Abstract
Ophiocordyceps sinensis is a fungus that infects Hepialidae caterpillars, mummifying the larvae and producing characteristic fruiting bodies (stromata) that are processed into one of the most valued traditional Chinese medicines (TCM). The product commands a very high price due to a high demand but a very limited supply. Adulteration with other fungi is a common problem and there is a need to test preparation for the presence of the correct fungus. In the current study, a PCR-based approach for the identification of O. sinensis based on a segment of the internal transcribed spacer (ITS) region was developed. The segments is 146-bp in size and is likely to be amplified even in materials where processing led to DNA fragmentation. Primer development was based on the alignment of sequence data generated from a total of 89 samples of O. sinensis and potential adulterants as well as sequences date from 41 Ophiocordyceps species and 26 Cordyceps species available in GenBank. Tests with primer pair, DCF4/DCR4, demonstrated generation of an amplicon from DNA extracted from O. sinensis stromata, but not from extracts derived from adulterants. Species-specific primer pairs were also developed and tested for detection of the common adulterants, Cordyceps gunnii, Cordyceps cicadae, Cordyceps militaris, Cordyceps liangshanensis and Ophiocordyceps nutans. The collection of primers developed in the present study will be useful for the authentication of preparation claiming to only contain O. sinensis and for the detection of fungi used as adulterants in these preparations.
Keywords: Ophiocordyceps sinensis, species-specific primers, identification, mini-barcode, DNA degradation
Introduction
Ophiocordyceps sinensis (syn. Cordyceps sinensis) is an extremely rare and precious traditional Chinese medicine (TCM) with multiple medicinal values (Wang and Yao, 2011; Quan et al., 2014). This medicinal material is mainly collected in alpine regions over 4,000 m around mountain snowlines on the Tibetan Plateau, the neighboring provinces of the Tibetan autonomous prefectures and the high Himalayas (Yi et al., 2011). As reported in the New Compilation of Materia Medica, O. sinensis is beneficial to the kidney. Treatment with O. sinensis has also been claimed to have curative effects on several conditions, including erectile dysfunction, bronchial diseases, diabetes, cough and cold, jaundice (Ashok Kumar and Kailash Chandra, 2011; Sirisidthi et al., 2015). The main bioactive components found in O. sinensis are adenosine, cordycepin, cordymin, cordysinin, ergosterol, guanosine, myriocin, melanin, lovastatin, and sitosterol (Hui-Chen Lo et al., 2013). Due to strict environmental requirements, O. sinensis collected in the field is considered much more pharmacologically valuable than which obtained through culture; however, at present, most of the natural materials is collected by local farmers who do not have the expertise to differentiate between O. sinensis and related species. According to one recent study, even some of the O. sinensis materials used for study purposes may contain mycelium from other related species (Dong and Yao, 2010). The increasing rate of adulterated O. sinensis preparations not only harms consumers and the reputation of Traditional Chinese Medicine (Qin et al., 2011) but also hampers scientific research on this product.
The present identification of O. sinensis relies mostly on morphological characteristics, even though this approach has long been controversial. The genus Ophiocordyceps was officially defined by Sung et al. (2007) and Chen et al. (2013) and includes O. sinensis and similar species distributed within the alpine regions such as O. gansuënsis, O. crassispora, O. kangdingensis, O. multiaxialis, O. nepalensis, and others. It is difficult to distinguish these species morphologically (Shrestha et al., 2010) and it is even difficult to differentiate O. sinensis from the closely related adulterants, such as Cordyceps gunnii, Cordyceps cicadae, Cordyceps militaris, Cordyceps liangshanensis and Ophiocordyceps nutans. Chemical methods have also been applied to authenticate O. sinensis (Hu et al., 2015; Zhang et al., 2015); however, these method required relative large amounts of sample material. Genetic methods such as analysis of internal transcribed spacer sequences (ITS) and random amplified polymorphic DNA (RAPD)-derived molecular markers have also been used to identify O. sinensis (Lam et al., 2015). These methods have focused on detection of O. sinensis in untreated fungal material rather than processed materials where DNA degradation or fragmentation can occur (Meissner et al., 2007; Shadi et al., 2011). Therefore, in the present study, a method with specific-species primers was developed in order to increase the probability of detection of O. sinensis and common fungal adulterants even in processed samples.
