Abstract
Objective(s):
Chronic myelogenous leukemia (CML) is a chronic myeloproliferative disorder characterized by the accumulation of myeloid cells with a chromosomal translocation known as the Philadelphia chromosome. In this study, we investigated the roles of miR-570 in CML development.
Materials and Methods:
Expression of miR-570 in CML samples and cell lines was determined by qRT-PCR. Glucose uptake and ATP concentration detection assays were used to analyze cell glucose metabolism. MTT and western blot assays were performed for cell proliferation and apoptosis, respectively. The targets of miR-570 were predicted by bioinformatics and confirmed using luciferase activity, qRT-PCR and western blot assays.
Results:
The expression levels of miR-570 were significantly reduced in CML clinical samples and cells. Overexpression of miR-570 inhibited cell proliferation, promoted apoptosis, and suppressed glucose metabolism in CML cells. Insulin receptor substrates (IRS) 1 and IRS2 were identified as direct targets of miR-570. IRS1 or IRS2 were knocked down in K562 cells. Loss of IRS1/2 expression led to suppressed cell proliferation, elevated apoptosis, and decreased glucose metabolism in CML cells, which is consistent with their roles as miR-570 targets.
Conclusion:
MiR-570 directly targeted IRS1 and IRS2 in CML, suppressing cell proliferation and glucose metabolism. MiR-570 may provide a strategy for CML therapy.
Keywords: Chronic Myelogenous Leukemia, IRS1, IRS2, miR-570
Introduction
Chronic myelogenous leukemia (CML) is a chronic myeloproliferative disorder characterized by the accumulation of myeloid cells with a chromosomal translocation known as the Philadel-phia chromosome (1). This disease affects about 1 in 60,000 individuals, and is predicted to result in 6,000 new diagnoses each year in the United States (2). The pathological mechanism of CML has been shown to involve a fusion event between the breakpoint cluster region protein (BCR) gene on chromosome 22 and the abelson murine leukemia viral oncogene homolog 1 (ABL-1) gene on chromosome 9, leading to constitutive activation of ABL-1, which in turn inhibits DNA repair and activates a number of signaling pathways to induce myeloid cell prolifera-tion (3).
MicroRNAs (miRNAs) are a class of 19-22 nucleotide ribonucleic acids (RNAs), which have recently emerged as novel regulators of cellular activities. They regulate gene expression in a post-transcriptional manner via binding to the 3’-untranslated region (3’-UTR) of target mRNA, resulting in mRNA degradation or translational inhibition (4). Over the past decade, a body of literature points to a major role for miRNAs in cancer initiation, progression, and metastasis (5). It has been shown that miRNAs may function as oncogenes in gain-of-function, or alternatively, as tumor suppressors in loss-of-function models. These results have been substantiated in human cancers in which deregulation of miRNA expression has been repeatedly reported. Importantly, it has been demonstrated that the development of CML is also
regulated by microRNAs. Overexpression study of miR-424 has shown that it inhibited gene expression of BCR/ABL by binding to the coding and 3’-UTR regions to sensitize CML to imatinib (6). The downregulation of miR-362-5p has also been shown to decrease the growth by activating the function of (GADD45α)-mediated JNK1/2 and P38 signaling (7).
Previous studies by other groups have established that miR-570 participates in the regulation of multiple human cancer types, including gastric (80, liver (9), and lung (10) cancers.
Patients
Fresh blood samples were collected from 15 chronic myelogenous leukemia (CML) patients at the Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang China. The samples were immediately snap-frozen in liquid nitrogen, and stored at -80 °C until RNA extraction. The study was approved by ethical committee of the Second Affiliated Hospital of Harbin Medical University, and each participant provided written informed consent. The clinical characteristics of 15 CML were listed in Table 1.
Table 1.
