Abstract
Nijmegen breakage syndrome (NBS) is a rare autosomal recessive disorder characterized by predisposition to hematopoietic malignancy, cell-cycle checkpoint defects, and ionizing radiation sensitivity. NBS is caused by a hypomorphic mutation of the NBS1 gene, encoding nibrin, which forms a protein complex with Mre11 and Rad50, both involved in DNA repair. Nibrin localizes to chromosomal sites of class switching, and B cells from NBS patients show an enhanced presence of microhomologies at the sites of switch recombination. Because nibrin is crucial for embryonic survival, direct demonstration by targeted deletion that nibrin functions in class switch recombination has been lacking. Here, we show by cell-type-specific conditional inactivation of Nbn, the murine homologue of NBS1, that nibrin plays a role in the repair of γ-irradiation damage, maintenance of chromosomal stability, and the recombination of Ig constant region genes in B lymphocytes.
Keywords: Nijmegen breakage syndrome, B lymphocytes, DNA repair
The nuclear complex of Mre11, Rad50, and nibrin (also known as Nbs1 or p95) is relevant for chromosomal stability, meiotic recombination, and telomere maintenance in eukaryotic cells (1–3), and the targeted inactivation of Mre11 (4), Rad50 (5), or nibrin (3) causes embryonic lethality in the mouse. In humans, two related chromosome instability disorders, Nijmegen breakage syndrome (NBS) and Ataxia-Telangiectasia-like disorder (ATLD) are caused by mutation of the genes encoding nibrin and Mre11, respectively. More than 90% of NBS patients are homozygous for a 5-bp deletion in exon 6 of the NBS1 gene. This mutation leads to a premature termination of translation that results in a 27-kDa amino-terminal peptide and a 70-kDa carboxyl-terminal peptide because of an alternative initiation of translation (6). Because the mutations in the NBS patients and of MRE11 in ATLD are hypomorphic, it is difficult to deduce the exact role of nibrin and Mre11 in DNA repair (7). NBS patients are characterized by predisposition to hematopoietic malignancy, cell-cycle checkpoint defects, and ionizing radiation sensitivity of fibroblasts and lymphoblastoid cells. A characteristic, variable deficiency of serum IgG and IgA with normal IgM levels is observed (8, 9). Individual Sμ–Sα switch-recombination junctions of Ig class-switched B lymphocytes from NBS and ATLD patients show a preponderance of microhomologies at the site of recombination (9, 10). This observation could be explained by the direct involvement of nibrin in the recombination of Ig constant region genes, by impaired survival or proliferation of activated B lymphocytes, or by impaired T cell help. In two murine models for NBS with hypomorphic mutations, in which Nbn exons 2 and 3 or Nbn exons 4 and 5 are replaced by a neo gene, the total IgG levels in the serum are indistinguishable from normal controls (11, 12); however, a defect in the T cell-dependent B cell activation is observed (11). Nevertheless, an argument in favor of a direct involvement of nibrin in switch recombination is the colocalization of nibrin foci with the IgH locus in activated B lymphocytes (13).
Here, we show by conditional inactivation of Nbn in murine B lymphocytes, that nibrin plays a role in Ig class-switch recombination.
Materials and Methods
Animals. Mice with a targeted mutation of the Nbn gene of 129/Sv mice were generated as described in detail elsewhere (14), by insertion of lox P sequences upstream and downstream of exon 6 of Nbn (Nbnlox-6) in E 14.1 ES cells, injection of NbnWT/lox-6 and NbnWT/Δ6 E 14.1 cells, the later being obtained from the former by transient expression of Cre (pMC-Cre), into C57BL/6 blastocysts and mating of the resulting chimera to 129/Sv mice. Offspring with NbnΔ6/Δ6 genotypes was completely absent when Nbnlox-6/Δ6 mice were interbred, indicating that Nbn is functionally inactivated by deletion of exon 6 (14).
For Cre-mediated deletion of exon 6 of Nbn in Nbnlox-6/lox-6 B lymphocytes, Nbnlox-6/lox-6 mice were crossed with heterozygous CD19-Cre mice of C57BL/6 background, a kind gift of K. Rajewsky (Harvard Medical School, Boston) (15). In all experiments, heterozygous NbnWT littermates served as controls. Animals were maintained and bred in specific-pathogen-free facilities.
CD19-Cre efficiency was determined by quantitative multiplex PCR, specific for the floxed exon 6, deleted exon 6 and WT Nbn alleles, as described elsewhere in detail (14), using the primers flox-forward (5′-GCTTGGCTCAAGTAGTACTG-3′), del-/WT-forward (5′-ATAAGACAGTCACCAC-3′), and the corresponding, fluorescein-conjugated reverse primer (5′-fluorescein-TTATGTCACTGAGGACCTCC-3′). PCR products were separated on a 10% polyacrylamide gel, and quantified in a vistra FluorImager SI (Amersham Pharmacia Bioscience, Freiburg, Germany) and image quant software (Amersham Pharmacia Bioscience).