Materials and methods
Collection of samples and its sequences
A total of 89 samples of O. sinensis and its adulterants (C. gunnii, C. cicadae, C. militaris, C. liangshanensis, O. nutans) were collected from Qinghai Province, Tibet and Sichuan Province. The details of these samples are listed in Table 1. A total of 131 confirmed ITS sequences of O. sinensis were available from previous studies (Chen et al., 2001; Liu et al., 2002; Zhang et al., 2009; Xiang et al., 2014). Additionally, all the known ITS sequences of the genera, Ophiocordyceps and Cordyceps, were downloaded, and published sequences (sequences in published articles) were selected for further study.
Table 1.
Information for the samples of O. sinensis and its counterfeits used in this study.
| Latin name | Voucher no. | Locality |
|---|---|---|
| Ophiocordyceps sinensis* | CSC-1 | Qamdo, Tibet China |
| CSC-2 | Qamdo, Tibet China | |
| CSC-3 | Qamdo, Tibet China | |
| CSC-4 | Qamdo, Tibet China | |
| CSC-5 | Qamdo, Tibet China | |
| CSC-6 | Qamdo, Tibet China | |
| CSC-7 | Qamdo, Tibet China | |
| CSC-8 | Qamdo, Tibet China | |
| CSC-9 | Qamdo, Tibet China | |
| CSC-10 | Qamdo, Tibet China | |
| CSC-11 | Qamdo, Tibet China | |
| CSC-12 | Qamdo, Tibet China | |
| CSC-13 | Qamdo, Tibet China | |
| CSC-14 | Qamdo, Tibet China | |
| CSC-15 | Qamdo, Tibet China | |
| CSC-16 | Qamdo, Tibet China | |
| CSC-17 | Qamdo, Tibet China | |
| CSC-18 | Qamdo, Tibet China | |
| CSC-19 | Qamdo, Tibet China | |
| CSC-20 | Qamdo, Tibet China | |
| CSC-21 | Qamdo, Tibet China | |
| CSC-22 | Qamdo, Tibet China | |
| CSC-23 | Qamdo, Tibet China | |
| CSC-24 | Qamdo, Tibet China | |
| CSC-25 | Qamdo, Tibet China | |
| CSC-26 | Qamdo, Tibet China | |
| CSC-27 | Qamdo, Tibet China | |
| CSC-28 | Qamdo, Tibet China | |
| CSC-29 | Qamdo, Tibet China | |
| CSC-30 | Qamdo, Tibet China | |
| CSN-1 | Yushu, Qinghai China | |
| CSN-2 | Yushu, Qinghai China | |
| CSN-3 | Golog Qinghai China | |
| CSN-4 | Golog Qinghai China | |
| CSN-5 | Golog Qinghai China | |
| CSN-6 | Yushu, Qinghai China | |
| CSN-7 | Yushu, Qinghai China | |
| CSN-8 | Qamdo Tibet China | |
| CSN-9 | Qamdo Tibet China | |
| CSN-10 | Nakchu, Tibet, China | |
| CSN-11 | Nakchu, Tibet, China | |
| CSN-12 | Nakchu, Tibet, China | |
| CSN-13 | Nakchu, Tibet, China | |
| CSN-14 | Nakchu, Tibet, China | |
| CSN-15 | Dege Sichuan China | |
| CSN-16 | Kangting Sichuan China | |
| CSN-17 | Kangting Sichuan China | |
| CSN-18 | Litang Sichuan China | |
| CSN-19 | Litang Sichuan China | |
| CSN-20 | Litang Sichuan China | |
| CSN-21 | Dege Sichuan China | |
| NQ-1 | Nakchu, Tibet China | |
| NQ-2 | Nakchu, Tibet China | |
| NQ-3 | Nakchu, Tibet China | |
| REG-1 | Ruoergai Sichuan China | |
| REG-2 | Ruoergai Sichuan China | |
| REG-4 | Ruoergai Sichuan China | |
| YS-1 | Yushu, Qinghai China | |
| YS-2 | Yushu, Qinghai China | |
| YS-3 | Yushu, Qinghai China | |
| WZ-1 | Unknown, China (market) | |
| WZ-2 | Unknown, China (market) | |
| WZ-3 | Unknown, China (market) | |
| WZ-4 | Unknown, China (market) | |
| WZ-5 | Unknown, China (market) | |
| WZ-6 | Unknown, China (market) | |
| WZ-7 | Unknown, China (market) | |
| WZ-8 | Unknown, China (market) | |
| WZ-9 | Unknown, China (market) | |
| WZ-10 | Unknown, China (market) | |
| WZ-11 | Unknown, China (market) | |
| WZ-12 | Unknown, China (market) | |
| WZ-13 | Unknown, China (market) | |
| WZ-14 | Unknown, China (market) | |
| WZ-15 | Unknown, China (market) | |
| WZ-16 | Unknown, China (market) | |
| WZ-17 | Unknown, China (market) | |
| WZ-18 | Unknown, China (market) | |
| Ophiocordyceps nutans | XC-1 | Changbai Mountain Nature Reserve Jilin China |
| XC-2 | Changbai Mountain Nature Reserve Jilin China | |
| Cordyceps gunnii | GN-1 | Chengtu, Sichuan China (market) |
| GN-2 | Xizang China (market) | |
