Abstract
Retroviral DNA integration occurs throughout the genome; however, local “hot spots” exist where a strong preference for certain sites over others are seen, and more global preferences associated with genes have been reported. Previous data from our laboratory suggested that there are fewer integration events into a DNA template when it is undergoing active transcription than when it is not. Because these data were generated by using a stably transfected foreign gene that was only weakly inducible, we have extended this observation by comparing integration events into a highly inducible endogenous gene under both induced and uninduced transcriptional states. To examine the influence of transcription on site selection directly, we analyzed the frequency and distribution of integration of avian retrovirus DNA into the metallothionein gene, before and after its induction to a highly sustained level of expression by addition of ZnSO4. We found a 6-fold reduction in integration events after 100-fold induction of transcription. This result implies that, despite an apparent preference for integration of retroviral DNA into transcribed regions of host DNA, high-level transcription can be inhibitory to the integration process. Several possible models for our observation are as follows. First, when a DNA template is undergoing active transcription, integration might be blocked by the RNA polymerase II complex because of steric hindrance. Alternatively, the integrase complex may require DNA to be in a double-stranded conformation, which would not be the case during active transcription. Last, transcription might lead to remodeling of chromatin into a structure that is less favorable for integration.
Keywords: retrovirus, alpharetrovirus, chromatin, RAV-1, metallothionein
Integration of the retroviral DNA genome into host-cell DNA is an essential step in the retrovirus replication cycle, permitting viral genomes to become permanently fixed as proviruses into the DNA of the host and to use host transcriptional machinery for the production of viral RNA (reviewed in ref. 1). Although retrovirus integration can occur throughout the genome, local “hot spots” for integration exist where a strong preference for particular sites over others can be demonstrated statistically (2, 3). Recent work with HIV and murine leukemia virus has implied that there is also a preference for integration into transcribed regions of the host genome (4, 5), in the case of murine leukemia virus, near transcriptional start sites. The basis for these preferences is unknown, but they may reflect interaction of the preintegration complex with specific proteins or with specific DNA sequences or structures that are associated with transcription (reviewed in refs. 6 and 7).
Because the studies cited above did not directly examine the effect of transcription per se on integration, and because our previous data have suggested that there may be fewer integration events into an artificially introduced DNA template when it is undergoing active transcription than when it is not (8), we decided to test the effect of transcription on integration directly by comparing integration events into a highly inducible endogenous gene under both induced and uninduced transcriptional states. The avian metallothionein (MT) gene has a low level of constitutive expression, yet it can be rapidly induced to a very high level by the addition of zinc to the tissue culture medium (9, 10). For studying retroviral integration preferences in vivo, we chose to use the avian retrovirus system rather than a mammalian system, such as mouse or human, for three reasons. First, avian cell lines are available that are free of closely related endogenous retroviruses, allowing for accurate quantitation of new integration events; second, mammalian genomes, unlike avian, contain a large fraction of repetitive DNA, which can influence our ability to accurately quantitate integration events and can make analyses generally much more difficult; and third, unlike the mammalian MT gene, which occurs in multiple isoforms, the well studied avian gene is present in only one copy per genome (ref. 10 and data not shown), which makes mapping integration events much more straightforward.
Materials and Methods
Cells and Viruses. RAV-1, a subgroup A Alpharetrovirus strain, was isolated from a molecular clone described elsewhere (11). Infections were done as follows. QT6 (12) cells were plated at 2 × 106 cells per 100 × 20-mm tissue-culture plate 16-18 h before infection with 3 ml of RAV-1 stock for 1 h at 37°C. Virus stocks used for infection were freshly obtained from chronically infected QT6 cells. Except where mentioned below, cells were harvested at 48 h after infection, and either RNA or DNA was extracted as described.
