308) or the full sequence (1273–1296) of C3f are shown. Unique iC3b peptides containing the newly formed C-terminus (1276–1281) and N-terminus (1299–1311) generated upon cleavage of C3f are depicted. A curve could not be fit for peptide 1309–1322 in iC3b (indicated with a connecting red dashed line).
The most profound increase in D-uptake was observed for CUB peptides in iC3b when compared to C3b, with extensive deuteration noted for all CUB intra-domain peptides even at the earliest time point (10 s) (Fig. 3D, Fig. S1). Calculated protection factors (PF) were ~1–2, indicating a fully disordered domain. Moreover, enhanced HX was observed for peptides connecting CUB with TED (region 1252–1272) and MG8 (1323–1341), likely because of a loss of intra-domain hydrogen-bonded amides in TED and MG8. In C3b, unlike iC3b, several ordered and slow-exchanging segments were detected in CUB with average D-uptake levels reaching <65% over the course of the experiment. The exchange profile of C3f-containing peptides revealed a loosely folded, dynamic region with deuteration levels reaching 90% (Fig. 3B).
Validation of CUB’s distinct conformation in iC3b
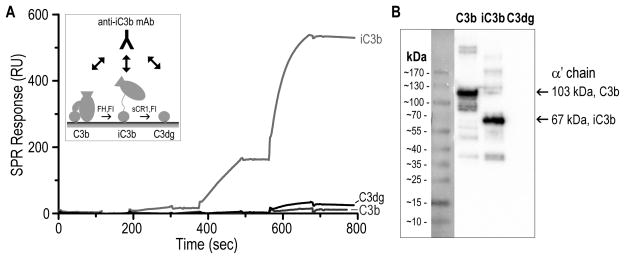
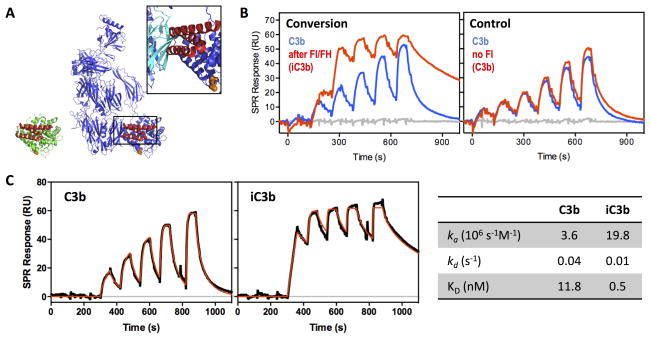
To further validate the proposed structural changes in iC3b, we used a commercial monoclonal anti-human iC3b antibody (clone 013III-1.1.6; hereafter termed anti-iC3b mAb) that had been described as detecting surface-bound iC3b but not C3b or C3dg (35). We used surface plasmon resonance (SPR) to confirm the specificity of this interaction; for this purpose, C3b was site-specifically captured on a sensor chip and converted to iC3b and C3dg via FI and cofactors. Consecutive injections of increasing concentrations of the anti-iC3b mAb showed strong binding and specificity of the antibody for surface-bound iC3b (Fig. 4A); no significant binding was detected for C3b, and the low residual binding observed for C3dg may result from the presence of trace iC3b molecules on the C3dg flow cell. The lack of significant C3dg:anti-iC3b mAb interaction was corroborated by western blotting (Fig. 4B).
Figure 4. Binding specificity of anti-iC3b mAb for C3-derived opsonins.
A. For evaluating the specificity of the anti-iC3b mAb towards surface-bound opsonins, C3b was captured on a SPR sensor chip in physiological orientation via its thioester moiety. Individual C3b surfaces were converted to iC3b and C3dg with factor I (FI) and cofactors FH and sCR1, respectively (45) (insert). A titration experiment was performed on the C3b, iC3b and C3dg surfaces using consecutive injections of increasing anti-iC3b mAb concentrations (0.01–10 μg/ml). Anti-iC3b mAb showed a significant binding affinity for iC3b when compared to C3b and C3dg. The residual binding to C3dg that was observed was most likely due to iC3b residues present on the C3dg surface. B. Purified C3b, iC3b and C3dg were subjected to gel electrophoresis under reducing and denaturing conditions. Under reducing conditions, C3b produces two individual bands corresponding to the αβ- and β-chains, connected via a disulfide bond. Proteins were subsequently analyzed by western blotting using the anti-iC3b mAb. Intense bands detected for C3b and iC3b correspond to the α′-chains of the proteins (103 kDa for C3b and 67 kDa for iC3b, produced upon cleavage of C3f). C3dg did not show any detectable interaction with the anti-iC3b mAb.