Characteristics of 15 CML patients
| Feature | N (%) |
|---|---|
| Gender | |
| Male | 9 (60.0) |
| Female | 6 (40.0) |
| Sokal score | |
| Low | 8 (53.3) |
| Intermediate | 4 (26.7) |
| High | 3 (20.0) |
| Subclassification | |
| Chronic | 14 (93.3) |
| Accelerated | 1 (6.7) |
| Blastic | 0 (0.0) |
| BCR-ABL1 mutation | |
| Yes | 6 (40.0) |
| No | 9 (60.0) |
| Splenomegaly | |
| Yes | 11 (73.3) |
| No | 4 (26.7) |
Cell lines, cell culture and cell transfection
Human CML cell lines K562 and LAMA-84 were cultured in DMEM with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), 100 U/ml penicillin and 100 mg/ml streptomycin. All cells maintained in 37 °C, saturated humidity incubator of 5% CO2.
Human miRNA and siRNA transfection to CML cells was performed using Lipofectamine® RNAiMAX Transfection Reagent according to manufacturer’s instructions. Briefly, CML cells (1×105/well) were seeded into 24-well plate. 10 pmol miRNA or siRNA was added into each well with 3 µl Lipofectamine® RNAiMAX Reagent in 50 µl Opti-MEM® Medium. The cells were subjected to further analysis after incubation for two days.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis
qRT-PCR was performed to detect the relative levels of miRNA and mRNA expression levels. Total RNA was extracted using RNA extraction Kit (Qiagen, Santa Clarita, CA, USA) according to the manufacturer’s instructions. Reverse transcription was performed with use of a PrimeScriptTM RT reagent Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. qRT-PCR was carried out using the SYBR Premix ExTaq real-time PCR Kit (Takara, Dalian, China). PCR was carried out in triplicate and analyzed using the ABI Prism 7500HT fast realtime PCR system (Applied Biosystems, Foster City, CA, USA). The relative quantification
values for each gene were calculated by the 2---Ct method using U6 or β-ACTIN as an internal control. The primers for peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC1α), phos-phoenolpyruvate carboxykinase 1 (PCK1) and ATP-binding cassette transporter A1 (ABCA1) were used as previous described (11). The IRS1 and IRS1 primers were used as followed: IRS1 forward 5’-TATGCCAGC-ATCAGTTTCCA-3’ reverse 5’-TTGCTGAGGTCATTTAGG-TCTT-3’; IRS2 forward 5’-TTCTTGTCCCACCACTTGAA-3’ reverse 5’-CTGACATGTGACATCCTGGTG-3’.
3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetra zolium bromide (MTT) assay
K562 cells were transfected with either miRNA (human miR-570, control miRNA or their inhibitors) or siRNA (human IRS1, IRS2 or control siRNA). After 24 hr transfection, cells were seeded into 96-well plate and continue cultured for 0, 24, 48 and 72 hr, respectively. At each time point, 20 µl MTT reagent (5 mg/ml, Sigma, St Louis, MO, USA) was added into each culture well and incubated for 4 hr at 37°C. Afterwards, the supernatant was removed and 150 µl dimethyl sulfoxide (DMSO, Sigma) was added to each well. After shaking for 10 min, absorbance at the wavelength of 570 nm was measured on a microplate reader.
Luciferase reporter assay
The 3’-UTR sequences of the IRS1 and IRS2 genes were amplified by PCR using complementary DNA (cDNA) reverse transcribed from K562 cell mRNA as the template, and were subcloned into the pMIR-REPORT luciferase vector to generate the IRS1 and IRS2 3’-UTR luciferase reporter plasmids. Plasmids and miRNA were co-transfected into HEK293T cells.
After 48 hr of transfection, cells were lysed with extraction buffer. Luciferase activity assays were performed using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA) following the manufacturer’s instructions.
Western blot analysis
Total K562 cell proteins were extracted using radioimmunoprecipitation assay (RIPA) buffer (200 µl, 50 mM Tris pH 8.0, 150 mM NaCl, 1% Nonidet P40 and 0.1% SDS) after treatment. The concentrations of the proteins were detected by using BCA protein assay Kit (Beyotime Biotech, Beijing, China). Lysates with equal protein were isolated by 10 % SDS-PAGE and transferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). Membranes were blocked for 1 hour in 5% non-fat milk in Tris-buffered saline with Tween 20 (TBST) and incubated with primary antibodies against cleaved Caspase 3, anti-Caspase 3, IRS1, IRS2 and β-ACTIN (Santa Cruz, Santa Cruz, CA, USA) overnight at 4 °C, followed by the incubation with appropriate horseradish peroxidase (HRP)-conjugated secondary antibody at optimized concentration. Signal was detected with an enhanced chemiluminescence (ECL) system (Beyotime Biotech).
