Abstract
Objective(s):
Pseudomonas aeruginosa is a Gram-negative and aerobic rod bacterium that displays mucoid and non-mucoid phenotype. Mucoid strains secrete alginate, which is the main agent of biofilms in chronic P. aeruginosa infections, show high resistance to antibiotics; consequently, the biological disruption of mucoid P. aeruginosa biofilms is an attractive area of study for researchers. Alginate lyase gene (algl) is a member of alginate producing operon which by glycosidase activity produces primer for other enzymes in this cluster. Also this activity can destroy the extracellular alginate; therefore this enzyme participates in alginate production and destruction pathway. Alginate lyase causes detachment of a biofilm by reducing its adhesion to the surfaces, and increases phagocytosis and antibiotic susceptibility. In this study, alginate lyase was purified in just one step and its properties were investigated.
Materials and Methods:
The purification was done by affinity chromatography, analysed by SDS-PAGE, and its effect on P. aeruginosa biofilms was surveyed by micro titer plate assay and SEM. The substrate specificity of the enzyme was determined by PCR.
Results:
Alginate lyase from isolate 48 was purified in one step. It is more thermally resistant than alginate lyase from Pseudomonas aeruginosa PAO1 and poly M, poly G and poly MG alginate were the substrate of this enzyme. Moreover, it has an eradication effect on biofilms from P. aeruginosa 48 and PAO1.
Conclusion:
In this study an alginate lyase with many characteristics suitable in medicine such as thermal stability, effective on poly M alginate, and bacterial biofilm destructive was introduced and purified.
Keywords: Alginate lyase, Pseudomonas aeruginosa, Affinity chromatography, Biofilm destruction
Introduction
Pseudomonas aeruginosa is known as one of the most important opportunistic human pathogens that causes infection in the respiratory system, urinary tract, digestive system, blood, wounds and burns, as well as arthritis and infection in immunosuppressed patients such as patients with severe burns and cancer, and its eradication is very difficult due to antibiotic resistance (1). Most infections are caused by the formation of biofilms; therefore, the elimination of the bacteria and its biofilm is necessary to treat these diseases. Mucoid strains can produce biofilms, which are defined as communities of attached microorganisms on a surface that play a significant role in infectious disease. The biofilm shows high rate of resistance to antibiotics and its matrix plays a crucial role in antibiotic resistance (2-4).
Alginate is a linear polymer consisting of β (1-4)-linked D-mannuronate (M) and L-guluronate (G) residues, which are the most important components of the P. aeruginosa biofilm known as a widespread virulence factor. Alginate provides some properties, such as mucoid biofilm structure and virulence factor, and plays a role in the persistence of lung infections; therefore, it has long been considered as an important target for cystic fibrosis (CF) treatment (5). Alginate blocks are polymers consisting of either poly M, poly G or poly MG. Some alginate blocks are acetylated, and observations have shown that the over production of acetylated alginate changes the structure and morphology of biofilms (6). These changes increase biofilm resistance to antimicrobial agents. In addition, alginate acts as a barrier to the penetration of some antibiotics; in other words, alginate production decreases the uptake and early bactericidal effect of aminoglycosides and alginate prevents the diffusion of positively charged hydrophilic drugs (3).
Alginate lyase (EC 4.2.2.3) has been considered as a research subject for many years. Researchers have shown that alginate lyase treatment can reduce viscosity in cultures of clinical isolates and in CF sputum; it dissociates biofilms from abiotic surfaces and enhances phagocytosis and killing of P. aeruginosa by human immune cells, and in addition, improves the efficacy of various antibiotics (7). Thus, alginate lyase can eradicate P. aeruginosa biofilms together with some antibiotics such as gentamicin (8-10).
In this study, rapid purification of alginate lyase from the clinical P. aeruginosa was conducted and its effect on P. aeruginosa biofilm formation, biofilm elimination and its other characteristics, such as substrate specificity and thermal stability, were studied.