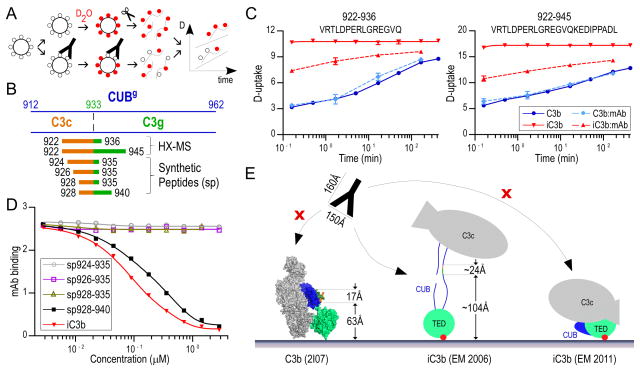
Next, we sought to determine the epitope-bearing region using HX-MS. The anti-iC3b mAb was incubated with C3b and iC3b in slight molar excess, and the peptidic D-uptake profiles of C3b and iC3b in presence of the anti-iC3b mAb were compared to respective D-uptake curves of free proteins in solution (Fig. 5A). Most HX profiles were similar for the proteins alone and in complex with anti-iC3b mAb, except for two overlapping peptides (922–936, 922–945) in CUBg of iC3b spanning the third cleavage site of FI that generates fluid-phase C3c and C3dg (Fig. 5B). These segments are structured in C3b and unstructured in free iC3b but displayed strong protection in the presence of the anti-iC3b mAb (−3 D for 922–936; −6 D for 922–945 at 10 s, Fig. 5C). By subtracting the D-uptake between these two peptides, we deduced that the epitope may involve three residues of the C3dg segment (937–945); however, considering that the anti-iC3b did not bind purified C3dg (Fig. 4), we inferred that the epitope must extend into the C3c segment of iC3b (Fig. 5B).
Figure 5. Validation of neo-epitopes on iC3b using an anti-iC3b mAb.
A. HX-MS workflow for the analysis of C3b and iC3b with and without anti-iC3b mAb. Protein molecules are shown as large circles, water and deuterated water molecules as small black and red circles, respectively, and mAb as a Y-shape cartoon. Incubation of the proteins with the anti-iC3b mAb is expected to protect the epitope-bearing region, resulting in less D-uptake. B. CUBg domain boundaries and peptides bearing the epitope region. The N-terminal of C3g is indicated with a dashed line. Peptides detected by HX-MS to have lower deuterium content in iC3b:anti-iC3b mAb, and synthesized peptides are indicated as linear stretches; these span the C3c/C3g fragments, and respective parts of their sequences are colored accordingly. C. D-uptake plots of two peptides (922–936 and 922–945) that were found to be more protected in iC3b in the presence of the mAb; their profile was unaffected in C3b:anti-iC3b mAb. D. Competition ELISA using anti-iC3b mAb and synthesized peptides. Anti-iC3b mAb was mixed with increasing concentrations of synthesized peptides or purified iC3b (reference), and the mixtures were transferred to wells of an ELISA plate coated with iC3b. A decrease in signal indicates ligand binding to the mAb, preventing an interaction of the mAb with coated iC3b. E. Schematic of anti-iC3b mAb interactions with opsonins C3b and iC3b. The epitope-bearing region (928–940) as defined by HX-MS and ELISA experiments is depicted on the C3b crystal structure [PDB: 2I07 (4)]. This partially structured region in C3b and steric hindrance provided by the structure itself and the surface prevent C3b:anti-iC3b mAb interactions and constitute the antibody specific for iC3b. The anti-iC3b mAb (“mAb” in the figure) has been drawn in ~1:2.5 scale; typical dimensions are depicted. The CUB and TED domains are featured in blue and lime green, respectively, C3c and C3g segments in orange and green, and the thioester bond in red.