Glucose uptake assay
Glucose uptake in K562 cells was measured using a fluorescent D-glucose derivative 2-[N-(7-nitrobenz-2-oxa-1,3-diaxol-4-yl)amino]-2-deoxyglucose (2-NBDG, Sigma). Briefly, after 30 min of glucose starvation, K562 cells were incubated for 20 min in the presence of 2-NBDG. Cells were then washed and the fluorescence was analyzed using a fluorescence microplate reader.
ATP concentration detection
ATP concentration was measured by luciferase activity assay using a luciferase kit from Sigma following the manufacturer’s protocol.
Statistical analysis
The data were presented as mean±standard deviation (SD). Experimental data were analyzed by Student’s t-test or one-way analysis of variance (ANOVA), and statistical analysis was performed with SPSS 16.0 software. Data were considered to be statistically significant when P<0.05.
Results
MiR-570 is downregulated in CML clinical samples and cell lines
In order to investigate the involvement of miR-570 in CML development, we first analyzed the expression level of miR-570 in blood samples from 15 CML patients and 8 healthy participants. The expression levels of miR-570 were significantly reduced in the blood samples from CML patients, compared to those in the blood samples from healthy participants (Figure 1A). Furthermore, gene expression data from Gene Expression Omnibus (GEO) DataSets GSE64011 was analyzed (12). The author compared miRNA expression between CML bone marrow cells and peripheral blood samples of healthy control participants. MiR-570 expression was decreased in bone marrow cells from CML patients, compared to those from healthy control participants (Figure 1B). In addition, we examined miR-570 expression in two CML cell lines, K562 and LAMA-84 cells. The expression levels of miR-570 were found significantly lower in both of the two CML cell lines compared to those in healthy blood pool sample (Figure 1C). Taken together, these results indicated that miR-570 was downregulated in both CML clinical samples and in CML cells, suggesting that miR-570 could be involved in the regulation of CML development.
Figure 1.
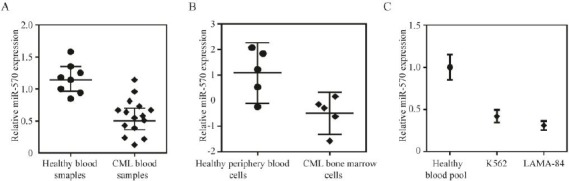
MiR-570 is downregulated in chronic myelogenous leukemia (CML) clinical samples and cell lines
(A) Expression levels of miR-570 in blood samples of 15 CML patient and 8 healthy volunteer as determined by qRT-PCR (P<0.001). U6 was used as an internal control. (B) Expression levels of miR-570 in bone marrow cells (CD34+) from five CML patients compared with those in periphery blood cells (CD34+) from five healthy participants as determined by analysis of Gene Expression Omnibus (GEO) DataSets GSE64011 (P<0.05). (C) Expression levels of miR-570 in K562 and LAMA-84 CML cells compared with those in healthy blood cells as determined by qRT-PCR (P<0.05)
MiR-570 regulates the proliferation, apoptosis and glucose metabolism of CML cells