| GN-3 | Hubei China (market) | |
| Cordyceps militaris | Y-1 | Hubei China (market) |
| Cordyceps cicadae | CH-1 | Hengduan Mountains Sichuan China |
| CH-2 | Bozhou Anhui China (market) | |
| CH-3 | Mopan Jiangsu China (market) | |
| Cordyceps liangshanensis | LS-1 | Sichuan Chian (market) |
| LS-2 | Sichuan Chian (market) |
DNA extraction, amplification, and sequencing
A total of 20–30 mg stromata of specimens were rinsed with 75% ethanol and milled using a ball-milling machine (Restch, Germany). Genomic DNA was extracted from the resulting powders using a Tiangen Plant DNA Kit (Tiangen Biotech, China). The ITS regions were amplified using an LA Taq polymerase chain reaction (PCR) kit (Takara Biotech Inc.) with the universal primer pairs 5F (5′-GGAAGTAAAAGTCGTAACAAGG-3′)/4R (5′-TCCTCCGCTTATTGATATGC-3′; Li et al., 2013). The PCR mixture contained 0.1 μL of LA Taq (5 U μL−1), 2.5 μL of 10 × LA Taq PCR buffer II (Mg2+ Plus), 1 μL of dNTP mixture (2.5 mM each), 0.6 μL of each primer (10 μM), and 1 μL (~120 ng) of genomic DNA in a total volume of 25 μL. The samples were amplified using a GeneAmp® PCR system 9700 (Applied Biosystems, Foster City, CA) under the following conditions: initial denaturation at 97°C for 1 min, followed by 30 cycles of denaturation at 97°C for 1 min, annealing at 48°C for 1 min, extension at 72°C for 3 min, and a final elongation step at 72°C for 7 min (Liu et al., 2001).
Sequence analysis and primer pairs design
The sequences were edited and assembled manually using CodonCode Aligner 5.1.4 (CodonCode Co., USA). Analysis of the ITS sequences database was conducted using CodonCode Aligner software to search species-specific motifs. Potential primers were designed and analyzed using Primer 6.0 software (Glantz, 2005) according to the species-specific motifs. All of the O. sinensis ITS sequences were aligned with MEGA (Lewis et al., 2013) software to verify the specificity of the primers for DNA from O. sinensis and its adulterants (C. gunnii, C. cicadae, C. militaris, C. liangshanensis, O. nutans).
Preparation of O. sinensis decoction and DNA extraction
Each sample was rinsed with 75% ethanol and was then milled using a ball-milling machine (Retsch, Germany); 40 mg of each milled sample was used for the genomic DNA extraction with the Tiangen Plant DNA Kit (Tiangen Biotech, China). Sterilized O. sinensis raw materials (stroma) were boiled in 500 mL double-distilled water for 60 and 90 min. The decoction was then dried on a stove by boiling, and 40 mg of the dried decoction was used for DNA extraction with the Tiangen Plant DNA Kit (Tiangen Biotech, China).
DNA amplification to verify the primer specificity and utility
PCR was performed on DNA extracted from O. sinensis decoctions and its adulterants. The reaction was carried out in 25 μL volumes comprised of 2 μL dNTP mixture (2.5 mmol/L), 1.0 μL primers DCF4 /DCR4 (2.5 μmol/L), 4 μL template DNA (~30 ng), 2.5 μL 10 × PCR Buffer (Tiangen Biotech, China), 8 μL Taq DNA polymerase and 6.5 μL sterilized water subject to the following conditions: initial denaturation at 94°C for 3 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 48°C for 30 s, extension at 72°C for 50 s, and a final elongation step at 72°C for 5 min. DNA from boiled materials was also amplified with ITS universal primer pairs to determine if the DNA was still suitable for amplification of larger sequences. PCR with DNA from adulterants was carried out is in 25-μL volumes comprised of 2 μL dNTP mixture (2.5 mmol/L), 2.0 μL primers (2.5 μmol/L), 2 μL template DNA (~30 ng), including pure DNA of adulterants and DNA mixture of O. sinensis and each target DNA for the specific primer (at a ratio of 1:1), 2.5 μL 10 × PCR Buffer (Tiangen Biotech, China), 8 μL Taq DNA polymerase and 6.5 μL sterilized water subject to the same conditions.