Lacking further overlapping peptides in this area, we proceeded to validate and refine the epitope by synthesizing a series of overlapping peptides of variable lengths, including residues spanning the C3c-C3g interface (Fig. 5B), and screening them for binding activity by competitive ELISA (Fig. 5D). Increasing concentrations of the peptides were incubated with the anti-iC3b mAb and then added to an iC3b-coated plate. Of this peptide panel, only peptide 928–940 showed competing activity for the iC3b:anti-iC3b mAb interaction comparable to that of soluble iC3b (Fig. 5D). This specificity was confirmed in a direct ELISA, in which anti-iC3b mAb was added to plates coated with the peptides, confirming that the epitope-bearing region lies on the C3c-C3g-connecting segment (residues 928–940) of iC3b (Fig. S2). Of note, and in contrast to the SPR analysis, a strong interaction between the anti-iC3b mAb and C3b was observed in the direct ELISA (Fig. S2), likely caused by partial denaturation of C3b when directly coated on plastic surfaces and consistent with the antibody’s gain in immunoreactivity toward C3b after protein denaturation in western blotting (Fig. 4B). However, the antibody may have residual reactivity against solution-phase C3b, since the proposed epitope lies on the outer surface of the CUB domain in C3b, which is partially folded and solvent-exposed (Fig. S3, (4)). Whereas the anti-iC3b mAb did not bind thioester-immobilized C3b (Fig. 4A), marked binding activity for solution-phase C3b was seen in a reversed SPR assay in which the anti-iC3b mAb was captured via its Fc segment (Fig. S3). These results suggest that the orientation of C3-derived opsonins may contribute to anti-iC3b mAb selectivity. For C3b, the distance of epitope-encompassing residues E928 and I940 from a surface was estimated at ~63 Å and ~80 Å, respectively (Fig. 5E). A disordered CUB domain in iC3b, with no internal hydrogen bonding as evidenced by HX-MS, would position these residues up to ~104–128 Å above the surface (~2 Å per amino acid (16)) and provide the space required for the iC3b:anti-iC3b mAb interaction to occur. In contrast, the folded CUB domain suggested by one EM study (17) should position the epitope closer to the surface, or even occlude it, and would hinder the iC3b:anti-iC3b mAb interaction (Fig. 5E).
The CUB-TED region in iC3b is distant from the MG core
Beyond effects on the domain itself, a disordered CUB in iC3b is expected to influence the orientation and accessibility of the opsonin. Indeed, all available X-ray structures of C3b show the TED in close proximity to the MG core, with CUB acting as a positioning element (4, 9–11, 14). The MG-CUB-TED arrangement is considered an important functional element of C3b, although some studies suggest higher flexibility in this interface (29, 30). To determine whether the MG core contacts the TED domain, we compared the exchange profiles of C3b and iC3b peptides lying on the interface between the CUB-TED region and MG core. Disorganization of the CUB domain is thought to alter solvent accessibility in MG2, which is in close contact with CUB and at the immediate interface between MG1 of the core and TED. Indeed, overlapping MG2 peptides (143–158, 143–164, 159–176, 165–176, 187–196, 187–199) containing residues that are engaged in polar contacts with CUB residues (P152:R957, E175:R915, Y187:A1294) in the C3b structure showed higher D-uptake in iC3b. Moreover, peptides lying on the TED-MG1 interface (35–47, 74–87, 94–100 of MG1; 1012–1024, 1012–1025, 1012–1026 of TED) showed higher D-uptake in iC3b, with TED peptide differences predominating (Fig. 3A, Fig. S1). Only subtle differences in these TED peptides were detected in a separate HX-MS experiment in which iC3b was compared to a C3d construct expressed in E. coli (data not shown) (22). The results suggest that in iC3b, TED is indeed distant from MG1, making TED’s solvent accessibility more similar to that of C3d than C3b. Of note, the exchange profile of peptide 74–83, characteristic of the major polymorphic form C380G, was similar for the two proteins and almost reached full deuteration levels during the experiment; in contrast, 74–87 (C380R) was more protected in C3b (−1 D in C3b at 10s vs. iC3b), indicating that for both proteins, the MG1-TED interface in C380G is more like iC3b in C380R.
We used SPR to further validate the distal MG1-TED interface in iC3b by conducting ligand binding studies with the bacterial inhibitor Efb. The immune evasion protein Efb from S. aureus recognizes a unique binding site on TED/C3d (22). In C3b, binding of the inhibitory domain of Efb (i.e., Efb-C) to TED is sterically restricted by the proximal MG1 domain due to the arrangement of the of the TED-CUB-MG system; Efb-C can still bind but with a slow association rate (Fig. 6A, (30)). Injection of an Efb-C derivative onto surface-immobilized C3b and iC3b showed stronger binding to iC3b (Fig. 6B), with differences from C3b in both kinetic association (~5-fold higher) and dissociation (~4-fold lower) rates (Fig. 6C). The higher association rate observed for iC3b is likely due to enhanced accessibility of the Efb-C binding site on the TED domain following CUB disorganization, whereas the slower dissociation rate suggests less steric disturbance and/or displacement of the TED:Efb-C complex by the MG core. The differential binding of Efb-C to C3b and iC3b thereby supports the finding that disordering of CUB in iC3b has consequences for the arrangement of MG1 and TED.