In order to investigate the roles of miR-570 in regulating the biological functions of CML cells, the miR-570 was overexpressed and knocked down in K562 cells by transfection of miR-570 plasmid and miR-570 inhibitor, respectively. The overexpression and the knockdown efficacies were confirmed by qPCR analysis (Figure 2A). MTT cell proliferation assay was then performed on these K562 cells. It was found that overexpression of miR-570 significantly inhibited the proliferation of K562 cells, whereas knockdown of miR-570 promoted K562 cell proliferation (Figure 2B). Furthermore, the apoptosis of K562 cells induced by serum free culture was examined. Overexpression of miR-570 led to both increased cleavage of Caspase-3 (Figure 2C) and increased cell death (Figure 2D). In contrast, knockdown of miR-570 resulted in reduced Caspase-3 cleavage (Figure 2C) and decreased cell death (Figure 2D). Since substantial changes in glucose metabolism have been demonstrated in the development of CML (13), we also sought to examine whether miR-570 affected glucose metabolism in CML cells. The 2-NBDG glucose uptake assay showed that miR-570 overexpression significantly reduced glucose uptake in K562 cells, whereas miR-570 knockdown stimulated glucose uptake (Figure 3A). Importantly, the catabolism of glucose in K562 cells was also affected by miR-570. Overexpression of miR-570 reduced ATP generation, while knockdown of miR-570 increased ATP generation in K562 cells (Figure 3B). In addition, the expression of genes associated with glucose metabolism, PGC1α, PCK1 and ABCA1 was examined (11). It was found that the expression levels of these genes were suppressed by miR-570 overexpression, and were induced by miR-570 knockdown (Figure 3C). Collectively, these results indicated that miR-570 inhibited cell proliferation, stimulated cell apoptosis, and suppressed glucose metabolism in CML cells.
Figure 2.
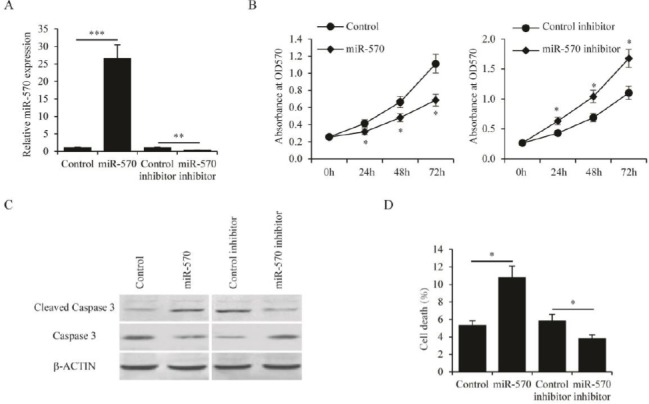
MiR-570 regulates proliferation and apoptosis of chronic myelogenous leukemia (CML) cells
(A) Expression levels of miR-570 as determined by qRT-PCR in K562 cells transfected with control plasmid, miR-570 expressing plasmid, control inhibitor, or miR-570 inhibitor. (B) MTT assay performed in K562 cells transfected with control plasmid, miR-570 expressing plasmid, control inhibitor, or miR-570 inhibitor at 0, 24, 48, and 72 hr after transfection. (C) Protein levels of cleaved Caspase 3 and full length Caspase 3 as determined by western blot analysis in K562 cells transfected with control plasmid, miR-570 expressing plasmid, control inhibitor, or miR-570 inhibitor. Cells were cultured in medium without serum. (D) Percentage of cell death in K562 cells transfected with control plasmid, miR-570 expressing plasmid, control inhibitor, or miR-570 inhibitor. Cells were cultured in medium without serum. All experiments were repeated in triplicate, mean±SEM, *P<0.05, **P<0.01 and ***P<0.001
Figure 3.
