Abstract
Ranked among the top10 infectious causes of death worldwide, group A Streptococcus (GAS) causes small- and large-scale outbreaks, depending on the trigger as transmission of a GAS strain or expansion of predominant clones. In China, GAS infections other than scarlet fever are not notifiable. In Shanghai, an epidemiological investigation was initiated after two successive severe pneumonia cases with one death in a digital factory, from where outbreaks are less widely reported. The investigation was performed using emm typing, pulsed-field gel electrophoresis (PFGE) typing, superantigen profiling, and genome analysis. This enabled characterization of relatedness among the outbreak isolates and identification of the mobile genetic elements present. Among 57 patients with respiratory symptoms investigated in the factory, emm5 GAS strains were isolated from 8 patients. The eight GAS infection cases comprising one fatal severe pneumonia case, six influenza-like illness cases, and one pharyngitis case. Two risk factors were identified: adult with an age of 18–20 years and close contact with a GAS patient or carrier. GAS attack rate was 14.0% (8/57), and GAS carriage rate was probably around 2.7% (14/521) based on surveys in two nearby districts. All the 10 outbreak associated isolates were assigned to emm5 and sequence type ST-99 (emm5/ST-99), harbored superantigen genes speC, speG, and smeZ, and were assigned to two similar PFGE patterns (clones). Among the outbreak associated isolates, all carried ermA with resistance to erythromycin and inducible resistance to clindamycin, and eight (80%) carried a tetM gene with resistance to tetracycline. Among the 14 carriage isolates, 12 were emm12/ST-36, and 2 were emm1/ST-28, all with superantigen genes speC, speG, ssa, and smeZ. All the carriage isolates harbored ermB and tetM with resistance to erythromycin, clindamycin, and tetracycline. Genome analysis showed the two outbreak clones were closely related and possessed new prophages carrying virulence gene sdc and antibiotic resistance genes of ermA and tetM, which were not found in the emm5 reference strain Manfredo. This is the first report of a GAS outbreak in this type of workplace. The outbreak was caused by two closely related emm5 clones that differed from the predominant emm types circulating in China.
Keywords: Streptococcus pyogenes, emm5, factory, outbreak, pulsed-field gel electrophoresis (PFGE), whole genome sequence
Introduction
Streptococcus pyogenes, one of the top10 infectious causes of death worldwide (Excler and Kim, 2016), is also known as group A Streptococcus (GAS). GAS is responsible for at least 517,000 deaths each year (Carapetis et al., 2005). It can cause a broad spectrum of diseases, from superficial infections (e.g., pharyngitis and impetigo) to invasive infections (e.g., necrotizing fasciitis and streptococcal toxic shock syndrome), and to immune-mediated diseases (e.g., acute rheumatic fever and rheumatic heart disease).
Group A Streptococcus is well established as a cause of outbreaks, which can occur not only in a school, a military camp or a hospital but also throughout a city or even a country (Wasserzug et al., 2009; Chen et al., 2012; Dooling et al., 2013; Ben Zakour et al., 2015; Cornick et al., 2017). Small-scale GAS outbreaks can be caused by transmission of a GAS strain to vulnerable individuals, whether by person-to-person contact in crowded settings or from a common source (Walker et al., 2014). By means of recognizing the outbreak strain and comparing it with isolates from contacts and the environment, we can analyze the outbreak source, mode of transmission and risk factors; subsequently the appropriate prevention measures can be implemented (Raymond et al., 2005). Large-scale GAS outbreaks can occur with the emergence of predominant clones either as a result of the horizontal acquisition of DNA encoding toxins or antibiotic resistance determinants, or through more subtle genetic changes (Walker et al., 2014). We can take the ongoing outbreak of scarlet fever in China for an example. Scarlet fever, the only notifiable disease among all the GAS infections in China (Yang et al., 2013b), has been epidemic in Hong Kong and mainland China since 2011, with an incidence of 21.7–31.4/100,000 population (Lau et al., 2012; Yang et al., 2013b). This large outbreak is caused mainly by emm12 GAS, and typically infects children attending kindergarten and school (97%) (Chen et al., 2012). The original emergence of the scarlet fever emm12 clones in Hong Kong is thought to have been associated with acquisition of toxicity and multidrug resistance introduced by newly identified mobile genetic elements (MGEs) ICE-emm12 variants, ΦHKU.vir, and ΦHKU.ssa (Davies et al., 2015). The integrative and conjugative element (ICE) ICE-emm12 carries the macrolide-resistance gene ermB and tetracycline-resistance gene tetM; the prophage ΦHKU.vir encodes superantigens SSA and SpeC, and the DNase Spd1 (Tse et al., 2012); ΦHKU.ssa encodes superantigen SSA. Moreover, variants of ICE-emm12 and ΦHKU.vir were found to be transferred horizontally between emm1 and emm12 strains. This conferred enhanced virulence and multidrug resistance in emm1 strains and elevated their proportion in scarlet fever isolates (Ben Zakour et al., 2015). Against this background, it was considered appropriate to analyze whether the outbreak strains carry MGEs associated with toxin production and antimicrobial resistance.