Deletion of Nbn Exon 6 by tat-Cre-Fusion Protein in Vitro. Recombinant tat-Cre fusion protein was purified from extracts of Escherichia coli bacteria transformed with a vector encoding a His-tat-nuclear localization sequence-Cre fusion protein as described (16), except that all buffers contained 500 mM NaCl. The vector was a kind gift of F. Edenhofer (University of Bonn, Bonn) and K. Rajewsky. The purified tat-Cre protein was stored in 500 mM NaCl/20 mM Hepes (pH 7.4) at 80 μM at -70°C. For Cre-loxP-mediated deletion of exon 6 of Nbn in vitro, 2.5 × 106 B lymphocytes of genotypes as indicated, purified by magnetic depletion of CD43+ cells from spleen cells (see below), were incubated for 1 h at 37°C in 1 ml of serum-free RPMI medium 1640, with 1.5 μM tat-Cre fusion protein added. For control, (untreated) cells were incubated alike, adding 0.5 M NaCl/20 mM Hepes (pH 7.4) instead of the tat-Cre stock solution. Cells were then washed with PBS with 0.5% BSA and activated (see below).
Activation of B Lymphocytes in Vitro and by Cytometric Analysis. B lymphocytes were isolated from murine spleens by magnetic depletion of other cells with CD43-specific magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). When indicated, the cells were then pretreated with tat-Cre fusion protein as described above.
For activation of splenic B cells for antibody class switching, the cells were stimulated with bacterial lipopolysaccharide (LPS; 40 μg/ml, Sigma–Aldrich, St. Louis) or LPS and IL-4 (X63-IL4 supernatant, 20 ng/ml final concentration of IL-4) as described (17). Cell division was tracked according to loss of carboxyfluorescein diacetate succinimidyl ester (CFSE) label (17). Cells were stained immediately after magnetic purification with 1 μM CFSE (Molecular Probes) in PBS for 2 min and 45 sec at room temperature. Cell numbers were determined by counting Trypan blue-stained cells in a Neubauer chamber, or cytometrically, by referencing their frequencies to defined numbers of TrueCount beads (Becton Dickinson). Viability was determined cytometrically according to light scatter and exclusion of propidium iodide (see below).
B lymphocytes in spleens of transgenic mice were analyzed cytometrically by staining spleen cells with anti-B220 conjugated to Cy5 (CD45R and RA3.6B2) and anti-IgM conjugated to phycoerythrin (R6–60.2, Pharmingen). To determine the frequencies of activated B cells expressing IgG1, CFSE-labeled, LPS/IL4-stimulated B cells were harvested on day 3 of culture and stained for expression of IgG1 on the cell surface, with digoxigenin-labeled monoclonal rat anti-mouse IgG1 [0.5 mg/ml; a kind gift of M. Assenmacher (Miltenyi Biotech) and H. Schliemann (German Rheumatism Research Center)] and anti-digoxigenin Fab conjugated to Cy5 (Roche, Mannheim, Germany). To determine the frequencies of activated B cells expressing IgG3, CFSE-labeled, LPS-activated B cells of day 3 cultures were stained for expression of IgG3 on the cell surface with biotinylated rat anti-murine IgG3 (R40–82) (Pharmingen) and streptavidin-Cy5 (Roche). Cells were analyzed by using a FACSCalibur cytometer (Becton Dickinson, Heidelberg) and cellquest and fcs-express software. Viable cells were gated according to forward and sideward light scatter, and exclusion of propidium iodide (1 μg/ml in PBS/BSA). All stainings were performed in the presence of 50 μg/ml rat monoclonal antibody against mouse CD16/32 (2.4G2).
Irradiation. To determine irradiation sensitivity, Nbnlox-6/Δ6 and Nbnlox-6/WT B cells were labeled with CFSE and treated with tat-Cre or control buffer, stimulated for 3 days with LPS/IL-4, harvested, aliquoted, and γ-irradiated, as indicated, in RPMI medium 1640 with 10% FCS on ice, in a Cs-137 irradiator (BIOBEAM 2000, MCP Medical International, Berlin). The cells were cultured for another 2 days in their original culture supernatant supplemented with another volume of fresh RPMI medium 1640 with 10% FCS before their numbers and CFSE label were analyzed cytometrically.
Detection of Deletion of Exon 6 of Nbn in Individual B Cells. Individual B cells were labeled as described above and sorted by fluorescence-activated single-cell sorting on a Cytomation MoFlow sorter (Cytomation, Freiburg, Germany) into wells of 96-well plates (Eppendorf). For detection of both floxed and deleted exon 6 Nbn genes by nested single-cell PCR, as described earlier (18), the primer combinations nbs-up1: 5′-TGCTGGCTATGTGAAGACTA-3′, nbs-down2: 5′-CTTCCAATAGCTGGTTCATC-3′, and nbs-down4: 5′-CCTGGGATGAAAGTGTGTTC-3′ were used for an initial amplification of genomic DNA. Floxed Nbn exon 6 was then amplified specifically with the primers nbs-up2 (5′-GATTCGTGAATGTAGTGCTG-3′) and nbs-down1 (5′-AGTGACTGATACCAAAAGGG-3′). Nbn genes with deleted exon 6 were amplified with the primers nbs-up2 (5′-GATTCGTGAATGTAGTGCTG-3′) and nbs-down3 (5′-AATACAGTGACTCCTGGAGG-3′).
Analysis of Nibrin Expression. Whole-cell extracts were prepared from Histopaque-purified cells (Sigma-Aldrich, Munich) at different time points of culture and total cell lysates equivalent to 5 × 105 cells were loaded onto a 10% SDS-polyacrylamide gel followed by electrophoresis and immunoblotting, as described (19). Nibrin and Mre11 proteins were detected with rabbit antibodies specific for the carboxyl terminus of Nbs1 (nibrin 15 CR) (20) and Mre11-specific rabbit antibodies (Novus Biologicals, Littleton, CO). Rabbit antibodies were visualized on the blot with horseradish peroxidase-conjugated goat anti-rabbit Fab-fragment, and ECL detection reagents (Amersham Pharmacia Bioscience).