Amplification and concentration measurement of diluted DNA
Pure O. sinensis DNA was two-fold serially diluted to different multiple to determine the minimum amount of DNA needed for production of amplicons that could be visualized by ethidium bromide staining of agarose gels.
Results
Development of unique primers for O. sinensis
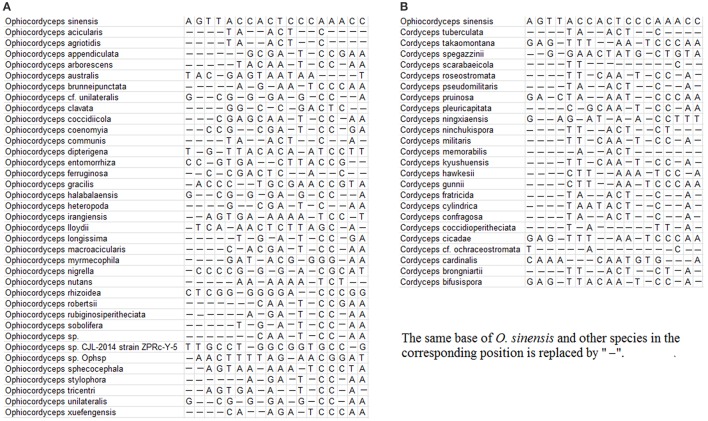
A total of 314 identified ITS sequences of O. sinensis, including 131 sequences generated in the previous study, were obtained with a length of ~500 bp after alignment; 112 published ITS sequences of 41 different species in Ophiocordyceps and 250 published ITS sequences of 26 species in Cordyceps were downloaded from the GenBank database. The search for primers specific to the fungal species of interest yielded the primer pairs listed in Table 2. The specificity of the primers DCF4/DCR4 for O. sinensis is illustrated in Figures 1A,B. There are at least 3 mismatches between the primers and the corresponding sequences from non- O. sinensis species.
Table 2.
Primers of O. sinensis and its counterfeits used for PCR amplification.
| Primer name | Species | Direction | Primer Sequences (5′–3′) | Amplicon size |
|---|---|---|---|---|
| DCF4 | O. sinensis | Forward | AGTTACCACTCCCAAACC | 146 |
| DCR4 | O. sinensis | Reverse | TGCTTGCTTCTTGACTGA | 146 |
| CCF | C. cicadae | Forward | TTACAACTCCCAACCCTTC | 209 |
| CCR | C. cicadae | Reverse | GATGCCAGAACCAAGAGAT | 209 |
| CGF | C. gunnii | Forward | TACCTATACTGTTGCTTCGG | 203 |
| CGR | C. gunnii | Reverse | GATGCCAGAACCAAGAGAT | 203 |
| CMF | C.militaris | Forward | TGAACATACCTATCGTTGCT | 167 |
| CMR | C.militaris | Reverse | ATGCCAGAGCCAAGAGAT | 167 |
| ONF | O. nutans | Forward | AACTCTCCAATTCTCTGTGA | 205 |
| ONR | O. nutans | Reverse | GCAATTCGCATTACTTATCG | 205 |
| CLF | C. liangshanensis | Forward | CAGCGGAGGGATCATTAC | 219 |
| CLR | C. liangshanensis | Reverse | GATGCCAGAACCAAGAGA | 219 |
Figure 1.
Alignment of DCF4 primer binding regions from congeners and common adulterants. (A) DCF4 binding regions from O. sinensis and other Ophiocordyceps species. (B) DCF4 binding regions of O. sinensis and other Cordyceps species.
Amplification with the species-specific primers DCF4/DCR4 and universal primers
The DNA in decoctions boiled for 60 or 90 min was amplified with the universal primer pair 5F/4R, as shown in Figure 2, it appears that the DNA extracted from the O. sinensis decoctions was possibly too fractured or otherwise degraded by boiling for 60 or 90 min to serve as template for amplification of the ITS sequence with the universal ITS primers. In contrast, PCR with the O. sinensis-specific primer pair yielded DNA that could be visualized after gel electrophoresis. As shown in Figure 3, primers DCF4/DCR4 were also used to amplify five common adulterants (C. gunnii, C. cicadae, C. militaris, C. liangshanensis, O. nutans), and no amplification product was seen with DNA obtained from them.