Quantitation of MT Gene Expression by Quantitative RT-PCR (qRT-PCR). Spliced and unspliced MT RNA was quantitated by qRT-PCR as follows. At various time points (15 min to 48 h) after induction by 100 μM ZnSO4, whole-cell RNA was harvested by washing each plate with PBS, scraping the cells into a 15-ml conical tube, and centrifuging, followed by freezing the cell pellet at 80°C. To extract RNA, the frozen pellet was processed by using TRIzol Reagent (Invitrogen) as recommended by the manufacturer. The RNA was then treated with Promega RQ1 DNase at a concentration of 1 unit/μg nucleic acid for 30 min at 37°C, followed by a phenol/chloroform extraction and EtOH precipitation. One-step qRT-PCR was performed as follows: 500 ng of RNA was added to a reaction mix containing 1× SYBR Green master mix (Applied Biosystems), 1.25 units of Multi-Scribe reverse transcriptase (Applied Biosystems), 0.4 units of RNase Inhibitor (Applied Biosystems), and 300 nM each primer, as described below (all primers were produced at the Tufts University Core Facility). The RNA was then amplified by using the GeneAmp 5700 sequence-detection system by using a one-step RT-PCR protocol as follows. cDNA was generated by annealing for 10 min at 25°C, followed by a 30-min incubation at 48°C. The resulting cDNA was then amplified in the same tube for 40 cycles of 15 sec at 95°C, followed by an extension step of 1 min at 60°C.
Primer 5′-TCAGGACTGCACTTGTGCTGC-3′ (sense) that detects sequences within the first exon of the quail MT coding region was used to detect either unspliced or spliced RNA species. The antisense primer used to quantitate unspliced RNA was 5′-ACCTGGGACAGGAAAGAAGC-3′, complementary to sequences in the first intron of the quail MT gene. A 102-bp product was generated by using this primer pair. Spliced RNA was detected by using antisense primer 5′-TGGCAGCAGCTGCACTTGCTT-3′, complementary to sequences in the third exon of the quail MT gene to produce a 177-bp product.
For each assay, quail MT RNA samples were run in triplicate and the values averaged after determination from a standard curve (described below). Relative quantitation was then determined from GAPDH sequences amplified for use as a calibrator. GAPDH products were generated by using primers 5′-TGCCATCACAGCCACACAGAAGAC-3′ (sense) and 5′-TGACTTTCCCCACAGCCTTAGCAG-3′ (antisense) that bind to nucleotides 547-610 and 711-688 of the chicken GAPDH gene respectively, generating a 130-bp product.
The following plasmids were serially diluted 10-fold to generate standard curves for real-time RT-PCR. pTopo/MT5-5 was used to quantitate unspliced MT RNA, pSP-cMT-6 was used for spliced MT RNA, and pTopo/gap8 was used to quantitate quail GAPDH. pTopo/Qgap8 and pTopo/QMT5-5 were created from cDNA generated with the Access RT-PCR System (Promega) by using the same primer pairs described above for generating GAPDH or unspliced quail qPCR products. In each case, the cDNA RT-PCR product was inserted into the pCR2.1-TOPO (Invitrogen) cloning vector according to the manufacturer's instructions. Plasmid pSP-cMT-6 containing quail MT cDNA sequences, was a generous gift from Glen Andrews (University of Kansas, Kansas City).
qPCR to Quantitate Viral DNA Products. RAV-1 DNA was quantitated by real-time PCR in the following way. Whole-cell DNA was extracted as described below for the in vivo integration assay. We added 1 μg of DNA to a reaction mix containing 1× SYBR Green master mix (Applied Biosystems), 0.5 units AmpErase uracil N-glycosylase (UNG) (Applied Biosystems), and 300 nM each of a primer pair complementary to conserved sequences in avian leukosis virus (ALV) gag (all primers were produced at the Tufts University Core Facility). The primers used were 5′-CAAGGCGTTTACTGCTTG-3′ (sense) and 5′-GCAGTGGACGCGCAATGT-3′ (antisense). These primers will effectively amplify all avian retroviral gag sequences used in this assay [RAV-1, Bryan high-titer strain present in the standard curve plasmid (see below) and ev-1 present in c33 cells]. The DNA was then amplified by using the GeneAmp 5700 sequence-detection system by an AmpErase UNG incubation for 2 min at 50°C, then AmpliTaq Gold activation for 10 min at 95°C, followed by 40 cycles of a 15-sec 95°C denaturation step, and a 1-min extension step at 60°C.