Materials and Methods
Bacterial strains and culture
A clinical mucoid P. aeruginosa (strain 48) was isolated from the sputum at Mostafa Khomeini Hospital, Tehran, Iran. P. aeruginosa PAO1 used as reference strain. Both of them were grown in YTG (0.5% yeast extract, 1% tryptone, and 0.2% glucose) at 37 ºC with shaking at 180 rpm for 10 hrs prior to extraction and for micro titer plate assay were grown in Tryptic Soy Broth (TSB), Merck, Germany containing 0.2% glucose.
Bacteria were collected by centrifugation at 28000 ×g for 25 min at 4 ºC, washed in 0.03 M Tris-HCl buffer (pH 7.5) containing MgCl2 with final concentration 0.2 M, and finally resuspended in 0.05 M Tris-HCl buffer (pH 7.3) containing MgCl2 with final concentration 0.2 M.
Alginate lyase extraction procedures
Crude enzyme prepared with a modification of the temperature shock method (11). Briefly, the bacterial suspension in 0.05 M Tris-HCl buffer (pH 7.3) containing MgCl2 with final concentration 0.2 M were vortexed and incubated at 37 ºC for 10 min followed by 15 min at 0 ºC. The bacteria were exposed to this alternating hot and cold temperature cycle three additional times, subsequently centrifuged at 8000 ×g for 15 min at 4 ºC. The supernatants containing alginate lyase were assayed for alginate lyase activity as described below.
Measurement of alginate lyase activity
Enzyme activity was measured by thiobarbituric acid assay (TBA) (12). The reaction mixture contained 100 µl of the alginate lyase preparation, 100 µl of assay buffer (0.03 M Tris-HCl, pH 8.5), containing 9 mM MgCl2 and 0.5 M NaC1 (in some experiments the buffer composition was the same but different concentrations of NaC1 were added), 50 µl of sodium alginate solution (in substrate specificity analysis this amount from each polyM, poly G or poly MG blocks were added) (2 mg/ml in assay buffer) was incubated at 37 ºC then 500 µl of 0.025 M priodric acid (HIO4) dissolved in H2SO4 0.125 M was added and the reaction mixtures were incubated at 37 ºC for 20 min. Then 1 ml of 2% sodium arsenite was added gently and the mixture incubated for 2 min at room temperature, then 2 ml of 0.3% TBA was added gently too. Finally the mixture was put in boiling water for 10 min, and the absorbance was measured at 548 nm. The results were shown in enzyme units (EU). One unit of enzyme activity was defined as the amount of enzyme that produced 1 nmole of β-formyl-pyruvate per min per ml and 0.2 absorbance at 548 nm indicates 10 unit of enzyme activity (13).
Residual alginate lyase enzyme activity at different temperatures
To investigate the stability of the enzyme at different temperatures and finding residual activity of the enzyme, the heat shock extracted enzyme was put for 1 hr at temperatures 37, 50, 60 and 80 ºC and then placed on ice until the temperature reached to zero. Finally, they were placed at 37 ºC and then the enzyme was added to the assay mixture for measuring the activity by TBA assay (14).
Survey the thermal stability of the alginate lyase enzyme at 80 ºC
Enzymes was incubated in 1, 2 and 4 hrs at 80 ºC, then placed on ice and after that was put at 37 ºC. After the temperature reached to 37 ºC, the assay mixture was added and the activity was measured by TBA assay (14, 15).
The effect of alginate lyase enzyme of P. aeruginosa 48 and PAO1 on the alginate blocks
Different blocks of sodium alginate (poly G, poly M and poly MG) were prepared (16). The effect of this extracted enzyme was evaluated quantitatively on the different sodium alginate blocks by TBA assay.
P. aeruginosa 48 biofilm formations in tubular reactors and alginate lyase extraction
In this method the biofilm composed at inner surface of the reactor (17) and the bacterial biomass was collected after 5 days, and the enzyme was extracted by heat shock method.