Figure 2.
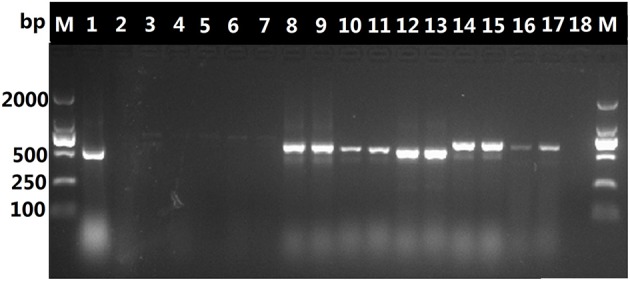
Amplification products generated with ITS universal primer pair 5F/4R and DNA from decoction and adulterants. Lane 1, pure O. sinensis DNA (~120 ng); lanes 2, 3, and 4, products from amplification of DNA (~30 ng) isolated from decoction boiled for 60 min; lanes 5, 6, and 7, amplification products from DNA (~30 ng) isolated from decoction boiled for 90 min; results for amplification with pure DNA (~30 ng) from C. liangshanensis (lanes 8–9), C. militaris (lanes 10-11), O. nutans (lanes 12–13), C. gunnii (lanes 14–15) and C. cicadae (lanes 16–17).
Figure 3.
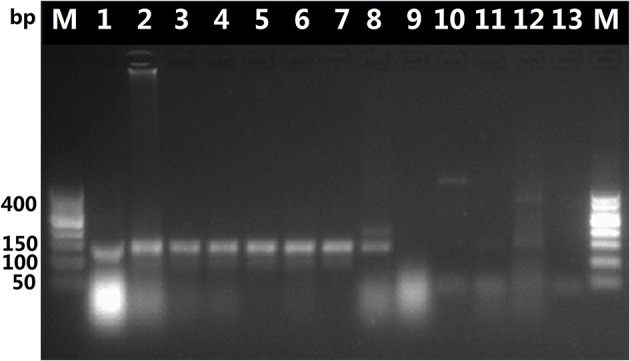
Results of PCR with primers DCF4/DCR4 with DNA isolated from decoction and adulterants. Lane 1, pure O. sinensis DNA (~120 ng); lanes 2, 3, and 4, amplification products from DNA (~30 ng) isolated from O. sinensis decoction boiled for 60 min; lanes 5, 6, and 7, amplification products from DNA (~30 ng) isolated from decoction boiled for 90 min; lanes 8, 9, 10, 11, and 12, results of PCR with pure DNA (~30 ng) isolated from adulterants C. liangshanensis, C. militaris, O. nutans, C. gunnii, and C. cicadae, respectively.
Amplification with specific primers for five common adulterants
In order to judge whether the sample were adulterated, five specific primer pairs (CCF/CCR, CGF/CGR, CMF/CMR, ONF/ONR, and CLF/CLR) for common adulterants were designed respectively, according to the conserved motifs obtained by aligning the ITS sequences of the targeted species. As shown in Figure 4, lanes 3, 7, 11, 15, and 19, none of the primers amplified DNA from O. sinensis, but they generated PCR products isolated from their respective target organism and in mixtures of O. sinensis and target organism DNA. We artificially mixed the DNA of O. sinensis and its adulterants, visual PCR products were obtained with each primer pair for each targeted species, as shown in Figure 4. The results showed that the method is suitable for the identification of the mixture of O. sinensis and its adulterants.
Figure 4.
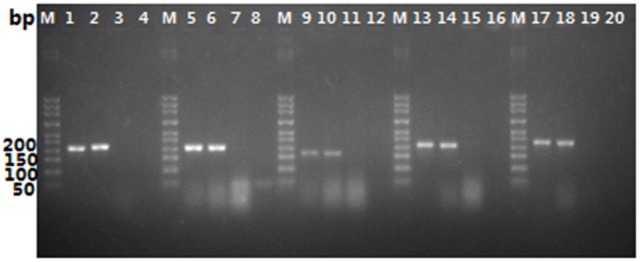
Ampification results for PCR with primer pairs CCF/CCR, CGF/CGR, CMF/CMR, ONF/ONR, and CLF/CLR. CCF/CCR (lane 1–4), CGF/CGR (lane 5–8), CMF/CMR (lane 9–12), ONF/ONR (lane 13–16), CLF/CLR (lane 17–20); lane1, 5, 9, 13, and 17, pure DNA isolated from targeted species (C. cicadae, C. gunnii, C. militaris, O. nutans, and C. liangshanensis, respectively); lane 2, 6, 10, 14, and 18, mixture of DNA isolated from O. sinensis and the targeted species (C. cicadae, C. gunnii, C. militaris, O. nutans, and C. liangshanensis, respectively) at a ratio of 1:1; lane 3, 7, 11, 15, and 19, pure DNA of O. sinensis; lane 4, 8, 12, 16, and 20, negative control.