Several molecular typing methods have been used to compare the outbreak strain with isolates from contacts and the environment in investigations of GAS outbreaks. One method is emm typing. The emm type has been used widely as an epidemiological marker in GAS surveillance and its distributions varied in high- and low-income countries (McMillan et al., 2013). The method is based on the variability of N-terminal region of the emm gene, the encoding gene of the matrix (M) protein which is a surface protein, vaccine antigen, and major virulence factor of GAS (McMillan et al., 2013). At least 248 emm types have been reported (Beall, 2014). Another method is pulsed-field gel electrophoresis (PFGE) typing, which has been considered to be among the “gold standard” tools used to investigate the clonal relatedness in epidemic surveys and to study outbreaks (Vitali et al., 2015). PFGE involves digesting the chromosomal DNA with restriction endonucleases that cleave infrequently, acquiring ‘fingerprint’ patterns by electrophoresis in pulsed field, and comparing the DNA restriction patterns of the isolates with one another to determine their relatedness (Tenover et al., 1995). The third method is superantigen profiling. Eleven superantigen-encoding genes have been reported in GAS, including three located in the chromosome (speG, speJ, and smeZ) and eight in prophages (speA, speC, speH, speI, speK, speL, speM, and ssa) (Sriskandan et al., 2007). They encode another important virulence factor of GAS: secreted pyrogenic exotoxin (SPE), which acts as a superantigen with pyrogenicity. It also has mitogenic activity for specific T cell subsets, which can increase host susceptibility to endotoxic shock and suppress immunoglobulin production (Commons et al., 2014). A single GAS strain usually harbors 4–6 superantigen genes, and these can provide a range of profiles for strain characterization. The last and the most powerful method is whole genome sequencing (WGS). On the one hand, phylogenetic analysis based on core genome genes (orthologous genes shared by all isolates) can disclose the relatedness between an outbreak strain and other isolates in the outbreak. On the other hand, genomic analysis can identify MGEs harbored in the GAS genome. The GAS genome is remarkable for its content of prophages, streptococcal phage-like chromosomal islands (SpyCIs), and other MGEs such as ICEs (Bessen et al., 2015).
This study is focused on the molecular investigation of a GAS outbreak in adults in a factory in China, where outbreaks are less widely reported. The investigation was triggered by the occurrence of two severe pneumonia cases in the factory during 9 days, including a fatal case. We not only characterized the relatedness among the outbreak strains and isolates from contacts but also identified the MGEs in the outbreak strains by means of emm typing, PFGE typing, superantigen profiling and whole genome analyzing. This report adds significantly to the limited data on adult GAS infections in China (Liu et al., 2014).
Materials and Methods
Study Setting
The factory producing mobile phone accessories is located at an industrial zone of Pudong New District of Shanghai. It covers an area of 520 acres, and employs over 70,000 workers aging from 16 to 45 years, who come from other provinces of China. The employees work and live in the industrial zone, where there are various facilities and shops. Over 3,000 workers work simultaneously on the same factory floor. Ten or twelve employees reside in a dormitory of about 28 square meters, sharing toilet facilities with other employees on the same floor. Locations where they work and live are centrally air-conditioned. There are seven departments in the manufactory, and each has a corresponding canteen equipped with an isolated kitchen. Only dining card consumption is accepted in all the seven canteens, and each employee has to consume in the assigned canteen corresponding to the department, for the card cannot be used in other canteens.
Invasive Case Definition and Case Finding
A confirmed invasive GAS case was defined as a patient in whom the illness was ascribed to GAS that was isolated from a normally sterile site (e.g., blood and pleural effusion), while a probable invasive GAS case had the bacteria isolated from a non-sterile site (e.g., sputum and throat). To identify more cases, we reviewed all medical records and laboratory reports of patients with respiratory symptoms in the factory from January 1, 2012 to October 9, 2013. Active surveillance was conducted on patients with signs or symptoms of respiratory infection by collecting sputum and throat specimens from October 10 to October 14. Each specimen was inoculated onto Columbia sheep blood agar plate, chocolate blood agar plate, and MacConkey agar plate (Oxoid, Basingstoke, United Kingdom) in order to isolate Staphylococcus aureus, S. pneumonia, S. pyogenes, Pseudomonas aeruginosa, Haemophilus influenza, and Klebsiella pneumoniae. A standardized data-abstraction form was used to collect demographic and clinical information of each patient.
Case-Control Study
To study the risk factors associated with GAS infection, including confirmed and probable cases, a case-control survey was performed. Control subjects were those patients in the factory with respiratory symptoms whose specimens were collected from October 10 to October 14 but without evidence of GAS colonization. We used a standardized data-abstraction form to obtain information including workshop, dormitory assignment, clinical presentation, medical and surgical histories. Statistical analysis was performed using SPSS (version 20.0; IBM), and statistical significance was assessed at p < 0.05.
Close Contact Investigation and GAS Carriage Surveys
We screened for GAS colonization among close contacts, who were defined as persons with experience of working and living with a confirmed GAS case. Environmental samples swabbed from the filter mesh at the air outlet of air conditioners, quilt, door handle, and washstand in the patients dormitory in the working and living rooms of the cases were also collected for GAS culture. We intended to conduct a GAS carriage survey among the healthy workers, but this was rejected by the administrator of the factory. We then performed a survey in two randomly selected districts of Shanghai by culturing throat swabs from October 15 to December 20 of 2013. The survey covered five age groups (<6 years, 7–14, 15–19, 20–60, and >60 years). The age group < 6 years were pre-school children, mostly in kindergarten. The age groups 7–14 years and 15–19 years were students in different grades, while 20–60 years and >60 years comprised residents in the community. Specimens were obtained by swabbing the posterior pharynx with ESwab Collection Kits (Copan, Brescia, Italia) and were inoculated onto Columbia sheep blood agar at 36°C with 5% carbon dioxide. A carrier was defined as a healthy person without any epidemiological relationship with a GAS case, but with GAS cultured from a non-sterile site.
Strain Identification and DNA Extraction
After culture for 24 h, β-hemolytic Gram positive cocci were tested by latex-agglutination with the Diagnostic Streptococcal Grouping Kit (Oxoid, Hampshire, United Kingdom). Isolates with the Lancefield group A antigen were identified by Vitek 2 system (bioMérieux, Marcy l′Etoile, France). Chromosomal DNA was extracted according to the protocol of Centers for Disease Control and Prevention1 (CDC).
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of 12 antimicrobial agents including erythromycin, clindamycin, and tetracycline were determined by broth dilution method and interpreted by the breakpoints according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) in 2015 (CLSI, 2015).
Detection of Resistance-Associated Genes and Mobile Genetic Elements
Macrolide resistance-related genes (ermB, ermA, and mef), tetracycline resistance-related genes (tetM and tetO), Tn916-Tn1545 transposon family (intTn and xis), and the novel MGEs ICE-emm12 and ΦHKU.vir, which were first identified in Hong Kong scarlet fever outbreaks in 2011, were screened by PCR as previously described (Perez-Trallero et al., 2007; Tse et al., 2012).