To determine the viral DNA copy number per cell, two separate standard curves were generated per assay, one generated with a 10-fold serial dilution of a plasmid and one generated by 10-fold serial dilutions of genomic sequences. Plasmid standard curves were generated by using dilutions of plasmid RCASBP(A) ΔP1 (a generous gift from Stephen Hughes, HIV Drug-Resistance Program, National Cancer Institute) to quantitate viral DNA sequences, or pTopo/Qgap8 (described above), to quantitate quail GAPDH sequences to act as a calibrator. Whole-cell standard curves were generated with cellular DNA (extracted as described below for the in vivo integration assay) from either C33 cells (a chicken cell line with the integrated provirus ev-1 present at one copy per genome) to quantitate viral copy number per cell or QT-6 cells to quantitate quail GAPDH sequences.
Quantitative Southern Blotting. Quantitative Southern blot analyses were done as follows. We ran 2-fold serial dilutions (5, 2.5, and 1.25 μg) of undigested cellular DNA (extracted as described below for the in vivo integration assay) on a 0.8-agarose gel. The following DNAs were used for each experiment: QT6 cells infected by RAV-1 induced with zinc to activate MT, RAV-1 infected QT6 cells not induced by zinc, C33 cells that have one genomic copy per cell of the endogenous retrovirus ev-1, and one well with 5 μg of uninfected QT6 cell extract. After electrophoresis, each gel was blotted onto an Immobilon-Ny+ membrane (Millipore, Billerica, MA), then hybridized to a radiolabeled probe according to the membrane manufacturer's instructions. Each probe was labeled with [α-32P]dCTP by using the Prime-It RmT Random Primer labeling kit (Stratagene, La Jolla, CA) as recommended by the manufacturer. Each blot was probed for viral sequences by using a 1,181 bp Msc1 restriction fragment containing RAV-1 gag sequences. The resulting blot was then exposed to a PhosphorImager screen overnight and the intensity of each band quantitated by using imagequant software (Molecular Dynamics, Amersham Biosciences). Each blot was then stripped in a 0.4 M NaOH solution at 45°C for 30 min, neutralized for 15 min with 0.1× SSC, 0.1% SDS and 0.2 M Tris·Cl, pH 7.5; then reprobed with quail MT sequences, which allowed us to normalize the amounts of integrated virus loaded in each lane.
The determination of the number of integrated proviruses per cell was done by a modification of a protocol described in ref. 3. Briefly, serial dilutions of undigested genomic DNA isolated from QT6 cells that were either uninfected, infected with RAV-1 and induced with 100 μM ZnSO4, or infected and uninduced was analyzed by electrophoresis on a 0.8% agarose gel next to serial dilutions of DNA isolated from chicken embryo fibroblast cells, which carry a single copy of an endogenous virus, ev-1 (13). By comparing the intensity of bands representing integrated RAV-1 proviral DNA to the equivalent lanes of chicken embryo fibroblast DNA, we were able estimate the number of proviruses per cell in both the zinc induced and uninduced cells.
PCR Assay to Detect in Vivo Integration. QT6 cells were infected with RAV-1 as described above, and the infection was allowed to continue for 24-48 h (enough time for integration to occur). PCR assays were then preformed on 20 μg of extracted DNA, which is an amount that is equivalent to ≈107 cells. At our estimated multiplicity of infection of approximately two proviruses per cell, we predicted ≈2 × 107 integration events in every 20 μg of DNA. Because the haploid quail genome is 109 bp, and we were analyzing a 500-bp region, we expected to detect one or two integration events per PCR.