Figure 6. Differential accessibility of Efb-C binding sites in C3b and iC3b.
A. Crystal structure of C3d:Efb-C (N138A mutant; PDB 3D5R; (27)) with C3d in green and Efb-C in red on the left and a model in which the TED domain of C3b (blue; PDB 2I07; (4)) is aligned with C3d. The insert shows the steric clash of Efb-C with the MG1 domain of C3b (highlighted in cyan). B. In agreement with the hypothesis of a disordered and more flexible CUB domain in iC3b, Efb-C binds more strongly to surface-immobilized iC3b than to C3b, as determined by SPR. In the semi-quantitative experiment, thioester-biotinylated C3b was captured on a streptavidin-coated sensor chip and a titration was performed with Efb-C (3–100 nM) before and after conversion of one C3b surface to iC3b to demonstrate that the observed increase in activity was indeed due to the iC3b transformation with loss of steric restriction around the Efb-C binding site. C. In order to quantitate the differential interaction profile, a kinetic titration was performed on the C3b and iC3b surfaces using consecutive injections of Efb-C at increasing concentration (6–100 nM), and the resulting SPR curves (black) were fitted to a Langmuir 1:1 binding model (red lines) to determine kinetic rate constants and binding affinity (table on the right). iC3b showed a >20-fold higher affinity for Efb-C when compared to C3b, caused by a ~5-fold higher association rate and a ~4-fold slower dissociation rate. Of note, a single amino acid mutant of Efb-C (i.e., N138A) that shares the same binding mode as wildtype Efb-C but features lower complex stability (KD of 19.8 vs. 1.5 nM; (27)) was used to facilitate the experiment and avoid harsh regeneration conditions.
Discussion
iC3b is a key immune effector whose high-resolution structure is not yet elucidated, and the two reported EM structures show conflicting results, with the CUB-TED-MG core arrangement being the focus of the discrepancy (16, 17). Here we employed HX-MS, a powerful technique for exploring conformational dynamics and structural perturbations of proteins in solution, to elucidate the structure of iC3b at peptide resolution. In contrast to orthogonal methods such as NMR, this technique can be applied to the large plasma proteins common to the complement cascade. The method’s accuracy relies critically on sequence coverage provided by the peptide pool from peptic proteolysis; we were able to improve on previous studies to reach >96% coverage (29, 30). As expected, we found that most of C3b and iC3b behaved similarly regarding HX, but we identified localized areas of increased exchange in iC3b in the MG1 and MG2 domains of the β-chain, and TED. By far the biggest increase in HX was detected for the CUB domain.
CUB domains are structural motifs of ~110 residues, widely found in functionally diverse, multi-domain proteins. They often share >25% sequence identity and conserved hydrophobicity patterns typical of antiparallel, complex β-sheets (36). The CUB in C3 is rather distinct, being composed of two distant sequence-intertwined sub-domains (CUBg and CUBf) separated by TED inserted in the L5 loop. Moreover, it lacks the four cysteine residues that are highly conserved in CUB domains and has only 4 of 17 conserved hydrophobic amino acid residues that define the CUB signature (37). Our HX-MS results for C3 CUB reveal regions that are structured in C3b, with protected amide groups within β-strands and dynamic regions that mainly include loops and C3f-containing peptides. The flexibility observed in C3f and adjacent loops is likely required to accommodate FI binding and later cleavage at R1281–S1282 and R1298–S1299 (20). The overall flexibility of the CUB in C3b may also account for the poor electron density of this domain in the C3 and C3b crystal structures that have required addition by modeling (2, 4, 9). Cleavage of C3f and generation of iC3b produced rapid HX in all the amide protons in CUB, with PF values suggesting random coil conformation (21). In contrast, PFs calculated for C3b amides varied significantly and were up to 104-fold higher, indicating conversion of CUB from an ordered to a highly disordered polypeptide state devoid of hydrogen bonds after C3f release (21). C3f forms part of CUB’s comparatively weak hydrophobic core, and its cleavage is therefore thought to disturb its overall folding, a concept supported by the lack of stabilizing disulfide bonds in the C3b CUB.