Analysis of Switch Recombination Junctions. Genomic DNA was prepared from day 3 LPS/IL-4 culture after proteinase K digestion. Sμ-Sγ1 junctions were amplified, cloned, and sequenced as described in refs. 21 and 22. The junctions were either amplified by using an Expand long template PCR system (Roche) with primers and conditions as described in ref. 22 and cloned with a TOPO-TA cloning kit (Invitrogen) before sequencing, or as described in ref. 21 with a nested PCR by using homemade Taq-polymerase and cloning into pBluescript after digestion with EcoRI. Switch junctions were identified by a blast search.
Karyotyping. Nbnlox-6/Δ6 B cells were incubated with either tat-Cre or with dialysis buffer as control and stimulated with LPS/IL-4. On day 3 of culture, cells were irradiated as indicated and cultured further for 3 h at 37°C. For the analysis of chromosomal breaks, 60 ng/ml colcemid was then added (KaryoMax colcemid solution, Gibco/BRL), and the cells were incubated for another 1 h. Metaphase spreads were prepared after hypotonic shock and stained with Giemsa by using standard technology (23). For assignment of chromosome breaks to individual chromosomes, metaphase spreads were prepared without colcemid, the chromosomes were partially digested with trypsin, and stained with Giemsa (23). Identification of chromosomes according to Giemsa-banding patterns was performed by following procedures in ref. 23.
Quantification of Iμ-Cμ and Iγ1-Cγ1 Transcripts. Total RNA was prepared with RNeasy (Qiagen, Hilden, Germany) from B cells activated for 2 days with LPS/IL-4, and transcribed into cDNA with a 1:1 ratio mixture of oligo(dT) and random hexamer primers (TaqMan reverse transcription reagent; Applied Biosystems). The cDNA was used for real-time PCR (LightCycler, FastStart DNA master SYBR green I kit, Roche Diagnostics, Penzberg, Germany). Specificity of the μ, γ1, and GAPDH primers (22) was confirmed by analysis of dissociation curves. The efficiency of amplification per round of PCR was 1.92, 1.95 and 1.96 for Iμ-Cμ, Iγ1-Cγ1, and GAPDH, respectively.
Detection of γ-H2AX Foci. Activated B cells were fixed for 10 min at room temperature in PBS with 4% formaldehyde, washed twice in PBS, and permeabelized with 0.1% Triton X-100 in PBS for 5 min on ice. The cells were washed again and blocked with 0.1% BSA/0.1% Triton X-100/PBS for 20 min at 37°C. γ-H2AX foci were stained with γ-H2AX-specific rabbit antibodies (Trevigen, Gaithersburg, MD) for 1 h at 37°C and then with Cy2-conjugated goat anti-rabbit IgG (Dianova, Hamburg) for 1 h at 37°C. DNA was stained with DAPI, 1 μM, in PBS for 5′ on ice. The cells were cytocentrifuged onto slides and analyzed by fluorescence microscopy (Zeiss Axioplan, Oberkochen, Germany).
Results
Nibrin-Deficient, Activated B Lymphocytes Show Increased Sensitivity to γ-Irradiation. Disruption of exon 6 of the Nbn gene by insertion of a neomycin-resistance gene (Nbnins6) leads to embryonic lethality (20). Here, we have analyzed mice with targeted insertion of LoxP sites flanking exon 6 of the Nbn gene (Nbnlox-6), allowing Cremediated conditional inactivation of Nbn in the germ-line or somatic cells (NbnΔ6; Fig. 1 A–C). Offspring with NbnΔ6/Δ6 genotypes was completely absent when Nbnlox-6/Δ6 mice were interbred, indicating that Nbn is functionally inactivated by deletion of exon 6 (14).
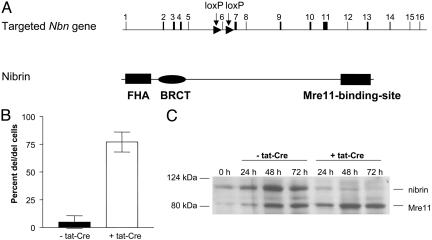
Fig. 1.
Generation of nibrin-deficient B lymphocytes. (A) Schematic representation of the targeted Nbn gene locus and the protein structure of nibrin. (B) Efficiency of tat-Cre-mediated Nbn exon 6 deletion. Nested single-cell PCR detects the deleted allele and the floxed exon 6 Nbn allele. Data represent the mean and SD of five individual cultures. (C) Expression of nibrin and Mre11 proteins in populations of LPS/IL4-activated Nbnlox-6/Δ6 B lymphocytes, not treated or treated with tat-Cre in vitro, at the indicated time points after activation. Cell extracts of 5 × 105 cells each were submitted to Western blot analysis, as described in Materials and Methods.