The PCR assay used to detect region-specific integration products was adapted from that used in previous studies in our laboratory (3, 8), and it was done as follows. Genomic DNA isolated from QT6 cells uninfected, infected and induced with 100 μg of ZnSO4, or infected and uninduced was diluted to 1 μg/ml, heated at 100°C for 5 min, and then placed in an 80°C heating block until addition to the PCR mixture. We added 20 μg of the DNA to the following PCR mixture to a final volume of 50 μl, 10 mM Tris·HCl (pH 8.3), 3 mM MgCl2, 50 mM KCl, 0.001% gelatin, 411 μM each deoxynucleoside triphosphate, 0.6 μM each primer (described below), and 5 units Taq polymerase (Sigma), then prewarmed to 80°C for 5 min. The reaction mixtures were transferred directly into a PCR machine preheated to 80°C; heated to 94°C for 5 min; and then amplified for 30 cycles at 94°C for 1 min, 65°C for 1.5 min, and 72°C for 2 min. For the final step in the last cycle, samples were heated to 72°C for 7 min. The entire PCR mixture was then purified by using a QIAquick PCR purification kit (Qiagen, Valencia, CA) according to the manufacturer's directions and elution in a final volume of 50 μl of 10 mM Tris·HCl (pH 8.3).
The PCR assay was done by using the following primers. To detect integrated proviruses present in a specific region of the genome, one primer was specific for viral sequences, and the second primer was specific for genomic sequences. The viral primer (U3-RAV, described in ref. 8, ATCGTCGTGCACAGTGCCTTT) primes from the antisense strand of RAV-1 U3 LTR sequences, and amplifies an integration product that includes the last 106 bp of the 5′ end of the integrated provirus. The genomic primer (CACCACCACGGCACTATAAAT) was specific for sequences near the transcription start site of the quail MT gene.
PCR products were visualized by extension of an end-labeled primer. Each entire 50-μl PCR was dried down and annealed with ≈0.2 pmol of an internally nested γ-32P-labeled primer (106 counts per reaction) in 1× buffer (40 mM Tris·HCl, pH 7.5/20 mM MgCl2/50 mM NaCl). Extension was carried out with Sequenase (United States Biochemical) for 30 min at 42°C. The internally nested primer bound to sequences that span the last 9 nt of the first exon and the first 12 nt of the first intron of the quail MT gene (GTGCTGCTGGTAAGTGGACTC). Samples were analyzed on a 4% polyacrylamide denaturing gel under standard conditions, with sequencing ladders run in parallel to provide size standards. The gels were then dried and exposed overnight at -80°C by using the BioMax MS intensifying screen and film system (Eastman Kodak).
Each integration event was analyzed by one or both of two different methods. First, the size of each primer extension product was determined, as compared with the sequencing ladder that was also run on the gel. The site of the corresponding integration event can be determined to within ≈3 bp by this method. In addition, many of the primer extension products were sequenced after extraction directly from a nondried polyacrylamide denaturing gel by the standard “crush-and-soak” method (14), followed by reamplification of the DNA by using the U3-RAV primer and nested primer extension primer described above. Sequencing of the primer extension products was also used for verification that the primer extension product analyzed represented a true integration product (see Table 2, which is published as supporting information on the PNAS web site).