Molecular Typing for GAS
All isolates were characterized by emm typing, multi-locus sequence typing (MLST), superantigen profiling, and PFGE typing with the restriction endonuclease SmaI (TaKaRa, Dalian, China) as previously described (Chen et al., 2012). PFGE images were analyzed with BioNumerics software package (version 6.0; Applied Maths, Austin, TX, United States) using the unweighted pair group method and an arithmetic averages (UPGMA) clustering algorithm. According to the Tenover criteria (Tenover et al., 1995), isolates with 0, 2–3, 4–6, and ≥7 bands different from the outbreak strain were designated, respectively, as genetically indistinguishable, closely related, possibly related, and different pattern. In other words, the isolates were part, probably part, possibly part, and not part of the outbreak, respectively. Isolates with genetically distinct PFGE pattern were assigned to different clusters.
Genome Sequencing and Analysis
For Illumina pair-end sequencing of each strain, at least 3 μg genomic DNA was used for sequencing library construction as described previously (Harris et al., 2010). After a library quality test, Raw sequencing data was generated by Illumina base calling software CASAVA v1.8.22 according to its corresponding manuscript. Sickle3 was applied to conduct reads data trimming with default parameters to get clean data in this study. We used SOAPdenovo4 (v2.01) to assemble genomes with multiple K-mer parameters and got the optimal results of the assembly. GapCloser software4 was subsequently applied to fill up the remaining local inner gaps and correct any single base polymorphism for the final assembly results. This emm5 Whole Genome Shotgun project has been deposited at GenBank under the accession NCTL00000000 and NCTM00000000. The versions described in this paper are NCTL01000000 and NCTM 01000000. The genome of Manfredo was set as a reference. Manfredo was the only emm5 strain genome present in GenBank (accession number AM295007) and was isolated from an acute rheumatic fever patient in 1952 in the United States (Holden et al., 2007). We also sequenced one emm1 isolate and three emm12 isolates in Shanghai, and performed the phylogenetic analysis by MEGA7 using the Neighbor-Joining method (Kumar et al., 2016). The accession numbers of the isolates involved in the phylogenetic analysis are shown in Supplementary Table S1.
Prophages were predicted using PHAST5 (Zhou et al., 2011). Superantigen-encoding genes (speA, speC, speG, speH, speI, speJ, speK, speL, speM, ssa, and smeZ), virulence genes (spd3, sdc, sdaB, sdaD, speB, spyCEP, scpA, Mac, and sic), antibiotic resistance-associated genes (ermA, ermB, mef, tetK, tetL, tetM, and tetO), serum opacity factor gene (sof), and streptococcal invasion locus (sil) were screened in the genomic sequences with the primers reported previously (Beall et al., 2000; Borek et al., 2012; Plainvert et al., 2014).
Ethical Aspects
All specimens from patients and close contacts were collected as part of the routine clinical management of patients, according to the national guidelines in China. Consequently, informed consent was not sought from patients and close contacts, while informed consent was obtained from all participants in the carriage survey. The study was approved by Shanghai Municipal Center for Disease Control and Prevention ethical review committee (No.: 2016-4). All subjects gave written informed consent in accordance with the Declaration of Helsinki.
Results
Descriptive Epidemiology
From October 1 to October 9 of 2013, there were 42 cases of pneumonia in the factory. Two severe cases aged 18–20 years were transferred to a grade three hospital and one of them was dead on October 9. From then it was mandatory for the medical clinic of the factory to report the daily number of new cases, new cases with fever (>38°C), and transferred cases. During October 1 to October 20, the total number of cases with fever was 2,562, accounting for 15–25% of the daily new cases, and 1,836 cases were transferred to the grade three hospital, including 17 cases of severe pneumonia. Specimens including pleural effusion, sputum, and throat swab were collected from patients with respiratory symptoms in the factory on October 9 (the number of patients was six, n = 6), October 11 (n = 31), and October 14 (n = 20). The GAS attack rate was 14.0% (8/57), with nine isolates from eight confirmed cases including one confirmed invasive (the dead case, two isolates from pleural effusion and throat swab, respectively) and seven confirmed non-invasive cases (Table 1). The GAS case fatality rate was 12.5% (1/8). No other respiratory pathogenic bacteria including S. aureus,S. pneumonia, P. aeruginosa, H. influenza, and K. pneumoniae were detectable by culture.
Table 1.
Clinical characterization of patients with emm5 group A Streptococcus (GAS) infection.
| Case | Age, y/Gender | Date of symptom onset | Clinical diagnosis | Outcome | Source of GAS culture | emm subtype | Department |
|---|---|---|---|---|---|---|---|
| 1 | 20/M | 2013/10/2 | Severe pneumonia, STSS∗ | Died | Pleural effusion, throat swab | 5.101 | F4 |
| 2 | 27/M | 2013/10/8 | Influenza-like illness | Survived | Sputum | 5.101 | F3 |
| 3 | 20/M | 2013/10/9 | Influenza-like illness | Survived | Throat swab | 5.101 | F4 |
| 4 | 18/F | 2013/10/10 | Pharyngitis | Survived | Throat swab | 5.101 | F4 |
| 5 | 19/M | 2013/10/13 | Influenza-like illness | Survived | Throat swab | 5.101 | F5 |
| 6 | 20/F | 2013/10/14 | Influenza-like illness | Survived | Throat swab | 5.101 | F4 |
| 7 | 18/M | 2013/10/14 | Influenza-like illness | Survived | Throat swab | 5.46 | F3 |
| 8 | 18/M | 2013/10/14 | Influenza-like illness | Survived | Throat swab | 5.101 | F3 |
∗STSS, streptococcal toxic shock syndrome.
The fatal GAS case (Patient 1) was a 20-year-old man who was transferred to hospital by ambulance for septic shock on October 9th. About a week before, he had developed fever, anergy, cough, and expectoration under no obvious pre-disposing causes, with the signs of stethalgia, breathlessness, cardiopalmus, and oliguria (urinary volume less than 250 ml per day). On examination, his temperature was 40.0°C, blood pressure was 110/70 mmHg, pulse rate was 155 beats per minute, and respiratory rate was 47 per minute. Chest computed tomography (CT) scan showed double pneumonitis and pleural effusion. Although appropriate treatment was performed, the patient died on the day of transfer to the hospital. The other seven probable GAS cases appeared between 8th October and 14th October, most of them showed fever (6/7), sore throat (5/7), headache (5/7), and nasal discharge (5/7). They all eventually recovered. The eight GAS case patients were aged from 18 to 27 years, and worked in three different departments (Table 1).