Sensitivity of the PCR method
To determine suitable concentration of DNA, the pure O. sinensis DNA was two-fold serially diluted to different ratio, from two to six times. Within the scope of dilution times from two to four, the amplification result of diluted DNA showed no obvious difference, as shown in Figure 5, ~8 ng of purified O. sinensis DNA in a 25-μL reaction volume were necessary for a band of the PCR products from primers DCF4/DCR4 to be visible in an ethidium bromide-stained gel.
Figure 5.

Amplification products generated with primer pair, DCF4/DCR4, and decreasing amount of O. sinensis DNA. Lane 1–6: Products generated in a 25-μL reaction mixture with ~120, 60, 30, 15, 8, and 4 ng of O. sinensis DNA.
Discussion
O. sinensis has a long history of use as a traditional medicine in China. Due to over exploitation in the past decades, O. sinensis has been listed as an endangered species. Naturally produced O. sinensis is worth more than gold and because of this high value, adulterants have emerged frequently in recent years, which leads to market instability and a decline in consumer confidence.
DNA-based identification has become important for the identification of medical plants (Ali et al., 2014) as this technique is convenient, generally accurate and usable by people without taxonomic knowledge. The ITS sequence has been recently selected as the official marker for fugal genetic identification by the Consortium for the Barcode of Life (Das and Deb, 2015). ITS sequences amplification was used to identify fungi from soils or water as an environmental DNA barcode (Bellemain et al., 2010). Dentinger et al. compared the suitability of cytochrome oxidase subunit I (CO1) gene and ITS sequences for mushrooms and fern allies identification and determined that ITS-based identification is superior (Dentinger et al., 2011). Our previous study focused on the identification of raw O. sinensis materials based on ITS sequences (Xiang et al., 2014); however, the suitability for use with processed TCMs was not determined. The current study demonstrated that it is possible to apply PCR-based methodology to determine the presence of O. sinensis DNA in TCMs. Therefore, in another previous study, we proposed a mini-barcoding technique using short barcodes with a relatively high identification specificity for TCM (Liu et al., 2016), demonstrating immediate relevance to both science, industry and consumers. Further studies on mini-barcoding for the identification of TCM are necessary and beneficial. Although ITS sequences are commonly used to identify fungi, the requirement for relatively intact DNA to obtain complete ITS amplicons can make this approach difficult when DNA is extracted from processed samples whose DNA might have been degraded. We hypothesized that amplification of shorter regions of ITS might be possible with DNA from processed samples since Lo et al. were able to amplify a 88-bp fragment from TCM material after it had been boiled for 120 min (Lo et al., 2015). The present study also showed that a 146-bp fragment could be amplified from DNA extracted from processed O. sinensis samples, whereas amplification of the entire ITS region was not possible. For the first time, a specific primer pair is proposed and is proved to be a very efficient tool for the identification of O. sinensis and its adulterants.
The specificity of this primer pair, allows authentication of O. sinensis materials by PCR and amplicon detection along without the need for sequencing. Therefore, analysis times and costs are reduced. The assays can potentially be further simplified and expedited by utilizing isothermal recombinase polymerase amplification and an amplicon detection method that does not involve gel electrophoresis (Del Río et al., 2014). The application of the O. sinensis-specific primer pair along with the five primer pairs targeting DNA from common adulterants should allow determining if a sample said to only contain O. sinensis actually also, or exclusively, contains adulterants added inadvertently or deliberately.
Conclusion
In this study, a species-specific primer pair that amplifies a 146-bp sequence unique to the ITS region of O. sinensis was established. Besides that, five specific primer pairs for common adulterated species were also established. The method developed in this study provides users with an easy authentication method and may make a major contribution to the detection of counterfeit products of O. sinensis in the markets. In conclusion, this method can greatly expand the molecular identification of DNA-degraded materials and result in the rapid authentication of O. sinensis and its common adulterants among all its congeners with high accuracy, specificity and low cost.