Purification of alginate lyase with an alginate-sepharose 6B affinity column
An alginate-sepharose 6B affinity column was prepared according to the procedure of Skjak-Braek and Larsen (18). Crude enzyme preparation was dialyzed against 0.05 M imidazole buffer (pH 6.8) and loaded onto alginate-sepharose 6B column. The column was washed with 0.05 M imidazole buffer, and bound proteins were eluted with 0.05 M imidazole buffers containing 0.1 M and 0.5 M NaC1 step by step. After that, the fractions with high absorbance at OD280 were dialyzed against heat shock buffer (Tris-HCl 0.05 M, MgCl2 0.2 M, pH 7.3) and were measured for lyase activity by TBA method and monitored by SDS/polyacrylamide gel electrophoresis by silver nitrate staining.
SDS polyacrylamide gel-electrophoresis
A Laemmli SDS polyacrylamide gel (15% separating gel, pH 8.8 and 4% stacking gel, pH 6.8) was prepared (19). The protein preparations were diluted in 5× sample buffer containing 0.125 M Tris-base (pH 6.8), glycerol, beta-mercaptoethanol and bromophenol blue as tracking dye. The samples were then loaded into the electrophoresis wells after boiling for 5 min. Electrophoresis was carried out in running buffer (0.025 M Tris-base, 0.192 M glycine and 0.1% SDS) at room temperature with 120 mV, until the tracking dye ran off the gel. The gel was stained with silver staining (20).
The effect of alginate lyase on the inhibition of biofilm production and elimination of P. aeruginosa biofilm by micro titre plate assay
Micro titre plate assay was used for assessment of P. aeruginosa biofilm production inhibition and elimination of produced biofilms. For the first purpose, P. aeruginosa 48 and P. aeruginosa PAO1 were cultured overnight on triptic soy agar (TSA, Merck, Germany), and then the bacterial suspension was prepared in TSB containing 0.2% glucose. 100 µl of this suspension and 50 µl of crude enzyme of P. aeruginosa strain 48 were loaded on micro titre plate and incubated at 37 ºC for overnight. After 24 hrs, the wells were washed three times by sterile normal saline. For the second purpose, just 100 µl of this suspension was loaded on micro titre plate and after 24- hr incubation at 37 ºC and three times washing by sterile normal saline, 50 µl of crude enzyme of P. aeruginosa 48 was added and incubated at 37 ºC overnight and then washed.
Then 200 µl of TSB containing 0.2% glucose and 50 µl TTC was added to all wells and incubated in the dark for 5 hrs at 37 ºC and shaking by 120 rpm. After the incubation, well contents were removed to new micro titre plate and absorbance was measured at 490 nm (21).
The effect of alginate lyase on biofilm production inhibition and elimination of P. aeruginosa biofilm by scanning electron microscopy (SEM)
In this method for sample preparation, 1 cm × 1 cm slides were placed in wells of micro titre plate. For biofilm production inhibition assessment, the bacterial suspension was adjusted to McFarland standard 0.5 in TSB containing 0.2% glucose and the same volume of the extracted alginate lyase enzyme was added. Then the mixture was loaded onto wells and incubated overnight at 37 ºC. For the analysis of P. aeruginosa biofilm elimination, the bacterial suspension was adjusted to McFarland standard 0.5 in TSB containing 0.2% glucose and was loaded onto wells and incubated overnight at 37 ºC. After incubation, the wells were washed by PBS buffer pH 7 three times and the same volume of the extracted alginate lyase enzyme was loaded on the wells and incubated overnight at 37 ºC. After that the wells were rinsed three times by sterile deionized water and 2.5% glutaraldehyde was added and located overnight in the dark at 4 ºC. Then it was rinsed by sterile deionized water, and for dehydration it left 7 min in 25%, 50% and 75% ethanol and twice in 90% ethanol and three times in absolute ethanol. After air drying in desiccators for 48 hr, finally coated by gold particle and observed by scanning electron micro-scopy (22).