To determine the role of Nbn in activation and differentiation of B cells, naïve B lymphocytes were isolated from the spleens of Nbnlox-6/Δ6 mice, and labeled with CFSE, to track the proliferative history of switched cells. Nbn exon 6 was deleted from the floxed alleles by incubation of the cells with tat-Cre fusion protein (16). The cells were then activated with bacterial LPS for 3 days, in the presence or absence of IL-4, to target switch recombination to IgG1 or IgG3, respectively (24). The efficiency of tat-Cre-mediated deletion of Nbn exon 6 was determined by single-cell PCR analysis for the deleted versus the floxed allele. In >75% of Nbnlox-6/Δ6 B cells, exon 6 was deleted from the floxed allele 3 days after tat-Cre treatment (Fig. 1B). Without tat-Cre, Nbn exon 6 was detectable in >95% of the cells, indicating the efficiency of the single-cell PCR. On the protein level, expression of p95/nibrin was significantly reduced on day 1 as compared with Mre 11, and was nearly absent on days 2 and 3 (Fig. 1C). As expected, tat-Cre-mediated inactivation of Nbn in activated B cells leads to an increased sensitivity of the cells to γ-irradiation-induced DNA damage on day 3 of culture, reflected by dose-dependent impairment of proliferation (Fig. 2A) and an enhanced chromosomal instability (Fig. 2B). However, the spontaneous chromosomal instability observed is not as drastic as observed by Reina-San-Martin et al. (25).
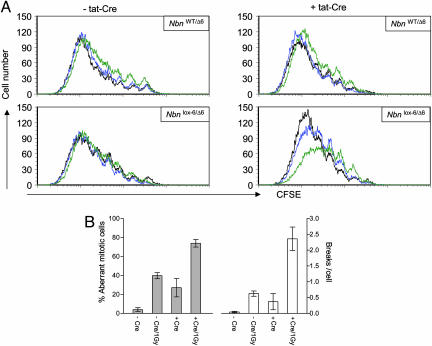
Fig. 2.
Radiation sensitivity and chromosomal instability of nibrin-deficient B lymphocytes. (A) Radiation sensitivity of Nbnlox-6/Δ6 and NbnWT/Δ6 B lymphocytes after deletion of exon 6 of Nbn by tat-Cre in vitro. Cells were labeled with CFSE and activated with LPS/IL4 for 3 days. Cells were then not irradiated intentionally (black line), or γ-irradiated with 1.5 Gy (blue) or 3.6 Gy (green line). Two days later, proliferation and survival of the cells was analyzed cytometrically. Proliferative history of surviving cells was determined according to loss of CFSE label and numbers of surviving cells relative to reference beads (TrueCount, Becton Dickinson), standardizing the numbers of recorded viable cells not stained for propidium iodide. (B) Chromosomal instability of LPS/IL4-activated Nbnlox-6/Δ6 B lymphocytes after treatment with tat-Cre in vitro. On day 3 of culture, metaphase spreads were prepared from cells 3 h after γ-irradiation (1 Gy). Metaphases were Giemsa-stained and scored for aberrant chromosome numbers and apparent chromosomal breaks (-Cre/-Gy, 102 metaphases analyzed; -Cre/+Gy, 59 metaphases; +Cre/-Gy, 236 metaphases; +Cre/+Gy, 138 metaphases).
Ig Class Switching Is Impaired by Inactivation of Nbn. In the absence of intentional irradiation, tat-Cre-mediated conditional inactivation of Nbn in LPS- or LPS/IL-4-activated B cells did not impair their survival, as indicated by the numbers of viable cells between days 1, 2, and 3 of culture (Fig. 5, which is published as supporting information on the PNAS web site) and the frequencies of dead cells according to propidium iodide staining (Table 3 and Fig. 6A, which are published as supporting information on the PNAS web site), nor was proliferation changed detectably, as measured by loss of CFSE-staining (Fig. 3A). Inactivation of Nbn did affect antibody class switching, in that the frequencies of activated B cells that had switched to IgG3 or IgG1, respectively, were reduced by >50% in cells of all generations in a given stimulation on day 3 (Fig. 3 B–D). The similar reduction in switched cells over all generations suggests that nibrin is directly involved in the process of switch recombination, rather than in the survival of switched cells. In support of this hypothesis, on day 3 of LPS/IL-4 culture, ≈80% of tat-Cre-treated Nbnlox-6/Δ6 B cells that had switched to IgG1 had deleted exon 6 of Nbn (data not shown).
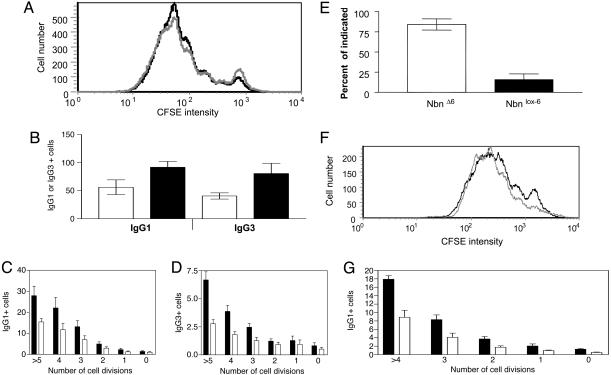
Fig. 3.