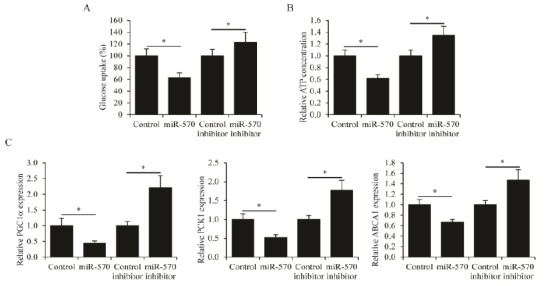
MiR-570 regulates glucose metabolism in chronic myelogenous leukemia (CML) cells
Levels of glucose uptake and (B) ATP concentrations in K562 cells transfected with control plasmid, miR-570 expressing plasmid, control inhibitor, or miR-570 inhibitor. (C) Expression levels of peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC1α), phosphoenolpyruvate carboxykinase 1 (PCK1) and ATP-binding cassette transporter A1 (ABCA1) as determined by qRT-PCR in K562 cells transfected with control plasmid, miR-570 expressing plasmid, control inhibitor, or miR-570 inhibitor. All assays were performed in three independent experiments, mean±SEM, *P<0.05
IRS1 and IRS2 are targets of miR-570
The molecular targets of miR-570 in CML cells were then identified. Using bioinformatics tools TargetScan (http://www.targetscan.org), IRS1 and IRS2 could be putative targets of miR-570 based on multiple complementary binding sites (Figure 4A). To confirm this prediction, the 3’-UTR sequence of human IRS1 and IRS2 were subcloned into pMIR-REPORT plasmids, then co-transfected these plasmids into HEK293T cells with either miR-570 plasmid or miR-570 inhibitor. Co-transfection with miR-570 plasmid significantly reduced the luciferase activity coupled to the 3’-UTR of both IRS1 and IRS2, whereas co-transfection with miR-570 inhibitor enhanced the luciferase activity coupled to the 3’- UTR of both IRS1 and IRS2 (Figure 4B). Furthermore, the expression levels of IRS1 and IRS2 in K562 cells transfected with either miR-570 plasmid or miR-570 inhibitor were examined. Consistent with the luciferase reporter assay results, the mRNA levels, as determined by qRT-PCR analysis (Figure 4C), and the protein levels, as determined by Western blot analysis (Figure 4D), of both IRS1 and IRS2 were suppressed by the transfection of miR-570 plasmid, and were promoted by the transfection of miR-570 inhibitor. In addition, from clinical samples, negative correlation between the expression levels of miR-570 and IRS1, and between those of miR-570 and IRS2 (Figure 5). Taken together, these results were all consistent with the hypothesis that both IRS1 and IRS2 were inhibitory targets of miR-570.
Figure 4.
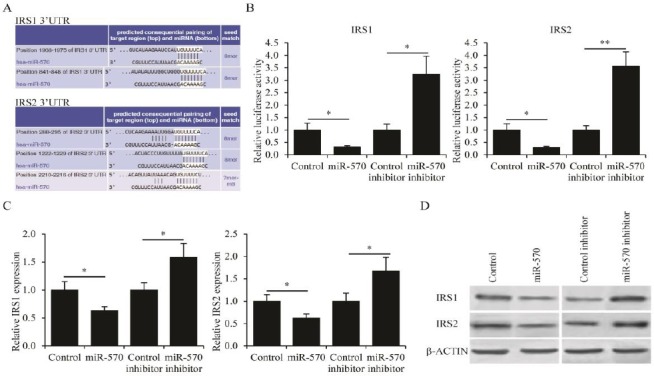
IRS1 and IRS2 are targets of miR-570
(A) Putative miR-570 binding sites in the 3’- untranslated region (UTR) of IRS1 and IRS2 mRNA. (B) Normalized luciferase activity in HEK293T cells co-transfected with IRS1 or IRS2 luciferase reporter plasmids, and control plasmid, miR-570 expressing plasmid, control inhibitor, or miR-570 inhibitor. (C) IRS1 and IRS2 mRNA levels as determined by qRT-PCR analysis and (D) IRS1 and IRS2 protein levels as determined by western blot in K562 cells transfected with control plasmid, miR-570 expressing plasmid, control inhibitor, or miR-570 inhibitor. All experiments were repeated in triplicate, mean±SEM, *P<0.05
Figure 5.