Our finding of a disordered CUB in iC3b is further corroborated by antibody recognition data. The presentation of an unstructured region of >50 consecutive residues offers an opportunity for new linear epitopes. Indeed, our results reveal that the neo-epitope recognized by a commercial anti-iC3b mAb (clone 013III-1.1.6 (35)) is linear, resides in the CUB, and is likely located in a sequence spanning C3c-C3g (residues 928–940) that harbors the third FI cleavage site (15). Although the epitope appears to be partially exposed in C3b, antibody accessibility is likely to be sterically unfavorable for surface-bound C3b opsonin. After conversion by FI, the epitope becomes fully exposed, as seen in the subsequent recognition of surface-bound iC3b. Further FI-mediated conversion to fluid-phase C3c and C3dg cleaves the epitope, explaining the lack of specificity for these downstream fragments. It must be noted, however, that the selectivity of clone 013III-1.1.6 for iC3b may not be absolute, since soluble or plastic-bound, partially denatured C3b may be recognized by the mAb (Figs. 4, S2).
Our results further show that CUB unfolding does not significantly affect the structure of the C3c or C3d/TED segments. TED is an α-barrel structure composed of an inner layer of six parallel helices surrounded by an outer layer of six more helices (8), and our data suggest a highly stable hydrogen-bonded network with PF values ranging from 102–105 over the course of the experiment. These data fit well with the available crystal structures of C3 fragments before iC3b generation (i.e., C3b) and after its degradation (i.e., C3c, C3d), which show few structural differences.
Importantly, our data support the hypothesis that CUB conversion translates into a changed arrangement of the C3c and C3d segments in iC3b with consequences for downstream signaling via CRs. The higher D-uptake of peptides exposed at the TED-MG1 and CUB-MG2 interfaces and the different binding affinities observed between Efb-C and C3b/iC3b can be explained by more dynamic conformational ensembles in iC3b. The available crystal structures of C3b position the TED and CUB domains adjacent to the MG core, held together by polar contacts (4, 9). This arrangement is corroborated by EM studies indicating that the intact MG1/TED interface is the most frequently occupied conformation (16). Conversely, other EM studies suggest that C3b occupies various conformations in solution, with the TED domain in different positions around the molecule (17, 38); small-angle X-ray scattering data feature TED in two different orientations, both adjacent to the MG core (30, 39), and X-ray and neutron scattering data even propose a uniform population with TED extending away from MG1 (40). Similarly, there are discrepancies in the EM-based data concerning the arrangement of iC3b; some studies suggest a highly flexible positioning after conversion of CUB (16), whereas one indicates that TED moves into a compact position at the MG core and the CTC domain, with CUB adjacent (17). The solution HX data in our study show areas of protection in surface-exposed regions of MG1, MG2, and TED in C3b, correlating well with the conformation in the crystal structure being at least partially occupied. Moreover, in iC3b, we find a complete loss of protection in CUB and no new areas of increased protection in TED, CTC or the MG core, corroborating the hypothesis that an extended and flexible, rather than a more compact, arrangement defines the predominant state of iC3b in solution. In this HX-supported model, some areas of TED would be masked in C3b and become more accessible to binding events in iC3b. Indeed, a disordered CUB has been proposed to alleviate TED of steric clashes and allow it to gain reactivity toward CR2 (12) and CR3 (13), both of which recognize major binding sites on the C3d segment. Despite the structural and functional similarity between CR3 and CR4, the major binding site for CR4 on iC3b was found to lie on MG3/MG4 rather than TED. Although this region is similar in the C3b and C3c crystal structures (2, 4), the selectivity of iC3b for CR4 is attributed to a different orientation of the MG3/MG4 interface that may make it more accessible to the receptor (17, 18). We obtained full coverage in this region, detecting 41 overlapping peptides (Fig. S1), but observed no differences between the D-uptake profiles of the peptides from C3b and iC3b, indicating similar hydrogen bonding and solvent accessibility in MG3/MG4 for C3b and iC3b. Although we cannot fully exclude the possibility that different H-bonds are formed within the same peptide, with the overall number of hydrogen bonds remaining constant, it nevertheless appears unlikely, since such changes would be reflected in at least slight D-uptake differences. Additional factors, such as a secondary binding site and/or greater accessibility and favorable positioning of the C3c segment for CR4 binding after CUB cleavage may contribute to the receptor’s selectivity for iC3b (18). Even in the case of CR3, the regained flexibility between TED and the MG core of iC3b may enable a dynamic arrangement that allows a more complex binding mode involving additional contact sites as previously postulated in literature (41).