Antibody class-switch recombination in B lymphocytes conditionally inactivated for Nbn in vitro or in vivo. (A) Proliferation according to loss of CFSE label of Nbnlox-6/Δ6 B lymphocytes analyzed on day 3 of LPS/IL-4 activation, and treated with tat-Cre (solid line) or not treated (shaded line) in vitro, before the onset of activation. Data shown are from a representative experiment. (B) Comparison of Nbnlox-6/Δ6 (white bars) and NbnWT/lox-6 (black bars) B cells switched to IgG1 upon activation by LPS/IL-4, or switched to IgG3 upon activation by LPS, in tat-Cre treated relative to cultures not treated with tat-Cre. Data represent the mean and SD of individual LPS (n = 6 for Nbnlox-6/Δ6 and n = 4 NbnWT/lox-6)orLPS/IL4 cultures (n = 7 for Nbnlox-6/Δ6 and n = 5 NbnWT/lox-6; P < 0.01, Mann–Whitney U test). Flow cytometric analysis of surface IgG1 (C) and surface IgG3 expression by CFSE-labeled Nbnlox-6/Δ6 B lymphocytes (D) treated with tat-Cre (white bars) or not treated (black bars), after 3 days of LPS/IL-4 and LPS stimulation, respectively. The frequencies of IgG1- or IgG3-expressing cells are shown separately for each population that has undergone the indicated number of cell divisions since the onset of activation, according to CFSE staining. Data represent the mean and SE of four individual LPS/IL4 or LPS cultures. (E) Efficiency of depletion of exon 6 of Nbn in vivo, in CD19-Cre Nbnlox-6/lox-6 B lymphocytes, detected by quantitative PCR specific for the deleted and nondeleted floxed Nbn alleles. (Data from three mice are represented.) (F) Proliferation of CD19-Cre Nbnlox-6/lox-6 (solid line) and CD19-Cre Nbnlox-6/WT B lymphocytes (shaded line) after activation with LPS for 3 days, according to loss of CFSE label. (G) Flow cytometric analysis of surface IgG1 expression on CFSE-labeled CD19-Cre Nbnlox-6/lox-6 (white bars) and CD19-Cre Nbnlox-6/WT B lymphocytes (black bars) stimulated with LPS/IL-4 for 3 days. The frequency of IgG1+ cells is shown for each population that had undergone the indicated number of cell divisions since the onset of activation, according to loss of CFSE label. Data represent the mean and SE of four individual LPS/IL4 cultures.
Sequences of switch recombination breakpoints, isolated from tat-Cre-treated Nbnlox-6/ins6 populations, show no statistically significant difference when compared with breakpoints of switched Nbnlox-6/ins6 cells not treated with tat-Cre (Table 1). Neither the location of breakpoints within the switch regions (Fig. 7, which is published as supporting information on the PNAS web site) nor the frequency and average length of microhomologies and insertions at the switch-recombination sites (Table 1) differed between tat-Cre treated and nontreated cells. The frequencies of mutations in the vicinity of switch junctions (± 10 bp) was higher in tat-Cre-treated Nbnlox-6/ins6 (3.3 × 10-2) than in nontreated cells (1.8 × 10-2) but was also not statistically significant (P = 0.108).
Table 1. Sμ–Sγ1 switch recombination junctions of tat-Cre-treated Nbnlox-6/ins6 cells.
| Cells | Percentage of junctions with ≥ 2-bp homology* | Average length of perfect match, bp | Average length of imperfect match,† bp | Percentage of junctions with ≥ 1-bp insertion | Average length of insertion, bp |
|---|---|---|---|---|---|
| Nbnins6/lox-6 (n = 19) | 63 | 2.3 | 3.4 | 11 | 1.5 |
| Nbnins6/lox-6, tat-Cre-treated (n = 41) | 46 | 2.9 | 3 | 24 | 1.2 |
Percentages of perfect matches over ≥ 2 bp: 47% of Nbn versus 22% of Nbn, tat-Cre-treated
Percentage of perfect matches over ≥ 2 bp matches, one mismatch
In an attempt to delete the Nbn gene before the onset of activation of B cells, Nbn was inactivated early in B cell ontogeny in vivo by CD19-Cre-mediated deletion of exon 6 in CD19-Cre/Nbnlox-6/lox-6 and CD19-Cre/Nbnlox-6/WT B cells, and splenic B cells from such mice were activated with LPS or LPS/IL-4 in vitro. Frequencies and absolute numbers of splenic B cells were not affected by CD19-mediated inactivation of Nbn (Fig. 8, which is published as supporting information on the PNAS web site). More than 80% of the Nbn alleles of CD19-Cre/Nbnlox-6/lox-6 B lymphocytes had been inactivated in vivo, corresponding to at least 60% of cells deficient for Nbn (Fig. 3E). On day 3 of culture, 70% of the floxed Nbn alleles of IgG1+ cells were deleted, as compared with 90% of IgG1- B cells (data not shown). When compared with CD19-Cre/Nbnlox-6/WT B cells, CD19-Cre/Nbnlox-6/lox-6 cells showed diminished proliferative capacity (Fig. 3F), and cell counts were ≈30–50% lower on day 3 of stimulation (Table 3). The frequencies of cells expressing IgG1 were consistently reduced by 50–70% in all generations of the activated B cells (Fig. 3G), confirming the role of nibrin in switch recombination.