Reviewing the medical records and microbiology laboratory reports of the factory from January to September in 2013, we did not find any evidence of GAS infection, but discovered another fatal case of severe pneumonia.
Case-Control Study
All the 8 GAS patients and 49 control subjects who had respiratory symptoms but without GAS colonization were included. By univariable comparison of demographic and clinical characteristics, case patients were more likely than control patients to be adults of 18–20 years old and to have a roommate or teammate with GAS infection or colonization (Table 2).
Table 2.
Univariate comparison of demographic and clinical characteristics of cases and controls.
| Demographic and clinical characteristics | Cases,N = 8 n (%) | Control,N = 49 n (%) | P-value |
|---|---|---|---|
| Age (18–20 year) | 7 (87.5) | 13 (26.5) | <0.05 |
| Male | 6 (75) | 40 (81.6) | 0.64 |
| Died during admission | 1 (12.5) | 0 (0) | 0.14 |
| Roommate or teammate with GAS colonization | 2 (25) | 0 (0) | <0.05 |
| Work overtime (≥10 h per day) | 8 (100) | 49 (100) | 1.00 |
| Signs and symptoms | |||
| Fever (≥38.0°C) | 7 (87.5) | 24 (49.0) | 0.06 |
| Cough | 5 (62.5) | 31 (63.3) | 1.00 |
| Pharyngalgia | 5 (62.5) | 33 (67.3) | 1.00 |
| Headache | 5 (62.5) | 21 (42.9) | 0.45 |
| Muscular stiffness | 3 (37.5) | 18 (36.7) | 1.00 |
| Anergy | 6 (75) | 23 (46.9) | 0.25 |
| Nasal discharge | 6 (75) | 17 (34.7) | 0.05 |
Close Contact Investigation
There were 11 roommates and 9 teammates of the dead case (Patient 1), among whom 4 close contacts developed respiratory symptoms and were administrated with antibiotics before the survey. We managed to collect throat specimens of 3 roommates of Patient 1 while the other 13 close contacts rejected requests. One close contact of Patient 1 was GAS culture-positive. Thirty-nine environmental samples were cultured and none was GAS-positive.
Carriage Survey
In the GAS carriage surveys conducted in other two districts, 14 (2.7%) of 521 healthy people were positive. The age groups and their frequencies of GAS carriage were ≤6 years old (5.4%, 4/74), 7–14 years old (7.4%, 9/122), 15–25 years old (0), 26–59 years old (0), and ≥60 years old (0.8%, 1/120).
Molecular Characterization of Outbreak Isolates
All the 10 outbreak associated isolates, including nine isolates from eight cases and one isolate from a close contact, were assigned to emm5 and MLST profile of ST-99 (emm5/ST-99). There were two emm subtypes: 5.101 (n = 9; GenBank accession numbers as KJ807822-KJ807831) and 5.46 (n = 1), which differed by only one nucleotide. They all harbored superantigen genes speC, speG, and smeZ. The outbreak associated isolates were assigned to two possible related PFGE patterns (clones) with only four bands different (Figure 1). Isolates from Patient 1, his roommate and Patient 5 showed indistinguishable pattern, but there was no epidemiological relationship between the two patients. Isolates from the other six patients were also identified as indistinguishable, among which two patients were teammates. Screening of antimicrobial resistance-associated genes showed that all the outbreak associated isolates carried ermA corresponding to their resistance to erythromycin (4–>128 μg/ml), and inducible resistance to clindamycin (0.25–0.5 μg/ml, D-zone test positive), eight (80%) isolates carried tetM gene corresponding to their resistance to tetracycline (16–32 μg/ml). All the outbreak associated isolates harbored MGEs intTn and xis and none harbored ICE-emm12 or ΦHKU.vir.
FIGURE 1.

PFGE patterns of group A Streptococcus (GAS) isolates from patients and healthy people in the investigation. According to the Tenover criteria (Tenover et al., 1995), isolates with 0, 2–3, 4–6, and ≥7 bands different from the outbreak strain were designated as genetically indistinguishable, closely related, possible related, and different pattern, respectively, which means the isolates were part, probably part, possibly part, and not part of the outbreak, respectively. PFGE, pulsed-field gel electrophoresis.
Molecular Characterization of Carriage Isolates
Among the 14 carriage isolates, which were isolated from asymptomatic carriers in the other two districts, 12 (86%) were assigned to emm12/ST-36, and 2 (14%) were emm1/ST-28. They all harbored speC, speG, ssa, and smeZ. Isolates of emm12 were assigned to three different PFGE patterns, among which two patterns differing in only one band were represented by 11 isolates (91.7%). The two emm1 carriage isolates were assigned to two PFGE patterns with one band different. All the carriage isolates harbored ermB and tetM, which was compatible with their resistance to erythromycin (>128 μg/ml), clindamycin (>128 μg/ml), and tetracycline (16–32 μg/ml). They all possessed MGEs intTn, xis, ICE-emm12, and ΦHKU.vir.
Whole Genome Sequence Analysis
Based on the PFGE data, two emm5 isolates (spy0390 from pleural effusion of Patient 1 and spy0392 from sputum of Patient 2) were selected as the representatives of the two outbreak clones to perform genome sequencing. The assembled spy0390 draft genome was comprised of 1,953,330 bp with G+C content of 38.5%, and spy0392 of 1,934,710 bp with G+C content of 38.4%. Comparison was performed among the two assembled sequences and one emm5 reference genome sequence (Manfredo), the genome of which comprised 1,841,271 bp with a G+C content of 38.6%.
Phylogenetic analysis was conducted with genomes of three emm5, three emm1, and six emm12 strains (Figure 2). There were three clades corresponding to each emm type. In the emm5 clade, outbreak isolates spy0390 and spy0392 showed a very close evolutionary relatedness and formed a distinct branch.