Negative correlation between miR-570 and IRS1/2 expression
(A) Correlation analysis performed between miR-570 expression levels and IRS1 mRNA expression levels in chronic myelogenous leukemia (CML) (n = 15, r = -0.542). (B) Correlation analysis performed between miR-570 expression levels and IRS2 mRNA expression levels in CML (n = 15, r = -0.747)
IRS1 and IRS2 regulate CML cell proliferation, apoptosis and glucose metabolism
IRS1 and IRS2 have been reported to function as oncogenes to promote cancer development (14-16). In this context, since IRS1 and IRS2 were demons-trated in this study to be target genes of miR-570, we decided to investigate whether IRS1 and IRS2 regulate the development of CML. To this end, the expressions of IRS1 or IRS2 in K562 cells were knocked down by siRNA transfection, and the efficacy of knockdown was confirmed by western blot analysis (Figure 6A). MTT cell proliferation assay was then performed on these cells. Knockdown of either IRS1 or IRS2 significantly decreased the proliferation of K562 cells (Figure 6B). It was also found that both IRS1 and IRS2 knockdown led to higher levels of cell apoptosis induced by no serum culture conditions (Figure 6C). In addition, K562 cells with either IRS1 or IRS2 knockdown exhibited significantly lower levels of glucose uptake (Figure 6D). These results suggested that knockdown of IRS1 and IRS2 could inhibit the growth and expansion of CML cells, and therefore supported the stimulatory roles for IRS1 and IRS2 in CML development. These results were consistent with the inhibitory function of miR-570 in regulating the development of CML.
Figure 6.
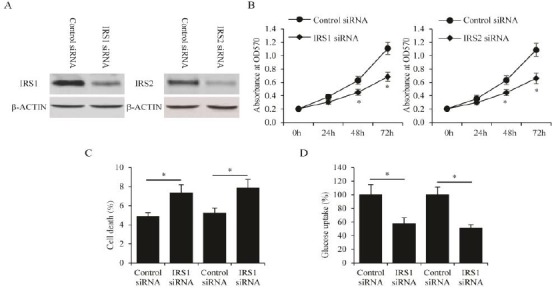
Knockdown of IRS1/2 suppresses chronic myelogenous leukemia (CML) cell proliferation and glucose metabolism
(A) Protein levels of IRS1 and IRS2 as determined by western blot in K562 cells transfected with siRNAs targeting IRS1 and IRS2, respectively. (B) MTT assay performed in K562 cells transfected with siRNAs targeting IRS1 or IRS2 at 0, 24, 48, and 72 hr after transfection. (C) Percentage of cell death in K562 cells transfected with siRNAs targeting IRS1 or IRS2. Cells were cultured in medium without serum. (D) Levels of glucose uptake in K562 cells transfected with siRNAs targeting IRS1 or IRS2. All assays were performed in three independent experiments, mean ± SEM, *P<0.05
Discussion
In this study, we explored the function of miR-570 in the regulation of CML development. High expression of B7-H1 indicated increased risk of death in malignant tumor patients. It was also found that miR-570 is bound to 3’UTR of B7-H1mRNA in gastric cancer (8) and inhibited liver cancer cells by targeting B7-H1 (9). However, miR-570 functions as an oncogene in lung cancer through inhibiting KLF9 (10). Although a number of studies have revealed the functional association between miR-570 and various types of cancers, our study represents the first effort to establish the involvement of miR-570 in hematopoietic malignancy.
In this study, the function of miR-570 on cell proliferation, cell apoptosis, and cellular glucose metabolism was examined. The expression levels of miR-570 were significantly reduced in both CML clinical samples and CML cells. Overexpression of miR-570 resulted in suppressed cell proliferation, elevated levels of apoptosis, and reduced glucose metabolism. Inhibition of miR-570, on the other hand, led to opposite cell behaviors. These aspects of cell behavior are among the key steps in the progression of cancer malignancy. Therefore, these results were consistent with our discovery that miR-570 were downregulated in CML.
IRS1/2 were identified as inhibitory targets of miR-570 by bioinformatics prediction, as confirm-ed the prediction with different approaches. First, a luciferase assay was established to monitor the activities of the 3’-UTR of IRS1 and IRS2. It turned out that in co-transfection assays, the presence of miR-570 significantly reduced 3’-UTR activities to drive luciferase expression. This observation was further substantiated by the discovery that endogenous IRS1 and IRS2, in both mRNA and protein levels, were suppressed by the overexpression
of miR-570, and were stimulated by miR-570 inhibition. In a third approach, an inverse correlation was found between the miR-570 and IRS1/2 expression levels in clinical human samples. Combination of these evidences indicated that IRS1 and IRS2 are bona fide direct targets of miR-570.