Alginate lyase gene evaluation
After DNA extraction by phenol- chloroform method (22), the degenerate primers D1 (5´-WBBAACAACCACTCVTACTGG-3´) and D2 (5´-BGHACARSAGGGTTCCAGCCA-3´) were used for M-specific lyases conserved region amplification (16).PCR was carried out for 35 cycles, each cycle with 1 min denaturation at 94 ºC, 45 Sec annealing at 62 ºC, and 1 min extension at 72 º C and the final elongation step at 72 ºC was for 10 min. PCR products were electrophoretically analyzed, isolated from gel and sequenced (Zist fanavaran company). The sequence was compared with conserved region for M-specific lyases by Bioedit software.
Results
Enzyme extraction
The alginate lyase enzyme was extracted from planktonic state of bacteria and its activity was assayed.
The effect of NaCl concentration on alginate lyase activity
Various concentrations of NaCl (0–0.5 M) were used in the assay buffer. After reviewing by the ANOVA program (one-way ANOVA, Tukey test and P<0.05), 0.5 M NaCl was chosen as the optimal concentration (Figure 1).
Figure 1.
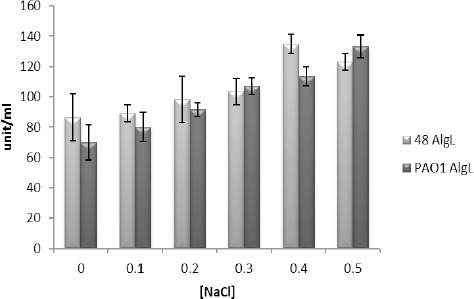
The alginate lyase activity from isolates PAO1 and 48 at different NaCl concentrations
Thermal stability of the alginate lyase
In this study, after incubation of the alginate lyase derived from P. aeruginosa PAO1 and P. aeruginosa 48 for 1 hr in the temperature range of 37–80 °C, the maximum amount of residual activity was found to be at 37 °C (Figure 2), but the enzyme had suitable stability at 80 °C. Thereafter, at different time points of 1–4 hr, the enzyme was incubated at 80 °C and after 4 hr, the activity of the enzyme extracted from PAO1 was 19.04% and from the strain 48 was 35.81% (Figure 3).
Figure 2.
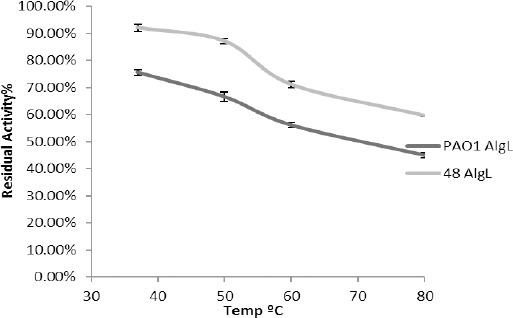
The residual activity of the alginate lyase from isolates PAO1 and 48 after 1 hr incubation at different temperatures
Figure 3.
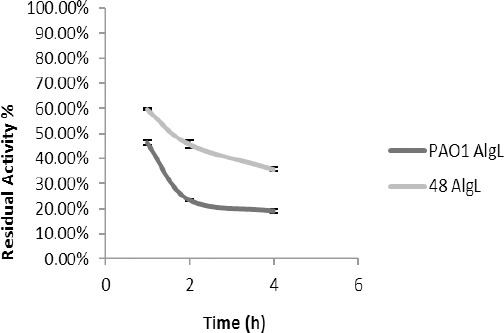
The residual activity of the alginate lyase from isolates PAO1 and 48 at 80 ºC at various times
Enzyme activity at 37 °C without incubation was considered as 100% and the results were analysed by ANOVA program (One-way ANOVA, Tukey test and P<0.05).