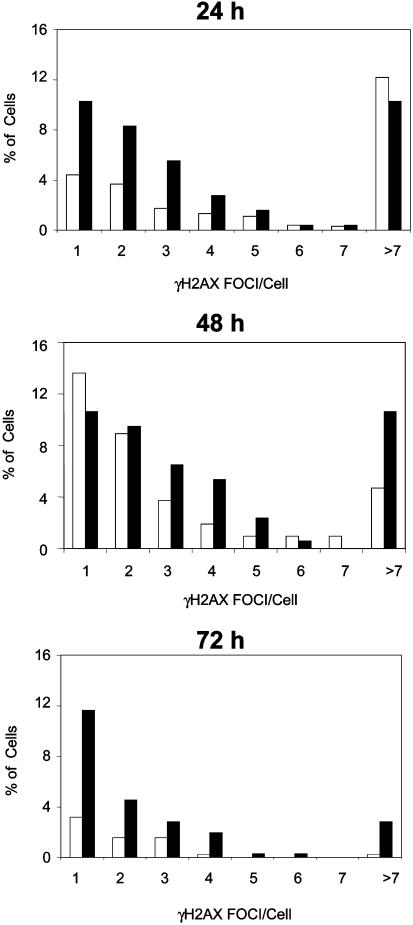
Inactivation of Nbn Affects Late but Not Early Stages of Switch Recombination. B lymphocytes isolated from CD19-Cre/NbnWT/ins6, CD19-Cre/NbnWT/lox-6, CD19-Cre/Nbnlox-6/lox-6, and CD19-Cre/Nbnlox-6/ins6 mice were activated with LPS/IL-4, and after 48 h, the Iμ–Cμ and Iγ1–Cγ1 switch transcripts were quantified by RT-PCR to determine the initial targeting of the Sμ and Sγ1 switch regions for switch recombination. Activated B cells of all genotypes showed no detectable differences in expression of switch transcripts (Table 2). The subsequent induction of DNA breaks in switch regions of activated B cells is reflected by the formation of γ-H2AX foci (13). This step was not impaired by inactivation of Nbn. Forty eight hours after onset of activation, >22% of CD19-Cre/Nbnlox6/WT B cells showed one or two γ-H2AX foci, as compared with 8% after 24 h, and 5% after 72 h. Less than 6% of the cells had three to seven foci at all times (Fig. 4). Of CD19-Cre/Nbnlox6/lox6 B cells, 18% after 24 h, 20% after 48 h, and 16% after 72 h showed one or two γ-H2AX foci, and 11% after 24 h, 15% after 48 h, and 6% after 72 h expressed three to seven foci, indicating an overall higher and persistent presence of DNA double-strand breaks, presumably, not only in switch regions but also in B cells lacking functional nibrin. Of 56 metaphase spreads derived from day 3 LPS/IL4-activated CD19-Cre/Nbnlox-6/lox-6 B cells, 8 metaphase spreads showed identifiable chromosome breaks. Four of the metaphase spreads could be attributed by differential Giemsa staining to the telomeric end of chromosome 12, i.e., the IgH locus (Fig. 9, which is published as supporting information on the PNAS web site). Metaphase spreads from activated Nbnlox-6/Δ6 B cells had not shown chromosome breaks at detectable frequencies (Fig. 2B).
Table 2. Induction of switch transcripts.
| Culture | Mouse Id. | Genotype | Iμ–Cμ transcripts versus GAPDH transcripts* | Iγ1–Cγ1 transcripts versus GAPDH transcripts* | Percent IgG1 cells† |
|---|---|---|---|---|---|
| 43 | CD19-Cre NbnWT/lox-6 | 24.3 | 4.1 | 9.4 | |
| I | 44 | CD19-Cre Nbnlox-6/ins6 | 20.7 | 4.1 | 3.2 |
| 42 | CD19-Cre Nbnlox-6/lox-6 | 21.8 | 4.3 | 4.3 | |
| 31 | CD19-Cre NbnWT/ins6 | 14.6 | 2.4 | 27.3 | |
| II | 35 | CD19-Cre Nbnlox-6/ins6 | 19.4 | 7.2 | 7.9 |
| 33 | CD19-Cre Nbnlox-6/lox-6 | 16.7 | 4.3 | 4.2 | |
| 51 | CD19-Cre NbnWT/lox-6 | 23.3 | 10.0 | 4.8 | |
| III | 52 | CD19-Cre Nbnlox-6/ins6 | 21.9 | 8.4 | 2.5 |
| 46 | CD19-Cre Nbnlox-6/lox-6 | 22.1 | 7.5 | 1.8 |
Id., identification.
RT-PCR analysis was performed on day 2 after LPS/IL-4 activation
IgG1 analysis was performed on day 3 after LPS/IL-4 activation
Fig. 4.
Generation and maintenance of γ-H2AX foci in conditionally Nbn-inactivated, activated B lymphocytes. γ-H2AX foci were stained with a specific antibody in CD19-Cre Nbnlox6/lox6 (black bars) and CD19-Cre Nbnlox6/WT B lymphocytes (white bars) after 24, 48, and 72 h of LPS/IL-4 activation. The samples were double-blinded and foci were scored by fluorescence microscopy. (WT/24 h, 976 cells; WT/48 h, 213 cells; WT/72 h, 379 cells; lox6/24 h, 253 cells; lox6/48 h, 169 cells; lox6/72 h, 352 cells.)
Discussion
The present data strongly suggest that nibrin is directly involved in the process of class-switch recombination of activated B lymphocytes. It acts downstream of switch transcript-induced targeting of switch regions (26, 27) and activation-induced cytidine deaminase (AID)-induced DNA breaks in the targeted switch regions (28). The nibrin/Mre11/Rad50 complex could be targeted to sites of action of AID by means of replication protein A, which can bind to Mre11 (29), and to AID (30). So far, the exact function of nibrin in class-switch recombination is not clear. Because nibrin by itself has no apparent enzymatic activity (7, 31), it has been speculated that it may be required to assemble Mre11 and Rad50 in the nucleus, targeting them to staggered DNA ends of switch regions, and perhaps, modulating their enzymatic activity (7, 31, 32). In vitro,the nibrin/Mre11/Rad50 complex can bind to single-stranded or double-stranded DNA, tether DNA ends (33), and process DNA as a 3′-5′ exonuclease, endonuclease, and helicase (34). In class-switch recombination, the nibrin/Mre11/Rad50 complex may be involved in modification, realignment, and ligation of switch-region breaks, acting in concert with the Ku70/80/DNA-Pkc and the DNA-ligase4/XRCC4 complexes (35).