IRS1 and IRS2 have been extensively studied for their involvement in cancer malignancy. Upon activation by insulin/insulin-like growth factor I receptors, IRSs can recruit intracellular proteins containing Src homology-2 domains and lead to the transduction of intracellular signaling pathways (17). Recent studies showed that constitutive activation of IRS1/2 was found in a variety of solid tumors, and that overexpression of IRS1/2 promoted tumorigenesis and metastasis (18-24). The activation of IRS1/2 in cancer development is consistent with our findings that knockdown of IRS1 and IRS2 inhibits the growth and expansion of CML cells, and is compatible with our hypothesis that miR-570 suppresses CML progression by inhibiting IRS1/2. It is important to note that, since our study is only focused on CML, the possibility that miR-570 regulates other cancer types by inhibiting other targets than IRS1/2 cannot be excluded.
Recent discovery of miRNAs as important regulators of gene expression and the common deregulation of miRNA expression in cancer malignancies shed a light on our understanding of CML. Future studies will be needed to determine whether miR-570 can be used to aid in diagnosis, prognosis, and determination of the optimal treatment options in CML. Furthermore, future studies need also address whether pharmacological activators for miR-570 can be used to improve the outcomes of CML patients.
Conclusion
In this study, miR-570 was found to function as an anti-cancer miRNA in CML, through inhibition of IRS1 and IRS2 expression; in addition, it regulated several processes of CML cell development including promotion of apoptosis, inhibition of cell prolifera-tion and glucose metabolism. The effect of IRS1/2 was further investigated on CML. Consistent with their roles as miR-570 targets, knockdown of IRS1/2 led to inhibition of cell malignance.
Conflict of interest
All authors declared no conflict of interest in this manuscript.
References
- 1.Nowell PC. The minute chromosome (Phl) in chronic granulocytic leukemia. Blut. 1962;8:65–66. doi: 10.1007/BF01630378. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller K D, Jemal A. Cancer statistics 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.McWhirter JR, Galasso DL, Wang JY. A coiled-coil oligomerization domain of Bcr is essential for the transforming function of Bcr-Abl oncoproteins. Mol Cell Biol. 1993;13:7587–7595. doi: 10.1128/mcb.13.12.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity:microRNA biogenesis pathways and their regulation. Nat Cell. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 5.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hershkovitz-Rokah O, Modai S, Pasmanik-Chor M, Toren A, Shomron N, Raanani P, et al. Restoration of miR-424 suppresses BCR-ABL activity and sensitizes CML cells to imatinib treatment. Cancer Lett. 2015;360:245–256. doi: 10.1016/j.canlet.2015.02.031. [DOI] [PubMed] [Google Scholar]
- 7.Yang P, Ni F, Deng RQ, Qiang G, Zhao H, Yang MZ, et al. MiR-362-5p promotes the malignancy of chronic myelocytic leukaemia via down-regulation of GADD45alpha. Mol Cancer. 2015;14:190. doi: 10.1186/s12943-015-0465-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Wang W, Li F, Mao Y, Zhou H, Sun J, Li R, et al. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet. 2013;132:641–648. doi: 10.1007/s00439-013-1275-6. [DOI] [PubMed] [Google Scholar]
- 9.Guo W, Tan W, Liu S, Huang X, Lin J, Liang R, et al. MiR-570 inhibited the cell proliferation and invasion through directly targeting B7-H1 in hepatocellular carcinoma. Tumour Biol. 2015;36:9049–9057. doi: 10.1007/s13277-015-3644-3. [DOI] [PubMed] [Google Scholar]