Substrate specificity of alginate lyase
To investigate the substrate specificity of the enzymes, the blocks of poly M and poly G containing sodium alginate were used in the TBA assay. We found that alginate lyases from the strains PAO1 and 48 are effective on all blocks. The enzyme from strain PAO1 showed no difference between the effects on the blocks containing poly M and poly G, but the enzyme from the strain 48 showed more activity towards the poly G block than the poly M block (like previous we used One-way ANOVA, Tukey test and P<0.05). Upon comparison between enzymes from strains 48 and PAO1, we concluded that there was no significant difference in their activities on poly G (Figure 4).
Figure 4.
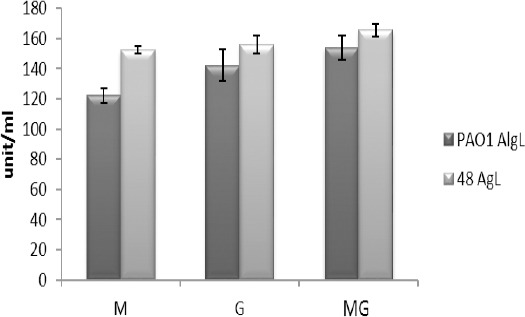
The alginate lyase assay of strain 48 and strain PAO1 using polyM, polyG and poly MG blocks of sodium alginate as substrates
Detection of poly M alginate lyase conserve region
In this study also, we used these primers to characterise the alginate lyase gene from strain 48. The results showed high sequence similarity from strain PAO1 and strain 48 to the conserved region in the poly M alginate lyase genes (Figure 5 and Figure 6).
Figure 5.

The alignment between the alginate lyase sequences of strain 48 with conserved sequence of PAO1 strain
Figure 6.
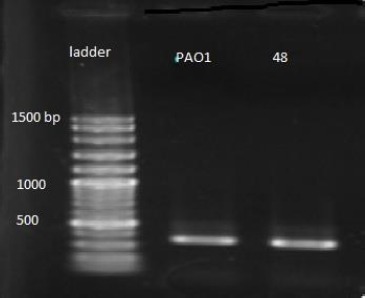
PCR analysis of poly M conserved region in strain PAO1 and strain 48
Alginate lyase purification
Crude preparations were fractionated on an alginate-sepharose 6B affinity column. The majority of alginate lyase activity was found when the column was eluted with 0.5 M NaCl (Table 1).
The fraction that had lyase activity was applied on SDS-polyacrylamide gels and was shown to be approximately 60 kDa (Figure 7).
Figure 7.
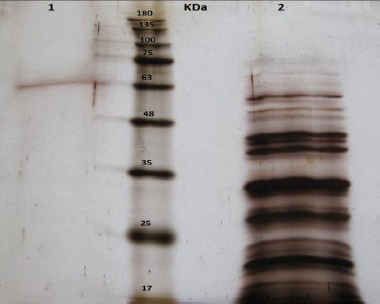
SDS-polyacrylamide gel electrophoresis of alginate lyase. Line 1: The purified enzyme by affinity chromatography; line 2: The crude extract
The effect of the enzyme extract of P. aeruginosa 48 on P. aeruginosa biofilm
In this study, the destruction of P. aeruginosa biofilm by the alginate lyase enzyme was studied using micro titer plates, and it caused a significant reduction in biofilm. According to present study, the enzyme extracted from strain 48 could destroy strain PAO1 biofilm to 42.08% and inhibited its formation up to 23.35%, and purified enzyme removed PAO1 biofilms to 19.42%. In addition, this enzyme was capable of destroying its biofilm up to 33.21% and inhibiting biofilm formation to 9.13%, and it could remove biofilm of strain 48 to 19.52% (Figure 8). Also, these results were confirmed by SEM (Figure 9). The ability of the extracted enzyme from strain 48 to destroy its own biofilm was 33.21% and it may contribute towards the treatment of CF patients (in all of this report we used one-way ANOVA, Tukey test and P<0.05). It should be noted that the cell extract involves other materials such as DNase that could play a role in destroying biofilm and this could be a reason for the lower effect of the pure enzyme on biofilm.