Finally, a disordered CUB and destabilized TED-CUB-MG positioning also explain the differential binding of C3 convertase components and regulators. For example, CUB is part of the binding site for FB on C3b, allowing its initial binding and conformational conversion to enable activation by FD (11, 42). Also, an ordered CUB in C3b contributes to the interaction with regulators so that they interfere with the binding of FB and its Bb fragment (decay acceleration activity) and/or form a common binding pocket for FI to enable the cleavage to iC3b and C3dg (cofactor activity) (10, 14). In this context, it may be presumed that binding of CR1/CD35, the only cofactor to mediate the third cleavage on CUB, to iC3b may be more complex than for C3b (20) and may involve an induced-fit process in which CR1 induces partial refolding of the CUB that would allow binding of FI and cleavage at position R932–E933 to generate C3c and C3dg.
It has to be noted that our findings do not exclude the potential coexistence of distinct conformers, including compact forms, of iC3b in solution or on surfaces. Minor or very short-lived states of the protein may not be detected by HX-MS due to limitations concerning sensitivity or temporal resolution of the method, respectively. Indeed, conformational dynamics has been attributed to several C3 fragments and may well contribute to the broad functional spectrum of C3-derived opsonins (3, 4, 16, 30). Particularly in the case of surface-attached iC3b, the local molecular environment may further influence the distribution of different iC3b conformers. For example, two distinct forms of surface-deposited iC3b with differential binding to complement receptors and mAb’s have been postulated previously (43) but not further characterized. Based on our experiments, the extended, flexible form of iC3b appears to be dominant at least in solution and may drive the adaptability of this immune effector.
Overall, this work sheds light on the structural characteristics of iC3b that have thus far remained elusive to crystallography and conflicting in EM studies (16, 17). Our HX-MS data, supported by antibody and ligand binding experiments, demonstrate that the CUB domain in iC3b is devoid of secondary structure, leading to high flexibility, destabilization of the relative positioning of domain segments, and greater accessibility of the TED/C3d domain when compared to C3b. Also, we found no evidence for significant conformational changes within the C3c and C3d segments themselves. The resulting model explains the selectivity of a unique commercial anti-iC3b mAb and functional differences between C3b and iC3b concerning convertase formation, regulator binding and receptor signaling. Thus, our structural characterization of the protein at a peptide resolution provides molecular insight into the protein’s activity and selectivity.
Supplementary Material
Acknowledgments
We thank Dr. Deborah McClellan for her editorial assistance.
Footnotes
This work was supported by grants from the National Institutes of Health (AI068730, AI030040), the National Science Foundation (no. 1423304) and the European Community’s Seventh Framework Programme (no. 602699; DIREKT)
Conflict of Interest Statement
The authors state no conflict of interest.
References
- 1.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janssen BJC, Huizinga EG, Raaijmakers HCA, Roos A, Daha MR, Nilsson-Ekdahl K, Nilsson B, Gros P. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 3.Ricklin D, Reis ES, Mastellos DC, Gros P, Lambris JD. Complement component C3 – The “Swiss Army Knife” of innate immunity and host defense. Immunol Rev. 2016;274:33–58. doi: 10.1111/imr.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janssen BJC, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 5.Roozendaal R, Carroll MC. Complement receptors CD21 and CD35 in humoral immunity. Immunol Rev. 2007;219:157–166. doi: 10.1111/j.1600-065X.2007.00556.x. [DOI] [PubMed] [Google Scholar]
- 6.Underhill DM, Ozinsky A. Phagocytosis of microbes: complexity in action. Annu Rev Immunol. 2002;20:825–852. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 7.Gaither TA, Vargas I, Inada S, Frank MM. The complement fragment C3d facilitates phagocytosis by monocytes. Immunol. 1987;62:405–411. [PMC free article] [PubMed] [Google Scholar]
- 8.Nagar B, Jones RG, Diefenbach RJ, Isenman DE, Rini JM. X-ray crystal structure of C3d: A C3 fragment and ligand for complement receptor 2. Science. 1998;280:1277–1281. doi: 10.1126/science.280.5367.1277. [DOI] [PubMed] [Google Scholar]