Conditional inactivation of Nbn in B cells activated in vitro and in vivo reduces the frequencies of switched cells by >50%. This effect is direct, rather than indirect, because the survival of the activated B cells is not affected, as for tat-Cre-mediated Nbn inactivation, or only slightly affected, as for CD19-Cre-mediated inactivation. The survival of switched B cells as reflected by propidium iodide uptake is not affected in either system. Further evidence that proliferation and survival of switched cells is not selectively impaired is provided by the observation that the reduction in frequencies and numbers of switched cells was similar for all generations of Nbn-inactivated B cells at the time of analysis. As with tat-Cre-mediated inactivation of Nbn in vitro, CD19-Cremediated deletion of Nbn exon 6 in vivo resulted in an ≈50% reduction in the frequencies of switched cells. This result makes it unlikely that all of the remaining switched cells had used residual functional nibrin for switch recombination although this result cannot formally be excluded for some cells. Because of technical limitations of the systems used here for conditional inactivation of the Nbn gene, which does not generate homogeneous populations of Nbn inactivated B cells, the present results give only a minimum estimate of the contribution of nibrin to switch recombination. It remains to be shown whether nibrin is critical for successful switch recombination, or whether alternative, nibrin-independent mechanisms do exist. The extended microhomologies observed at class switch junctions of NBS patients (9, 10) could be interpreted as evidence in favor of alternate pathways, i.e., homology-directed recombination, most likely mediated by the mismatch repair system. Even there, the nibrin/Mre11/Rad50 complex could be involved, because direct interaction of Mre11 and Mlh1 has been reported (36). Here, we do not find extended microhomologies in switch recombination junctions of switched, tat-Cre-treated Nbnlox-6/ins6 cells, suggesting that these microhomologies might, rather, reflect the hypomorphic action of mutated nibrin in switching NBS B cells than the action of a mismatch-repair system. The observed persistence of γ-H2AX foci and chromosome 12 breaks in activated nibrin-deficient B cells support the notion that nibrin is required for efficient repair of nonhomologous DNA break recombination of class-switch regions.
In addition to the important role that nibrin plays in class-switch recombination, it is also crucial for the survival of B lymphocytes. This role did not affect our analysis of switch recombination, which was focused on short-lived plasma blasts and performed generation-wise, but retarded proliferation and impaired survival of in vitro-activated CD19-Cre/Nbnlox-6/lox-6 B cells was observed. This finding was not surprising, because >20% of activated B cells show obvious chromosome breaks on day 3 of culture, breaks that are apparently not repaired efficiently, as reflected by their persistence through mitosis and by the persistence of γ-H2AX foci in activated B cells. The important role of nibrin in the repair of class-switch recombination breaks, as demonstrated here, may be related to the preponderance of B cell lymphoma among NBS patients, ≈40% of whom develop this malignancy in early childhood (37).
Supplementary Material
Acknowledgments
We thank F. Edenhofer for providing the His-tat-nuclear localization sequence-Cre fusion protein expression plasmid, K. Rajewsky for providing CD19-Cre mice, and Farah Hatam, Hyun-Dong Chang, Katharina Hein, and Uwe Niesner for discussion. We also thank L. Reiners-Schramm, H. Schliemann, T. Kaiser, K. Raba, and J. Radszewski for excellent technical assistance, and B. Reina-San-Martin, M. C. Nussenzweig, A. Nussenzweig, and S. Difilippantonio for discussions. This work was supported by SFB 577 of the Deutsche Forschungsgemeinschaft.
Abbreviations: NBS, Nijmegen breakage syndrome; LPS, lipopolysaccharide; CFSE, carboxyfluorescein diacetate succinimidyl ester.