- 10.Tong XD, Liu TQ, Wang GB, Zhang CL, Liu HX. MicroRNA-570 promotes lung carcinoma proliferation through targeting tumor suppressor KLF9. Int J Clin Exp Pathol. 2015;8:2829–2834. [PMC free article] [PubMed] [Google Scholar]
- 11.Ramirez CM, Goedeke L, Rotllan N, Yoon JH, Cirera-Salinas D, Mattison J A, et al. MicroRNA 33 regulates glucose metabolism. Mol Cell Biol. 2013;33:2891–2902. doi: 10.1128/MCB.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang LS, Li L, Chu S, Shiang KD, Li M, Sun HY, et al. MicroRNA-486 regulates normal erythropoiesis and enhances growth and modulates drug response in CML progenitors. Blood. 2015;125:1302–1313. doi: 10.1182/blood-2014-06-581926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Racil Z, Razga F, Drapalova J, Buresova L, Zackova D, Palackova M, et al. Mechanism of impaired glucose metabolism during nilotinib therapy in patients with chronic myelogenous leukemia. Haematologica. 2013;98:e124–126. doi: 10.3324/haematol.2013.086355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dearth RK, Cui X, Kim HJ, Hadsell DL, Lee AV. Oncogenic transformation by the signaling adaptor proteins insulin receptor substrate (IRS)-1 and IRS-2. Cell Cycle. 2007;6:705–713. doi: 10.4161/cc.6.6.4035. [DOI] [PubMed] [Google Scholar]
- 15.Geng Y, Ju Y, Ren F, Qiu Y, Tomita Y, Tomoeda M, et al. Insulin receptor substrate 1/2 (IRS1/2) regulates Wnt/beta-catenin signaling through blocking autophagic degradation of dishevelled2. J Biol Chem. 2014;289:11230–11241. doi: 10.1074/jbc.M113.544999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reuveni H, Flashner-Abramson E, Steiner L, Makedonski K, Song R, Shir A, et al. Therapeutic destruction of insulin receptor substrates for cancer treatment. Cancer Res. 2013;73:4383–4394. doi: 10.1158/0008-5472.CAN-12-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways:insights into insulin action. Nat Rev Mol Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 18.Bommer GT, Feng Y, Iura A, Giordano TJ, Kuick R, Kadikoy H, et al. IRS1 regulation by Wnt/beta- catenin signaling and varied contribution of IRS1 to the neoplastic phenotype. J Biol Chem. 2010;285:1928–1938. doi: 10.1074/jbc.M109.060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan BT, Lee AV. Insulin receptor substrates (IRSs) and breast tumorigenesis. J Mammary Gland Biol Neoplasia. 2008;13:415–422. doi: 10.1007/s10911-008-9101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dearth RK, Cui X, Kim HJ, Kuiatse I, Lawrence NA, Zhang X, et al. Mammary tumorigenesis and metastasis caused by overexpression of insulin receptor substrate 1 (IRS-1) or IRS-2. Mol Cell Biol. 2006;26:9302–9314. doi: 10.1128/MCB.00260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagle JA, Ma Z, Byrne MA, White M F, Shaw LM. Involvement of insulin receptor substrate 2 in mammary tumor metastasis. Mol Cell Biol. 2004;24:9726–9735. doi: 10.1128/MCB.24.22.9726-9735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q, Tang Q, Qin D, Yu L, Huang R, Lv G, et al. Role of microRNA 30a targeting insulin receptor substrate 2 in colorectal tumorigenesis. Mol Cell Biol. 2015;35:988–1000. doi: 10.1128/MCB.01242-14. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Meyer K, Albaugh B, Schoenike B, Roopra A. Type 1 insulin-like growth factor receptor/insulin receptor substrate 1 signaling confers pathogenic bctivity on breast tumor cells lacking REST, molecular and cellular biology. 2015;35:2991–3004. doi: 10.1128/MCB.01149-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng H, Zhang F, Lin X, Huang C, Zhang Y, Li Y, et al. MicroRNA-1225-5p inhibits proliferation and metastasis of gastric carcinoma through repressing insulin receptor substrate-1 and activation of beta-catenin signaling. Oncotarget. 2016;7:4647–4663. doi: 10.18632/oncotarget.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]