- 9.Wiesmann C, Katschke KJ, Yin J, Helmy KY, Steffek M, Fairbrother WJ, McCallum SA, Embuscado L, DeForge L, Hass PE, van Lookeren Campagne M. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature. 2006;444:217–220. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Wu YQ, Ricklin D, Janssen BJC, Lambris JD, Gros P. Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat Immunol. 2009;10:728–733. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forneris F, Ricklin D, Wu J, Tzekou A, Wallace RS, Lambris JD, Gros P. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science. 2010;330:1816–1820. doi: 10.1126/science.1195821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Elsen JMH, Isenman DE. A crystal structure of the complex between human complement receptor 2 and its ligand C3d. Science. 2011;332:608–611. doi: 10.1126/science.1201954. [DOI] [PubMed] [Google Scholar]
- 13.Bajic G, Yatime L, Sim RB, Vorup-Jensen T, Andersen GR. Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3. PNAS. 2013;110:16426–16431. doi: 10.1073/pnas.1311261110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forneris F, Wu J, Xue X, Ricklin D, Lin Z, Sfyroera G, Tzekou A, Volokhina E, Granneman JCM, Hauhart R, Bertram P, Liszewski MK, Atkinson JP, Lambris JD, Gros P. Regulators of complement activity mediate inhibitory mechanisms through common C3b-binding mode. EMBO J. 2016;35:1133–1149. doi: 10.15252/embj.201593673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison RA, Lachmann PJ. Novel cleavage products of the third component of human complement. Mol Immunol. 1980;17:219–228. doi: 10.1016/0161-5890(80)90074-7. [DOI] [PubMed] [Google Scholar]
- 16.Nishida N, Walz T, Springer TA. Structural transitions of complement component C3 and its activation products. PNAS. 2006;103:19737–19742. doi: 10.1073/pnas.0609791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alcorlo M, Martínez-Barricarte R, Fernández FJ, Rodríguez-Gallego C, Round A, Vega MC, Harris CL, de Cordoba SR, Llorca O. Unique structure of iC3b resolved at a resolution of 24 Å by 3D-electron microscopy. PNAS. 2011;108:13236–13240. doi: 10.1073/pnas.1106746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Yu Y, Mi LZ, Walz T, Springer TA. Molecular basis for complement recognition by integrin αXβ2. PNAS. 2012;109:4586–4591. doi: 10.1073/pnas.1202051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isenman DE. Conformational changes accompanying proteolytic cleavage of human complement protein C3b by the regulatory enzyme factor I and its cofactor H. Spectroscopic and enzymological studies. J Biol Chem. 1983;258:4238–4244. [PubMed] [Google Scholar]
- 20.Roversi P, Johnson S, Caesar JJE, McLean F, Leath KJ, Tsiftsoglou SA, Morgan BP, Harris CL, Sim RB, Lea SM. Structural basis for complement factor I control and its disease-associated sequence polymorphisms. PNAS. 2011;108:12839–12844. doi: 10.1073/pnas.1102167108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammel M, Sfyroera G, Ricklin D, Magotti P, Lambris JD, Geisbrecht BV. A structural basis for complement inhibition by Staphylococcus aureus. Nat Immunol. 2007;8:430–437. doi: 10.1038/ni1450. [DOI] [PubMed] [Google Scholar]
- 23.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak MY, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotech. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayne L. Chapter thirteen - Hydrogen exchange mass spectrometry. Methods Enzymol. 2016;566:335–356. doi: 10.1016/bs.mie.2015.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connelly GP, Bai Y, Jeng M-F, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 26.Sarrias MR, Franchini S, Canziani G, Argyropoulos E, Moore WT, Sahu A, Lambris JD. Kinetic analysis of the interactions of complement receptor 2 (CR2, CD21) with its ligands C3d, iC3b, and the EBV glycoprotein gp350/220. J Immunol. 2001;167:1490–1499. doi: 10.4049/jimmunol.167.3.1490. [DOI] [PubMed] [Google Scholar]