FIGURE 2.
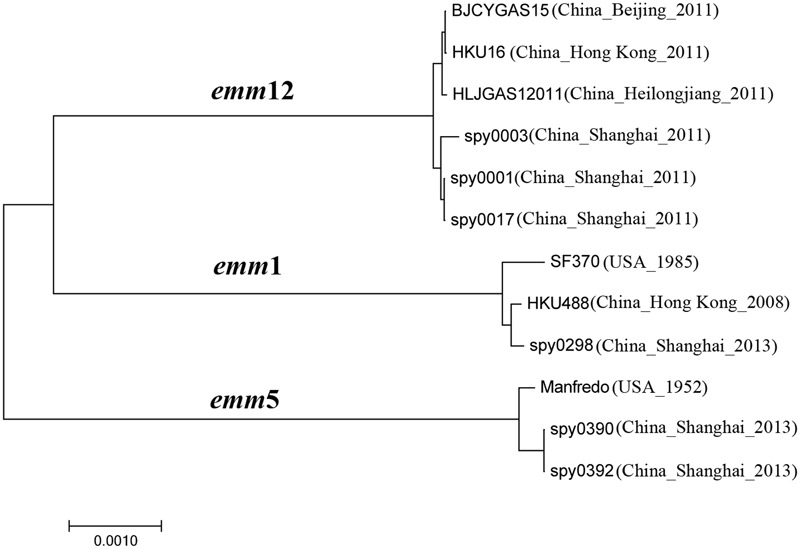
Phylogenetic analysis of Streptococcus pyogenes of emm1, emm5, and emm12 in Shanghai, China. The phylogenetic tree was generated in MEGA7 using the Neighbor-Joining method (Kumar et al., 2016). The optimal tree with the sum of branch length = 0.01685669 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and are in the units of the number of amino acid differences per site (Nei and Kumar, 2000). All positions containing gaps and missing data were eliminated. There were a total of 421,361 positions in the final dataset.
The three emm5 genomes shared 1,646 predicted genes, including superantigen coding genes speC, speG, and smeZ and other virulence genes: spd3, sdaB, speB, spyCEP, scpA, Mac, and sic. The two outbreak isolates shared 292 more predicted genes with each other than with Manfredo, including virulence gene sdc and antibiotic resistance genes of ermA and tetM, all of which were located within the predicted prophages (Figure 3).
FIGURE 3.
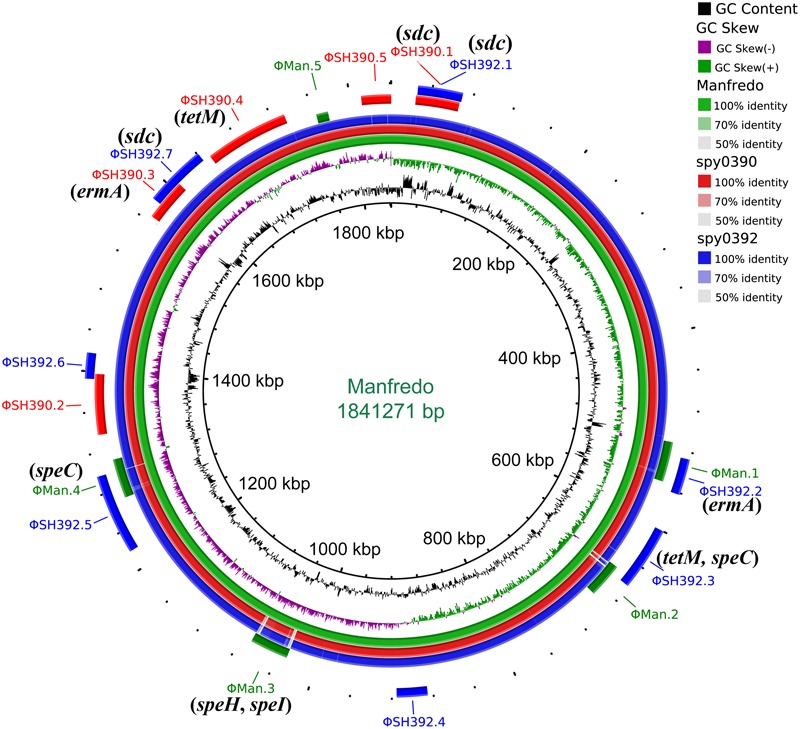
Circular genome map of Streptococcus pyogenes emm5 genome Manfredo with BLAST comparisons to the genomes of spy0390 and spy0392. The map was generated using BRIG (Alikhan et al., 2011). The innermost rings show G+C content (black) and G+C skew (purple/green) of Manfredo. The three outer rings show BLAST comparisons (using BLASTn and an E-value cutoff of 10.0) to the complete genome sequence of Manfredo (green) and the draft genome sequence of spy0390 (red) and spy0392 (blue). Legend shows percentage identity of BLASTn hits to the reference strain Manfredo. Labels around the outer ring refer to the insertion sites for predicted prophages with the corresponding color of the strain. Genes associated with antimicrobial resistance, superantigen, and virulence which are located within the prophages were labeled in parentheses.
Prophage Analysis of emm5
Five prophages were predicted to exist in spy0390 (ΦSH390.1 – ΦSH390.5), ranging in size from 41.9 to 84.1 kb, while seven prophages were predicted in spy0392 (ΦSH392.1 – ΦSH392.7), ranging from 25.1 to 78.1 kb (Supplementary Table S2). The macrolide-resistance gene ermA was located within the prophage ΦSH390.3 and ΦSH392.2, while tetracycline-resistance gene tetM was within ΦSH390.4 and ΦSH392.3 (Figure 3). The DNase coding gene sdc was present in the prophage ΦSH390.1, ΦSH392.1, and ΦSH392.7, therefore spy0392 possessed two copies of sdc (Figure 3). ΦSH390.1 and ΦSH392.1 were the same prophage, with the same insertion site and size. They shared 23.6% (10.3 kb/43.7 kb) nucleotide homology to ΦMan.3, which was one of the five prophages (ΦMan.1 – ΦMan.5) of Manfredo. The locations of ΦSH390.3 and ΦSH392.7 partially overlapped, but without any nucleotides in common. Other prophages in spy0390 and spy0392 had distinct locations but shared some associated nucleotides (Supplementary Table S2). ΦSH390.2 and ΦSH392.7 shared 38.2 kb nucleotides with Manfredo. ΦSH390.3 and ΦSH392.6 shared 13.2 kb nucleotides with Manfredo. ΦSH390.4 and ΦSH392.5 shared 17.2 kb nucleotides with Manfredo. ΦSH390.5 and ΦSH392.3 shared 19.5 kb nucleotides with Manfredo.