Figure 8.
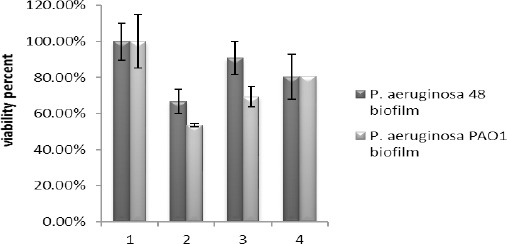
The influence of the enzyme extracts from strain 48 on Pseudomonas aeruginosa 48 and Pseudomonas aeruginosa PAO1 biofilm. 1) 24- hr biofilm 2) The elimination of 24- hr biofilm by enzyme extract from strain 48 3) The inhibition of biofilm formation by the enzyme extract from strain 48 4) The removal of 24- hrs biofilm by the purified enzyme (0.043 mg/ml)
Figure 9.
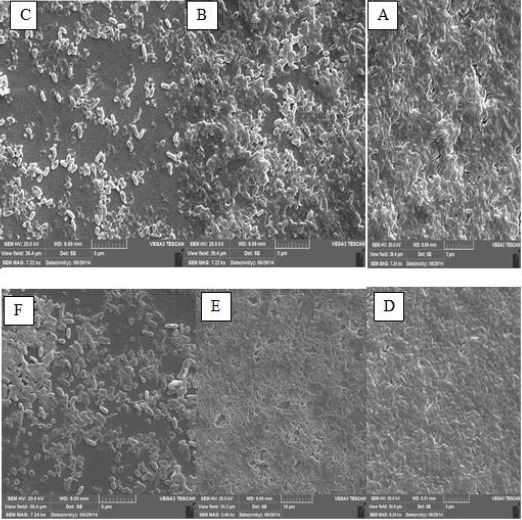
Representative SEM images of A) 24- hr biofilm from Pseudomonas aeruginosa PAO1, B) The removal of 24- hr PAO1 biofilm by the enzyme extract of strain 48, C) The prevention of PAO1 biofilm formation by the enzyme extract of strain 48, D) 24- hr P. aeruginosa 48 biofilm, E) The removal of 24- hr strain 48 biofilm by the enzyme extract of strain 48, F) The prevention of strain 48 biofilm formation by the enzyme extract of strain 48
Discussion
The main biological function of alginate lyase (AlgL) is to degrade alginates that fail to become exported out of the cell and thereby become stranded in the periplasmic space. At high levels of alginate synthesis in the absence of AlgL, such stranded polymers may accumulate in the periplasm to such an extent that the integrity of the cell is lost, leading to the observed toxic effects. During exponential growth in the absence of AlgL, alginate synthesis may be disturbed due to a moderate level of periplasmic swelling, while no direct toxicity is generated. At late stages of growth, on the other hand, both toxicity and inefficient alginate synthesis are observed due to the more extensive accumulation of polymer in the periplasm (24). Because of this property of AlgL, many researchers also use it in degradation of alginate moiety of P. aeruginosa biofilm.
AlgL has different properties in the various strains of a species of bacterium. The aim of this study was extraction of the clinical P. aeruginosa AlgL and characterization of it.
The optimum concentration of NaCl for the strain 48 and PAO1 AlgL activity was 0.5 M but Eftekhar and Schiller observed highest activity in a buffer containing 0.3 M NaCl for alginate lyase derived from P. aeruginosa WcM # 2 (11). Li and colleagues also found highest activity in a buffer containing 0.2 M NaCl for the enzyme derived from Pseudoalteromonas sp. SM0524 that represents the salt-resistant bacteria (25).