- 27.Haspel N, Ricklin D, Geisbrecht BV, Kavraki LE, Lambris JD. Electrostatic contributions drive the interaction between Staphylococcus aureus protein Efb-C and its complement target C3d. Protein Sci. 2008;17:1894–1906. doi: 10.1110/ps.036624.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlsson R, Katsamba PS, Nordin H, Pol E, Myszka DG. Analyzing a kinetic titration series using affinity biosensors. Anal Biochem. 2006;349:136–147. doi: 10.1016/j.ab.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 29.Schuster MC, Ricklin D, Papp K, Molnar KS, Coales SJ, Hamuro Y, Sfyroera G, Chen H, Winters MS, Lambris JD. Dynamic structural changes during complement C3 activation analyzed by hydrogen/deuterium exchange mass spectrometry. Mol Immunol. 2008;45:3142–3151. doi: 10.1016/j.molimm.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Ricklin D, Hammel M, Garcia BL, McWhorter WJ, Sfyroera G, Wu YQ, Tzekou A, Li S, Geisbrecht BV, Woods VL, Lambris JD. Allosteric inhibition of complement function by a staphylococcal immune evasion protein. PNAS. 2010;107:17621–17626. doi: 10.1073/pnas.1003750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirani S, Lambris JD, Müller-Eberhard HJ. Structural analysis of the asparagine-linked oligosaccharides of human complement component C3. Biochem J. 1986;233:613–616. doi: 10.1042/bj2330613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris CL, Heurich M, Cordoba SRd, Morgan BP. The complotype: dictating risk for inflammation and infection. Trends Immunol. 2012;33:513–521. doi: 10.1016/j.it.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chetty PS, Mayne L, Lund-Katz S, Stranz D, Englander SW, Phillips MC. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. PNAS. 2009;106:19005–19010. doi: 10.1073/pnas.0909708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke J, Itzhaki LS. Hydrogen exchange and protein folding. Curr Opin Struct Biol. 1998;8:112–118. doi: 10.1016/s0959-440x(98)80018-3. [DOI] [PubMed] [Google Scholar]
- 35.Tamerius JD, Pangburn MK, Müller-Eberhard HJ. Detection of a neoantigen on human C3bi and C3d by monoclonal antibody. J Immunol. 1985;135:2015–2019. [PubMed] [Google Scholar]
- 36.Bork P, Beckmann G. The CUB domain: A widespread module in developmentally regulated proteins. J Mol Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- 37.Romero A, Romao MJ, Varela PF, Kolln I, Dias JM, Carvalho AL, Sanz L, Topfer-Petersen E, Calvete JJ. The crystal structures of two spermadhesins reveal the CUB domain fold. Nat Struct Mol Biol. 1997;4:783–788. doi: 10.1038/nsb1097-783. [DOI] [PubMed] [Google Scholar]
- 38.Alcorlo M, López-Perrote A, Delgado S, Yébenes H, Subías M, Rodríguez-Gallego C, Rodríguez de Córdoba S, Llorca O. Structural insights on complement activation. FEBS J. 2015;282:3883–3891. doi: 10.1111/febs.13399. [DOI] [PubMed] [Google Scholar]
- 39.Ricklin D, Tzekou A, Garcia BL, Hammel M, McWhorter WJ, Sfyroera G, Wu YQ, Holers VM, Herbert AP, Barlow PN, Geisbrecht BV, Lambris JD. A molecular insight into complement evasion by the staphylococcal complement inhibitor protein family. J Immunol. 2009;183:2565–2574. doi: 10.4049/jimmunol.0901443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez E, Nan R, Li K, Gor J, Perkins SJ. A revised mechanism for the activation of complement C3 to C3b: a molecular explanation of a disease-associated polymorphism. J Biol Chem. 2014;290:2334–2350. doi: 10.1074/jbc.M114.605691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taniguchi-Sidle A, Isenman DE. Interactions of human complement component C3 with factor B and with complement receptors type 1 (CR1, CD35) and type 3 (CR3, CD11b/CD18) involve an acidic sequence at the N-terminus of C3 alpha’-chain. J Immunol. 1994;153:5285–5302. [PubMed] [Google Scholar]
- 42.Rooijakkers SHM, Wu J, Ruyken M, van Domselaar R, Planken KL, Tzekou A, Ricklin D, Lambris JD, Janssen BJC, van Strijp JAG, Gros P. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat Immunol. 2009;10:721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsson UR, Funke L, Nilsson B, Ekdahl KN. Two conformational forms of target-bound iC3b that distinctively bind complement receptors 1 and 2 and two specific monoclonal antibodies. Ups J Med Sci. 2011;116:26–33. doi: 10.3109/03009734.2010.528465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bajic G, Yatime L, Klos A, Andersen GR. Human C3a and C3a desArg anaphylatoxins have conserved structures, in contrast to C5a and C5a desArg. Protein Sci. 2013;22:204–212. doi: 10.1002/pro.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt CQ, Bai H, Lin Z, Risitano AM, Barlow PN, Ricklin D, Lambris JD. Rational engineering of a minimized immune inhibitor with unique triple-targeting properties. J Immunol. 2013;190:5712–5721. doi: 10.4049/jimmunol.1203548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.