Results
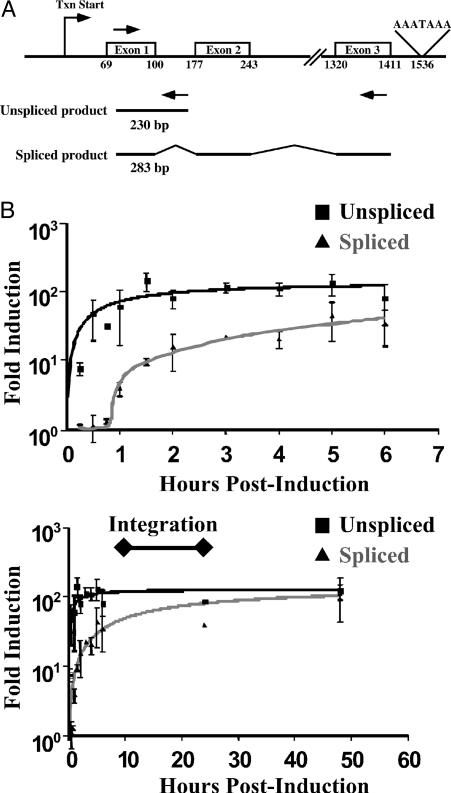
Expression of MT After Induction by Zinc in Quail Cells. Because it was desirable to use the well studied quail tumor (QT6) cell line (12), the structure of the quail MT gene was determined (see Fig. 5 and Supporting Materials and Methods, which are published as supporting information on the PNAS web site) and is shown in Fig. 1A, along with the primers used to quantitate MT expression. MT expression was quantitated at various time points after induction by using an assay based on real-time reverse transcriptase-mediated PCR (qRT PCR) (Fig. 1B). As shown in Fig. 1B Upper, RT-PCR using primers that specifically detected nascent (unspliced) transcripts revealed a rapid increase MT RNA levels as early as 15 min after zinc treatment, rising to ≈100-fold over the constitutive level by 1 h. By contrast, the increase in spliced transcripts lagged behind that of unspliced by ≈1 h. Importantly, the induction of expression of both unspliced and spliced transcripts lasted for ≥50 h (Fig. 1B Lower), and therefore, MT transcription remained at a high level throughout the period of integration.
Fig. 1.
Transcription of MT RNA with or without induction by zinc. QT6 cells were infected with the RAV-1 strain of ALV and either induced with 100 μM ZnSO4 or left uninduced. Total RNA was extracted at various times after infection and quantitated by real-time RT-PCR using SYBR Green fluorescence detection of the DNA product. (A) The quail MT gene showing primer pairs used to quantitate spliced and unspliced MT RNA by real-time RT-PCR. Arrows indicate the location of each primer. (B) Time course for induction of quail MT RNA as determined by the RT-PCR assay. The x axis represents time after induction by zinc, and the y axis represents fold increase in spliced or unspliced MT RNA relative to untreated cultures analyzed at the same time. Upper and Lower show the same data on two different time scales.
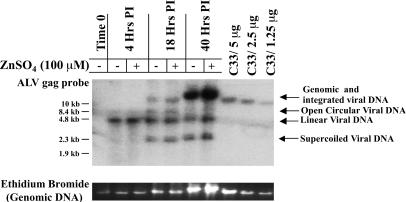
Zinc Induction Does Not Affect Viral Replication. For our analysis, it was important that zinc induction did not affect any viral replication processes such as entry, reverse transcription, or integration. To assess the effect of treatment with zinc on these events, we quantitated the amount of total and integrated viral DNA present after infection in zinc-induced and uninduced cells by using a real-time PCR assay (data not shown) to detect total viral DNA products and a quantitative blot hybridization analysis (Fig. 2) to distinguish among the various forms. As summarized in Table 1, the amount of total viral DNA was not significantly different between induced and uninduced infected cells (mean ratio, 1.03 ± 0.74), and there was a slight, but not significant, increase in total integrated DNA after induction (1.34 ± 0.44). This assay was performed on DNA from the six experiments that were used to map integration sites, as indicated below (Figs. 3 and 4). From these results, we conclude that zinc treatment during infection of QT6 cells does not affect viral replication processes such as entry, reverse transcription, or integration.
Fig. 2.
Effect of zinc induction on ALV DNA synthesis and integration. QT6 cells were infected with RAV-1 and either induced with 100 μM ZnSO4 or uninduced. Whole-cell DNA was extracted at the indicated times after infection. Integrated proviral DNA was quantitated by Southern blot hybridization using a probe specific for sequences within a highly conserved region of ALV pol. Arrows indicate the various forms of viral DNA. The bands representing the nonintegrated forms of ALV contain both 1-LTR and 2-LTR circles, because they were not distinguishable in this assay. Total DNA in the region of the gel (Genomic and integrated viral DNA) was visualized by staining with ethidium bromide. C33 cells are chicken embryo fibroblasts that contain a single endogenous provirus and provide a single-copy hybridization standard.