In this research after incubation at 80 °C for 4 hr, 35.81% of the strain 48 and 19.04% of the strain PAO1 enzyme activity was remained whereas, Xiao and colleagues investigated the thermal stability of the enzyme from a mucoid strain of P. aeruginosa after 1 hr of incubation at a temperature range 30–80 °C, and found that the highest thermal stability was at 30 °C and its activity was lost at 80 °C (14). Fu and colleagues also studied the enzyme from Vibrio sp. YKW-34, and observed that the enzyme was stable up to 70 °C but its activity reaches zero at 80 °C (26).
In general, there are great differences in the characteristics of alginate lyases, hence different substrate specificities have been reported for various alginate lyases. Farrell and colleagues worked on the substrate specificity of alginate lyase from P. aeruginosa, and during the study, it became clear that the enzyme is able to work on both polyM and poly G alginate (7), equally well; Jain and colleagues confirmed that the alginate lyase from P. aeruginosa has a bifunctional role in alginate transfer (27). Present study also showed that the enzymes of the strains PAO1 and 48 are effective on both blocks.
Xiao and colleagues designed primers to detect bacteria that produce polyM alginate lyase (14). The comparison between the sequences of poly M affective lyases has shown two conserved regions, ‘NNHSYW’ at the middle of protein sequence and ‘WLEPYCALY’ at the carboxyl end in strains 48 and PAO1.
In this study, the enzyme from strain 48 with significant properties was purified by a one-step procedures and it was shown that like the other investigations (8-10), it eliminates P. aeruginosa biofilms.
Conclusion
In this study, in one step, alginate lyase was purified. In addition, it was shown that this enzyme has the highest activity in buffer containing 0.5 M NaCl. It is thermally stable and it could eliminate the biofilm of P. aeruginosa and it could act on poly M alginate. All of these characteristics make the use of this enzyme important in the industry and in medicine.
Acknowledgment
The results described in this paper were part of student thesis and this survey has been supported by vice chancellor of Alzahra University, Tehran, Iran.
References
- 1.Pinard MA, Lotlikar SR, Boone CD, Vullo D, Supuran CT, Patrauchan MA, et al. Structure and inhibition studies of a type II beta-carbonic anhydrase psCA3 from Pseudomonas aeruginosa. Bioorg Med Chem. 2015;23:4831–4838. doi: 10.1016/j.bmc.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 2.O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;45:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 3.Høiby N, Johansen HK, Moser C, Song ZJ, Ciofu O, Kharazmi A. Pseudomonas aeruginosa and the biofilm mode of growth. Microb Infect. 2001;3:1–13. doi: 10.1016/s1286-4579(00)01349-6. [DOI] [PubMed] [Google Scholar]
- 4.Prasanna SS, Doble M. Medical biofilms–Its formationand prevention using organic molecules. J Indian Inst Sci. 2008;88:27–35. [Google Scholar]
- 5.Gacesa P. Bacterial alginate biosynthesis-recent progress and future prospects. Microbiology. 1998;144:1133–1143. doi: 10.1099/00221287-144-5-1133. [DOI] [PubMed] [Google Scholar]
- 6.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis:mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrell EK, Tipton PA. Functional Characterization of AlgL, an Alginate Lyase from Pseudomonas aeruginosa. Biochemistry. 2012;51:10259–10266. doi: 10.1021/bi301425r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatch RA, Schiller NL. Alginate Lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:974–977. doi: 10.1128/aac.42.4.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alipour M, Suntres ZE, Omri A. Importance of DNase and alginate lyase for enhancing free and liposome encapsulated aminoglycoside activity against Pseudomonas aeruginosa. J Antimicrob Chemother. 2009;64:317–325. doi: 10.1093/jac/dkp165. [DOI] [PubMed] [Google Scholar]