Discussion
To the best of our knowledge, this is the first report of a GAS outbreak occurring in an electronic component factory in China. Due to the low labor cost, this type of factory is common in China. In this study, we identified two emm5 clones by PFGE which were responsible for the outbreak. They shared the same emm subtype (except one isolate with one nucleotide different), sequence type, superantigen profile, and resistance to macrolides. The phylogenetic analysis suggested the two clones were closely related.
Two risk factors associated with GAS infections were discovered. One risk factor was having a roommate or teammate colonized with GAS. The two clones were found transmitted between roommates and teammates, respectively. GAS can be transmitted by direct contact with respiratory secretions or skin lesions of GAS-colonized people (Deutscher et al., 2011). We supposed the person-to-person contact facilitated the spread of the outbreak clone. Therefore, the isolation of GAS cases may be an effective measure to prevent similar GAS infections. Another risk factor was age ranging from 18 to 20 years. It is interesting to find that 87.5% (7/8) of GAS patients were at ages between 18 and 20 years old; by contrast, the GAS carriage was low in this age group (0/120) outside the industrial zone. Taking into account that the workers were non-indigenous and seldom go outside, they could be considered to be closed populations, just like soldiers on military bases, patients in long-term care facilities, and students in schools, who were reported to be particularly vulnerable to GAS infection outbreaks (Wasserzug et al., 2009; Chen et al., 2012; Dooling et al., 2013). The factory had an intensive workload, poor-hygiene, and crowded conditions that might facilitate GAS outbreaks. In our investigation, typical factors pre-disposing to invasive GAS infection, such as old age, skin lesions, and underlying medical conditions (Steer et al., 2012), were not observed in the population of the factory.
The source of the outbreak was not discovered though we have cultured the environmental samples of the patients. The possibility of foodborne source could not be ruled out. Taking into account patients in this outbreak were from three departments (Table 1), and the rule that employees in different departments go to different canteens for meal, and the PFGE patterns were without correlation with the department of the patients, we supposed the outbreak less likely caused by food contamination.
Outbreaks of GAS infections involving emm5 are uncommon worldwide. In this outbreak, emm5 GAS caused a fatal case of invasive infection and several superficial infection cases, with a case fatality rate of 12.5%. Sporadic infections caused by emm5 GAS are relatively common and it was reported to be one of the 10-most prevalent emm types in Poland (15%), South Korea (4%), Ethiopia (2%), and Israel (2%) (Smeesters et al., 2009). In Europe, emm5 caused 2% of invasive infections, with a case fatality rate of 30% (Luca-Harari et al., 2009). A significant association of emm5 with pharyngitis and acute rheumatic fever has been reported (Walker et al., 2014). emm5 strains were estimated to be the 19th most common cause of all GAS diseases in Asia, causing about 2.5% of pharyngeal diseases (Steer et al., 2009). However, the prevalence of emm5 strains in China is currently unknown and this study provides the first report of emm5-causing GAS infection outbreaks in China.
The emm5 clones found in this study were characterized as being different from the predominant emm clones in China. During 2011–2012, epidemics of over 100,000 scarlet fever cases were reported in mainland China (Ben Zakour et al., 2015), with emm12 as the dominant clones (Chen et al., 2012; Yang et al., 2013b). Compared with the scarlet fever emm12 clones, the emm5 outbreak clones were deficient in superantigen gene ssa, which was supposed to be an important trigger of the expansion of the scarlet fever clones in Hong Kong (Davies et al., 2015). Besides, the emm5 clones harbored the macrolide-resistance gene ermA, while the scarlet fever emm12 clones harbored ermB. The different characterizations between the scarlet fever emm12 clones and the emm5 outbreak clones should probably be attributed to their harboring different MGEs. The scarlet fever emm12 clones have ICE-emm12 carrying ermB as well as ΦHKU.vir and/or ΦHKU.ssa encoding superantigen SSA (Tse et al., 2012; Davies et al., 2015), while the emm5 outbreak clones only have ΦSH390.3 or ΦSH392.2 to carry ermA, without other elements associated with SSA. On the other hand, although tetM and speC genes are also present in the emm5 outbreak clones, they are located within prophages different from those in emm12 strains.
By genomic analysis, we found that the outbreak isolates possessed the virulence gene sdc which was located within prophage but this was not found in the emm5 reference strain Manfredo. In S. pyogenes, the gene sdc (also termed sdalpha; Borek et al., 2011) encodes Streptococcal DNase α (Sdα). This was first identified in 2001 in a M3 S. pyogenes strain isolated from a toxic shock-like syndrome patient (Hasegawa et al., 2002). Although the function of sdc is not well understood, several studies on DNase proteins in S. pyogenes have shown their roles in facilitating GAS to thwart the host innate immune response, for example degrading neutrophil extracellular traps to promote neutrophil survival and suppressing Toll-like receptor 9-mediated innate immune responses and macrophage bactericidal activity (Buchanan et al., 2006; Uchiyama et al., 2012). The acquisition of sdc gene might enhance the ability of emm5 strains to colonize host cells.
A deficiency of this study is that we were unable to analyze the GAS carriage of the healthy population of the factory. The manager of the factory refused to authorize us to conduct the survey, for diseases caused by invasive GAS infections are not notifiable in China. Another deficiency of this study is that no food samples were collected throughout the investigation. Consumption of contaminated food can also cause outbreaks of GAS infections (Kemble et al., 2013; Yang et al., 2013a). Although indirect proof has decreased the possibility of foodborne source, it is necessary for us to collect direct proof to rule it out.