Author contributions
JH designed this study. JH and LX provided experimental data. YL analyzed the raw data and drafted the manuscript. All authors helped to finish the manuscript and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No.81673552) and CAMS Innovation Fund for Medical Sciences (CIFMS, 2016-I2M-3-016).
References
- Ali M. A., Gyulai G., Hidvégi N., Kerti B., Al Hemaid F. M., Pandey A. K., et al. (2014). The changing epitome of species identification - DNA barcoding. Saudi J. Biol. Sci. 21, 204–231. 10.1016/j.sjbs.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok Kumar P., Kailash Chandra S. (2011). Traditional uses and medicinal potential of Cordyceps sinensis of Sikkim. J. Ayurveda Integr. Med. 2, 9–13. 10.4103/0975-9476.78183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellemain E., Carlsen T., Brochmann C., Coissac E., Taberlet P., Kauserud H. (2010). ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 10:189. 10.1186/1471-2180-10-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. Q., Ning W., Qu L. H., Li T. H., Zhang W. M. (2001). Determination of the anamorph of Cordyceps sinensis inferred from the analysis of the ribosomal DNA internal transcribed spacers and 5.8S rDNA. Biochem. Syst. Ecol. 29, 597–607. 10.1016/S0305-1978(00)00100-9 [DOI] [PubMed] [Google Scholar]
- Chen Z. H., Dai Y. D., Yu H., Yang K., Yang Z. L., Yuan F., et al. (2013). Systematic analyses of Ophiocordyceps lanpingensis sp. nov., a new species of Ophiocordyceps in China. Microbiol. Res. 168, 525–532. 10.1016/j.micres.2013.02.010 [DOI] [PubMed] [Google Scholar]
- Das S., Deb B. (2015). DNA barcoding of fungi using Ribosomal ITS Marker for genetic diversity analysis: a review. Int. J. Pure Appl. Biosci. 3, 160–167. Available online at: http://www.ijpab.com/form/2015%20Volume%203,%20issue%203/IJPAB-2015-3-3-160-167.pdf [Google Scholar]
- Del Río J. S., Yehia A. N., Acero-Sánchez J. L., Henry O. Y., O'Sullivan C. K. (2014). Electrochemical detection of Francisella tularensis genomic DNA using solid-phase recombinase polymerase amplification. Biosens. Bioelectron. 54, 674–678. 10.1016/j.bios.2013.11.035 [DOI] [PubMed] [Google Scholar]
- Dentinger B. T., Didukh M. Y., Moncalvo J. M. (2011). Comparing COI, and ITS as DNA barcode markers for mushrooms and allies (Agaricomycotina). PLoS ONE 6:e25081. 10.1371/journal.pone.0025081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C. H., Yao Y. J. (2010). On the reliability of fungal materials used in studies on Ophiocordyceps sinensis. J. Ind. Microbiol. 38, 1027–1035. 10.1007/s10295-010-0877-4 [DOI] [PubMed] [Google Scholar]
- Glantz S. A. (2005). Primer of Biostatistics: Statistical Software Program Version 6.0. New York, NY: McGraw-Hill Medical. [Google Scholar]
- Hu H., Xiao L., Zheng B., Wei X., Ellis A., Liu Y. M. (2015). Identification of chemical markers in Cordyceps sinensis by HPLC-MS/MS. Anal. Bioanal. Chem. 407, 8059–8066. 10.1007/s00216-015-8978-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui-Chen Lo C. H., Lin F. -Y., Hsu T. -H. (2013). A systematic review of the mysterious caterpillar fungus Ophiocordyceps sinensis in Dong-ChongXiaCao (冬蟲夏草 Dōng Chóng Xià Cǎo) and related bioactive ingredients. J. Trad. Complement. Med. 3, 16–32. 10.1016/S2225-4110(16)30164-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam K. Y., Chan G. K., Xin G. Z., Xu H., Ku C. F., Chen J. P., et al. (2015). Authentication of Cordyceps sinensis by DNA analyses: comparison of ITS sequence analysis and RAPD-derived molecular markers. Molecules 20, 22454–22462. 10.3390/molecules201219861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P. O., Kumar S., Tamura K., Nei M., Lewis P. O. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Song J., Xin T., Zhu Y., Shi L., Xu X., et al. (2013). DNA barcoding the commercial Chinese caterpillar fungus. FEMS Microbiol. Lett. 347, 156–162. 10.1111/1574-6968.12233 [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang L., Wang X., Chen X., Han J., Pang X. (2016). A nucleotide signature for the identification of american ginseng and its products. Front. Plant Sci. 7:319. 10.3389/fpls.2016.00319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. Y., Liang Z. Q., Liu A. Y., Yao Y. J., Hyde K. D., Yu Z. N. (2002). Molecular evidence for teleomorph-anamorph connections in Cordyceps based on ITS-5.8S rDNA sequences. Mycol. Res. 106, 1100–1108. 10.1017/S0953756202006378 [DOI] [Google Scholar]