- 10.Lamppa JW, Griswolda KE. Alginate lyase exhibits catalysis-independent biofilm dispersion and antibiotic synergy. Antimicrob Agents Chemother. 2013;57:135–147. doi: 10.1128/AAC.01789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eftekhar F, Schiller NL. Partial purification and characterization of a mannuronan-specific alginate lyase from Pseudomonas aeruginosa. Curr Microbiol. 1994;29:37–42. [Google Scholar]
- 12.Weissbach A, Hurwitz J. The formation of 2-keto-3- deoxyheptonic acid in extracts of Escherichia coli B.I. Identification. J Biol Chem. 1959;234:705–709. [PubMed] [Google Scholar]
- 13.Nguyen LK, Schiller NL. Identification of a slime polysaccharide depolymerase in mucoid strain of Pseudomonas aeruginosa. Curr Microbiol. 1989;18:323–329. [Google Scholar]
- 14.Xiao L, Han F, Yang Z, Lu X, Yu W. A novel alginate lyase with high activity on acetylated alginate of Pseudomonas aeruginosa FRD1 from Pseudomonas sp.QD03. World J Microbiol Biotechnol. 2006;22:81–88. [Google Scholar]
- 15.Miyake O, Ochiai A, Hashimoto W, Murata K. Origin and diversity of alginate lyases of families PL-5 and -7 in sphingomonas sp. strain A1. J Bacteriol. 2004;186:2891–2896. doi: 10.1128/JB.186.9.2891-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haug A, Larsen B, Smidsrød O. A study of the constitution of alginic acid by partial acid hydrolysis. Acta Chem Scand. 1966;20:183–190. [Google Scholar]
- 17.Sepehr Sh, Rahmani-Badi A, Babaie-Naiej H, Soudi MR. Unsaturated fatty acid, cis-2-decenoic acid, in combination with disinfectants or antibiotics removes pre-established biofilms formed by food-related bacteria. Plos One. 2014;9:1–9. doi: 10.1371/journal.pone.0101677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.SkjaK-BrK G, Larsen B. Purification of mannuronan C5-epimerase by affinity chromoatography on alginate-sepharose. Carbohydr Res. 1982;103:137–140. [Google Scholar]
- 19.Laemmeli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nat Pub Group(NPG) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Caetano-Anolles G, Bassam BJ. DNA amplification finger printing using arbitrary oligonucleotide primers. Appl Biochem Biotechnol. 1993;42:189–200. doi: 10.1007/BF02788052. [DOI] [PubMed] [Google Scholar]
- 21.Sabaeifard P, Abdi-Ali A, Soudi MR, Dinarvand R. Optimization of tetrazolium salt assay for Pseudomonas aeruginosa biofilm using micro titer plate method. J Microbiol Methods. 2014;105:134–140. doi: 10.1016/j.mimet.2014.07.024. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Hanna MN, Svensater G, Ellen RP, Cvitkovitch DG. Cell density modulates acid adaptation in streptococcus mutans:implications for survival in biofilms. J Bacteriol. 2001;183:6875–6884. doi: 10.1128/JB.183.23.6875-6884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol. 2001;2:2–4. doi: 10.1002/0471142727.mb0204s56. [DOI] [PubMed] [Google Scholar]
- 24.Bakkevig K, Sletta H, Gimmestad M, Aune R, Ertesvåg H, Degnes K, et al. Role of the Pseudomonas fluorescens alginate lyase (AlgL) in clearing the periplasm of alginates not exported to the extracellular environment. J Bacteriol. 2005;187:8375–8384. doi: 10.1128/JB.187.24.8375-8384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JW, Dong S, Song J, Li CB, Chen XL, Xie BB, et al. Purification and characterization of a bifunctional alginate lyase from Pseudoalteromonas sp.SM0524. Mar Drugs. 2011;9:109–123. doi: 10.3390/md9010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu XT, Lin H, Kim SM. Purification and characterization of a Na+/ K+dependent alginate lyase from turban shell gut vibrio sp.YKW-34. Enzyme Microb Technol. 2007;41:828–834. [Google Scholar]
- 27.Jain S, Ohman DE. Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa. Infect Immun. 2005;73:6429–6436. doi: 10.1128/IAI.73.10.6429-6436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