Conclusion
We described an emm5 GAS outbreak in a factory making digital telephone components. There was substantial morbidity and mortality. The outbreak was caused by two closely related emm5 clones, which were characterized as being different from the predominant emm types circulating in China. The extent of the outbreak and the case fatality rate of 12.5% among GAS case patients should serve to alert the factory managers and public health experts to the higher risk of serious respiratory illnesses in this setting.
Author Contributions
MC and JC conceived and designed the experiments. MLC, WW, and GW performed all experiments. HP, LZ, and XZ analyzed the data. LT, YC, and YZ assisted in antimicrobial susceptibility testing and molecular typing. YZ enrolled the patients. MLC and MC supervised the study and wrote the paper. All authors have read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Haifeng Tang and Lili Song for their work on the laboratory surveillance on influenza-like illness patients. We thank Delong Zhan for his assistance on GAS carriage survey.
Funding. This study was supported by grants from National Natural Science Foundation of China (81601801), Shanghai Rising-Star Program (17QA1403100), Natural Science Foundation of Shanghai (16ZR1433300), the 4th Three-year Action Plan for Public Health of Shanghai (GWTD2015S01, GWTD2015S02, and 15GWZK0101), and Young Medical Talent Training Program of Health System of Pudong New Area, Shanghai (PWRq2014-26).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01156/full#supplementary-material
References
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B. (2014). Streptococci Group A Subtyping Request form Blast 2.0 Server. Available at: https://www2a.cdc.gov/ncidod/biotech/strepblast.asp. [Google Scholar]
- Beall B., Gherardi G., Lovgren M., Facklam R. R., Forwick B. A., Tyrrell G. J. (2000). emm and sof gene sequence variation in relation to serological typing of opacity-factor-positive group A streptococci. Microbiology 146(Pt 5) 1195–1209. 10.1099/00221287-146-5-1195 [DOI] [PubMed] [Google Scholar]
- Ben Zakour N. L., Davies M. R., You Y., Chen J. H., Forde B. M., Stanton-Cook M., et al. (2015). Transfer of scarlet fever-associated elements into the group A Streptococcus M1T1 clone. Sci. Rep. 5:15877 10.1038/srep15877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen D. E., McShan W. M., Nguyen S. V., Shetty A., Agrawal S., Tettelin H. (2015). Molecular epidemiology and genomics of group A Streptococcus. Infect. Genet. Evol. 33 393–418. 10.1016/j.meegid.2014.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borek A. L., Obszanska K., Hryniewicz W., Sitkiewicz I. (2012). Detection of Streptococcus pyogenes virulence factors by multiplex PCR. Virulence 3 529–533. 10.4161/viru.21540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borek A. L., Wilemska J., Izdebski R., Hryniewicz W., Sitkiewicz I. (2011). A new rapid and cost-effective method for detection of phages, ICEs and virulence factors encoded by Streptococcus pyogenes. Pol. J. Microbiol. 60 187–201. [PubMed] [Google Scholar]
- Buchanan J. T., Simpson A. J., Aziz R. K., Liu G. Y., Kristian S. A., Kotb M., et al. (2006). DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16 396–400. 10.1016/j.cub.2005.12.039 [DOI] [PubMed] [Google Scholar]
- Carapetis J. R., Steer A. C., Mulholland E. K., Weber M. (2005). The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5 685–694. 10.1016/S1473-3099(05)70267-X [DOI] [PubMed] [Google Scholar]
- Chen M., Yao W., Wang X., Li Y., Wang G., Zhang X., et al. (2012). Outbreak of scarlet fever associated with emm12 type group A Streptococcus in 2011 in Shanghai, China. Pediatr. Infect. Dis. J. 31 e158–e162. 10.1097/INF.0b013e31825874f3 [DOI] [PubMed] [Google Scholar]
- CLSI (2015). “Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement,” in CLSI Document M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Commons R. J., Smeesters P. R., Proft T., Fraser J. D., Robins-Browne R., Curtis N. (2014). Streptococcal superantigens: categorization and clinical associations. Trends Mol. Med. 20 48–62. 10.1016/j.molmed.2013.10.004S1471-4914(13)00181-0 [DOI] [PubMed] [Google Scholar]
- Cornick J. E., Kiran A. M., Vivancos R., Van Aartsen J., Clarke J., Bevan E., et al. (2017). Epidemiological and molecular characterization of an invasive Group A Streptococcus emm32.2 outbreak. J. Clin. Microbiol. 55 1837–1846. 10.1128/jcm.00191-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M. R., Holden M. T., Coupland P., Chen J. H., Venturini C., Barnett T. C., et al. (2015). Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat. Genet. 47 84–87. 10.1038/ng.3147 [DOI] [PubMed] [Google Scholar]
- Deutscher M., Schillie S., Gould C., Baumbach J., Mueller M., Avery C., et al. (2011). Investigation of a Group A streptococcal outbreak among residents of a long-term acute care hospital. Clin. Infect. Dis. 52 988–994. 10.1093/cid/cir084 [DOI] [PubMed] [Google Scholar]
- Dooling K. L., Crist M. B., Nguyen D. B., Bass J., Lorentzson L., Toews K. A., et al. (2013). Investigation of a prolonged Group A Streptococcal outbreak among residents of a skilled nursing facility, Georgia, 2009-2012. Clin. Infect. Dis. 57 1562–1567. 10.1093/cid/cit558 [DOI] [PubMed] [Google Scholar]
- Excler J. L., Kim J. H. (2016). Accelerating the development of a group A Streptococcus vaccine: an urgent public health need. Clin. Exp. Vaccine Res. 5 101–107. 10.7774/cevr.2016.5.2.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S. R., Feil E. J., Holden M. T., Quail M. A., Nickerson E. K., Chantratita N., et al. (2010). Evolution of MRSA during hospital transmission and intercontinental spread. Science 327 469–474. 10.1126/science.1182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa T., Torii K., Hashikawa S., Iinuma Y., Ohta M. (2002). Cloning and characterization of the deoxyribonuclease sd alpha gene from Streptococcus pyogenes. Curr. Microbiol. 45 13–17. 10.1007/s00284-001-0042-4 [DOI] [PubMed] [Google Scholar]