References
- 1.Varon, R., Vissinga, C., Platzer, M., Cerosaletti, K. M., Chrzanowska, K. H., Saar, K., Beckmann, G., Seemanova, E., Cooper, P. R., Nowak, N. J., et al. (1998) Cell 93, 467-476. [DOI] [PubMed] [Google Scholar]
- 2.Tauchi, H., Kobayashi, J., Morishima, K., van Gent, D. C., Shiraishi, T., Verkaik, N. S., vanHeems, D., Ito, E., Nakamura, A., Sonoda, E., et al. (2002) Nature 420, 93-98. [DOI] [PubMed] [Google Scholar]
- 3.Zhu, J., Petersen, S., Tessarollo, L. & Nussenzweig, A. (2001) Curr. Biol. 11, 105-109. [DOI] [PubMed] [Google Scholar]
- 4.Xiao, Y. & Weaver, D. T. (1997) Nucleic Acids Res. 25, 2985-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo, G., Yao, M. S., Bender, C. F., Mills, M., Bladl, A. R., Bradley, A. & Petrini, J. H. (1999) Proc. Natl. Acad. Sci. USA 96, 7376-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maser, R. S., Zinkel, R. & Petrini, J. H. (2001) Nat. Genet. 27, 417-421. [DOI] [PubMed] [Google Scholar]
- 7.Lee, J. H., Ghirlando, R., Bhaskara, V., Hoffmeyer, M. R., Gu, J. & Paull, T. T. (2003) J. Biol. Chem. 278, 45171-45181. [DOI] [PubMed] [Google Scholar]
- 8.van Engelen, B. G., Hiel, J. A., Gabreels, F. J., van den Heuvel, L. P., van Gent, D. C. & Weemaes, C. M. (2001) Hum. Immunol. 62, 1324-1327. [DOI] [PubMed] [Google Scholar]
- 9.Pan, Q., Petit-Frere, C., Lahdesmaki, A., Gregorek, H., Chrzanowska, K. H. & Hammarstrom, L. (2002) Eur. J. Immunol. 32, 1300-1308. [DOI] [PubMed] [Google Scholar]
- 10.Lahdesmaki, A., Taylor, A. M., Chrzanowska, K. H. & Pan-Hammarstrom, Q. (2004) J. Biol. Chem. 279, 16479-16487. [DOI] [PubMed] [Google Scholar]
- 11.Kang, J., Bronson, R. T. & Xu, Y. (2002) EMBO J. 21, 1447-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams, B. R., Mirzoeva, O. K., Morgan, W. F., Lin, J., Dunnick, W. & Petrini, J. H. (2002) Curr. Biol. 12, 648-653. [DOI] [PubMed] [Google Scholar]
- 13.Petersen, S., Casellas, R., Reina-San-Martin, B., Chen, H. T., Difilippantonio, M. J., Wilson, P. C., Hanitsch, L., Celeste, A., Muramatsu, M., Pilch, D. R., et al. (2001) Nature 414, 660-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demuth, I., Frappart, P. O., Hildebrand, G., Melchers, A., Lobitz, S., Stockl, L., Varon, R., Herceg, Z., Sperling, K., Wang, Z. Q. & Digweed, M. (2004) Hum. Mol. Genet. 13, 2385-2397. [DOI] [PubMed] [Google Scholar]
- 15.Rickert, R. C., Roes, J. & Rajewsky, K. (1997) Nucleic Acids Res. 25, 1317-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peitz, M., Pfannkuche, K., Rajewsky, K. & Edenhofer, F. (2002) Proc. Natl. Acad. Sci. USA 99, 4489-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kracker, S. & Radbruch, A. (2004) Methods Mol. Biol. 271, 149-159. [DOI] [PubMed] [Google Scholar]
- 18.Novobrantseva, T. I., Martin, V. M., Pelanda, R., Muller, W., Rajewsky, K. & Ehlich, A. (1999) J. Exp. Med. 189, 75-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Towbin, H., Staehelin, T. & Gordon, J. (1979) Proc. Natl. Acad. Sci. USA 76, 4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumon-Jones, V., Frappart, P. O., Tong, W. M., Sajithlal, G., Hulla, W., Schmid, G., Herceg, Z., Digweed, M. & Wang, Z. Q. (2003) Cancer Res. 63, 7263-7269. [PubMed] [Google Scholar]
- 21.Ehrenstein, M. R., Rada, C., Jones, A. M., Milstein, C. & Neuberger, M. S. (2001) Proc. Natl. Acad. Sci. USA 98, 14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reina-San-Martin, B., Difilippantonio, S., Hanitsch, L., Masilamani, R. F., Nussenzweig, A. & Nussenzweig, M. C. (2003) J. Exp. Med. 197, 1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiener, F., Babonits, M., Bregula, U., Klein, G., Leonard, A., Wax, J. S. & Potter, M. (1984) J. Exp. Med. 159, 276-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radbruch, A., Muller, W. & Rajewsky, K. (1986) Proc. Natl. Acad. Sci. USA 83, 3954-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reina-San-Martin, B., Nussenzweig, M. C., Nussenzweig, A. & Difilippantonio, S. (2005) Proc. Natl. Acad. Sci. USA 102, 1590-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hein, K., Lorenz, M. G., Siebenkotten, G., Petry, K., Christine, R. & Radbruch, A. (1998) J. Exp. Med. 188, 2369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung, S., Rajewsky, K. & Radbruch, A. (1993) Science 259, 984-987. [DOI] [PubMed] [Google Scholar]
- 28.Rush, J. S., Fugmann, S. D. & Schatz, D. G. (2004) Int. Immunol. 16, 549-557. [DOI] [PubMed] [Google Scholar]
- 29.Robison, J. G., Elliott, J., Dixon, K. & Oakley, G. G. (2004) J. Biol. Chem. 279, 34802-34810. [DOI] [PubMed] [Google Scholar]
- 30.Chaudhuri, J., Khuong, C. & Alt, F. W. (2004) Nature 430, 992-998. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J. H. & Paull, T. T. (2004) Science 304, 93-96. [DOI] [PubMed] [Google Scholar]
- 32.Paull, T. T. & Gellert, M. (1999) Genes Dev. 13, 1276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Jager, M., van Noort, J., van Gent, D. C., Dekker, C., Kanaar, R. & Wyman, C. (2001) Mol. Cell 8, 1129-1135. [DOI] [PubMed] [Google Scholar]
- 34.Paull, T. T. & Gellert, M. (2000) Proc. Natl. Acad. Sci. USA 97, 6409-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang, J. & Dynan, W. S. (2002) Nucleic Acids Res. 30, 667-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Her, C., Vo, A. T. & Wu, X. (2002) DNA Repair (Amst) 1, 719-729. [DOI] [PubMed] [Google Scholar]
- 37.The International Nijmegen Breakage Syndrome Study Group (2000) Arch. Dis. Child 82, 400-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.