- Liu Z. Y., Yao Y. J., Liang Z. Q., Liu A. Y., Pegler D. N., Chase M. W. (2001). Molecular evidence for the anamorph—teleomorph connection in Cordyceps sinensis. Mycol. Res. 105, 827–832. 10.1017/S095375620100377X [DOI] [Google Scholar]
- Lo Y. T., Ming L., Shaw P. C. (2015). Identification of constituent herbs in ginseng decoctions by DNA markers. Chin. Med. 10, 1–8. 10.1186/s13020-015-0029-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner C., Bruse P., Mueller E., Oehmichen M. (2007). A new sensitive short pentaplex (ShoP) PCR for typing of degraded DNA. Forensic Sci. Int. 166, 121–127. 10.1016/j.forsciint.2006.04.014 [DOI] [PubMed] [Google Scholar]
- Qin S., Zhao J., Liu X., Hua C., Shu Y., Yue G. (2011). Current market state investigation and strategy of Cordyceps sinensin (Berk.) Sacc. Lishizhen Med. Mater. Med. Res. 22, 1236–1237. [Google Scholar]
- Quan Q. M., Wang Q. X., Zhou X. L., Li S., Yang X. L., Zhu Y. G., et al. (2014). Comparative phylogenetic relationships and genetic structure of the caterpillar fungus Ophiocordyceps sinensis and its host insects inferred from multiple gene sequences. J. Microbiol. 52, 99–105. 10.1007/s12275-014-3391-y [DOI] [PubMed] [Google Scholar]
- Shadi S., Xin Z., Janzen D. H., Winnie H., Jean- François L., Jacobus L. M., et al. (2011). Pyrosequencing for mini-barcoding of fresh and old museum specimens. PLoS ONE 6:e21252 10.1371/journal.pone.0021252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B., Zhang W., Zhang Y., Liu X. (2010). What is the Chinese caterpillar fungus Ophiocordyceps sinensis (Ophiocordycipitaceae)? Mycol. Int. J. Fungal Biol. 1, 228–236. 10.1080/21501203.2010.536791 [DOI] [Google Scholar]
- Sirisidthi K., Kosai P., Jiraungkoorskul W. (2015). Antihyperglycemic activity of Ophiocordyceps sinensis: a review. Indian J. Agric. Res. 49, 400–406. 10.18805/ijare.v49i5.5801 [DOI] [Google Scholar]
- Sung G.-H., Hywel-Jones N. L., Sung J.-M., Luangsa-ard J. J., Shrestha B., Spatafora J. W. (2007). Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 57, 5–59. 10.3114/sim.2007.57.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. L., Yao Y. J. (2011). Host insect species of Ophiocordyceps sinensis: a review. Zookeys 127, 43–59. 10.3897/zookeys.127.802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L., Son J., Xin T., Zhu Y., Shi L., Xu X., et al. (2014). DNA barcoding the commercial chinese Caterpillar Fungus, in The 14th National Conference on Traditional Chinese Medicine and Nature Medicine Paper Abstract (Beijing: ), 156–162. [Google Scholar]
- Yi L., Wang X. L., Lei J., Yi J., Hui L., Jiang S. P., et al. (2011). A survey of the geographic distribution of Ophiocordyceps sinensis. J. Microbiol. 49, 913–919. 10.1007/s12275-011-1193-z [DOI] [PubMed] [Google Scholar]
- Zhang J., Wang P., Wei X., Li L., Cheng H., Wu Y., et al. (2015). A metabolomics approach for authentication of Ophiocordyceps sinensis by liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. Food Res. Int. 76, 489–497. 10.1016/j.foodres.2015.07.025 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xu L., Shu Z., Liu X., An Z., Mu W., et al. (2009). Genetic diversity of Ophiocordyceps sinensis, a medicinal fungus endemic to the Tibetan Plateau: Implications for its evolution and conservation. BMC Evol. Biol. 9:290. 10.1186/1471-2148-9-290 [DOI] [PMC free article] [PubMed] [Google Scholar]