- Holden M. T., Scott A., Cherevach I., Chillingworth T., Churcher C., Cronin A., et al. (2007). Complete genome of acute rheumatic fever-associated serotype M5 Streptococcus pyogenes strain manfredo. J. Bacteriol. 189 1473–1477. 10.1128/JB.01227-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble S. K., Westbrook A., Lynfield R., Bogard A., Koktavy N., Gall K., et al. (2013). Foodborne outbreak of group a Streptococcus pharyngitis associated with a high school dance team banquet–Minnesota, 2012. Clin. Infect. Dis. 57 648–654. 10.1093/cid/cit359 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau E. H., Nishiura H., Cowling B. J., Ip D. K., Wu J. T. (2012). Scarlet fever outbreak, Hong Kong, 2011. Emerg. Infect. Dis. 18 1700–1702. 10.3201/eid1810.120062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. M., Zhao J. Z., Li B. B., Yang J. Y., Dong X. G., Zhang J. J., et al. (2014). A report on the first outbreak of a single clone group A Streptococcus (emm-type 89) tonsillopharyngitis in China. J. Microbiol. Immunol. Infect. 47 542–545. 10.1016/j.jmii.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Luca-Harari B., Darenberg J., Neal S., Siljander T., Strakova L., Tanna A., et al. (2009). Clinical and microbiological characteristics of severe Streptococcus pyogenes disease in Europe. J. Clin. Microbiol. 47 1155–1165. 10.1128/JCM.02155-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan D. J., Dreze P. A., Vu T., Bessen D. E., Guglielmini J., Steer A. C., et al. (2013). Updated model of group A Streptococcus M proteins based on a comprehensive worldwide study. Clin. Microbiol. Infect. 19 E222–E229. 10.1111/1469-0691.12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Kumar S. (2000). Molecular Evolution and Phylogenetics. New York, NY: Oxford University Press. [Google Scholar]
- Perez-Trallero E., Montes M., Orden B., Tamayo E., Garcia-Arenzana J. M., Marimon J. M. (2007). Phenotypic and genotypic characterization of Streptococcus pyogenes isolates displaying the MLSB phenotype of macrolide resistance in Spain, 1999 to 2005. Antimicrob. Agents Chemother. 51 1228–1233. 10.1128/AAC.01054-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plainvert C., Dinis M., Ravins M., Hanski E., Touak G., Dmytruk N., et al. (2014). Molecular epidemiology of sil locus in clinical Streptococcus pyogenes strains. J. Clin. Microbiol. 52 2003–2010. 10.1128/JCM.00290-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J., Schlegel L., Garnier F., Bouvet A. (2005). Molecular characterization of Streptococcus pyogenes isolates to investigate an outbreak of puerperal sepsis. Infect. Control Hosp. Epidemiol. 26 455–461. 10.1086/502567 [DOI] [PubMed] [Google Scholar]
- Smeesters P. R., McMillan D. J., Sriprakash K. S., Georgousakis M. M. (2009). Differences among group A streptococcus epidemiological landscapes: consequences for M protein-based vaccines? Expert Rev. Vaccines 8 1705–1720. 10.1586/erv.09.133 [DOI] [PubMed] [Google Scholar]
- Sriskandan S., Faulkner L., Hopkins P. (2007). Streptococcus pyogenes: Insight into the function of the streptococcal superantigens. Int. J. Biochem. Cell Biol. 39 12–19. 10.1016/j.biocel.2006.08.009 [DOI] [PubMed] [Google Scholar]
- Steer A. C., Lamagni T., Curtis N., Carapetis J. R. (2012). Invasive group A streptococcal disease: epidemiology, pathogenesis and management. Drugs 72 1213–1227. 10.2165/11634180-000000000-000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steer A. C., Law I., Matatolu L., Beall B. W., Carapetis J. R. (2009). Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect. Dis. 9 611–616. 10.1016/S1473-3099(09)70178-1 [DOI] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse H., Bao J. Y., Davies M. R., Maamary P., Tsoi H. W., Tong A. H., et al. (2012). Molecular characterization of the 2011 Hong Kong scarlet Fever outbreak. J. Infect. Dis. 206 341–351. 10.1093/infdis/jis362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama S., Andreoni F., Schuepbach R. A., Nizet V., Zinkernagel A. S. (2012). DNase Sda1 allows invasive M1T1 Group A Streptococcus to prevent TLR9-dependent recognition. PLoS Pathog. 8:e1002736 10.1371/journal.ppat.1002736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali L. A., Gherardi G., Petrelli D. (2015). Pulsed field gel electrophoresis of group A streptococci. Methods Mol. Biol. 1301 129–138. 10.1007/978-1-4939-2599-5_12 [DOI] [PubMed] [Google Scholar]
- Walker M. J., Barnett T. C., McArthur J. D., Cole J. N., Gillen C. M., Henningham A., et al. (2014). Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin. Microbiol. Rev. 27 264–301. 10.1128/CMR.00101-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserzug O., Valinsky L., Klement E., Bar-Zeev Y., Davidovitch N., Orr N., et al. (2009). A cluster of ecthyma outbreaks caused by a single clone of invasive and highly infective Streptococcus pyogenes. Clin. Infect. Dis. 48 1213–1219. 10.1086/597770 [DOI] [PubMed] [Google Scholar]
- Yang P., Peng X., Yang J., Dong X., Zhang M., Wang Q. (2013a). A probable food-borne outbreak of pharyngitis after a massive rainstorm in Beijing, caused by emm89 group A Streptococcus rarely found in China. Int. J. Infect. Dis. 17:e471 10.1016/j.ijid.2012.12.018S1201-9712(13)00020-9 [DOI] [PubMed] [Google Scholar]
- Yang P., Peng X., Zhang D., Wu S., Liu Y., Cui S., et al. (2013b). Characteristics of group A Streptococcus strains circulating during scarlet fever epidemic, Beijing, China, 2011. Emerg. Infect. Dis. 19 909–915. 10.3201/eid1906.121020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Liang Y., Lynch K. H., Dennis J. J., Wishart D. S. (2011). PHAST: a fast phage search tool. Nucleic Acids Res. 39 W347–W352. 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