Table 1. Effect of Zn++ treatment on ALV DNA synthesis and integration.
| Total DNA*
|
Integrated DNA†
|
|||
|---|---|---|---|---|
| Experiment | Without Zn induction | With Zn induction | Without Zn induction | With Zn induction |
| 1 | 3.6 | 2.6 (0.7) | 1.8 | 1.9 (1.1) |
| 2 | 2.0 | 2.6 (1.3) | 2.5 | 2.3 (0.9) |
| 3 | 4.0 | 2.4 (0.6) | 3.1 | 4.1 (1.3) |
| 4 | 1.0 | 2.6 (2.6) | 1.8 | 2.2 (1.2) |
| 5 | 3.8 | 1.6 (0.4) | ND | ND |
| 6 | 1.6 | 1.0 (0.6) | 0.9 | 2.0 (2.2) |
DNA was extracted and analyzed 24–48 hours after infection. Raw values were normalized to the single endogenous provirus ev-1 in uninfected chick embryo fibroblasts. The induced value relative to uninduced value is given in parentheses. DNAs from the experiments shown in Figs. 3 and 4 were analyzed. ND, not determined.
Assayed by real-time PCR using primers from within the pol gene
Assayed by quantitative Southern blot analyses of hybridized to a 32P-labeled ALV gag probe (Fig. 2)
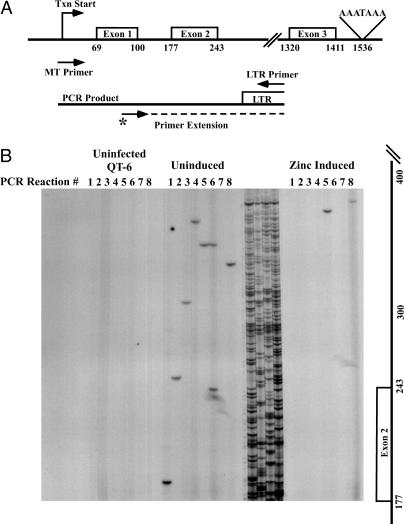
Fig. 3.
Effect of zinc induction on viral DNA integration. (A) Region within the quail MT gene where integration products were mapped in this assay system. Arrows indicate the positions of quail MT specific and viral LTR-specific PCR primers. The arrow marked with an asterisk indicates the location of the labeled primer-extension primer. (B) Representative data from one of six in vivo integration assays used to generate the histogram in Fig. 4. After amplification of eight parallel PCRs, each using the primers shown in A initiated with 20 μg of DNA from uninfected (Left), RAV-1 infected and uninduced (Center), or infected and Zinc-induced (Right) cells, specific amplification products were detected by extension of the 32P-end-labeled primer followed by electrophoresis in denaturing (sequencing) polyacrylamide gels. The sequencing ladder between the uninduced and induced lanes provided molecular size markers.
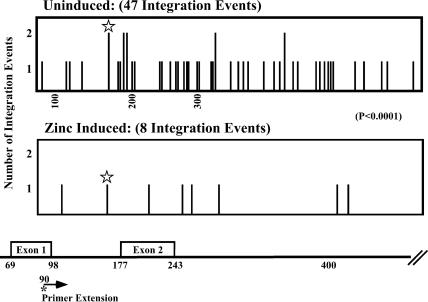
Fig. 4.
Location of integration events into the MT gene. PCR products generated in six separate experiments as described in Fig. 3 were used to determine the frequency of retroviral integration sites used at each base pair within the quail MT gene under both zinc-induction and uninduced conditions. The number of events observed at each site is plotted at the position of the site on the gene (Bottom). The arrow indicates the location within of the primer extension primer. The asterisk indicates the apparent use of the same site in both uninduced (twice) and zinc-induced cells.
Actively Transcribed Transcripts Are a Disfavored Target for Integration. Retroviral DNA integration into a particular gene or region of the DNA is a very rare event. Therefore, we have developed a sensitive assay system that detects integration of viral DNA in a specific region of the genome (Fig. 3A) (3, 8). This assay was used to map and quantitate integration events into the MT gene and to compare the number and pattern of these events during active transcription of a gene, as compared with its uninduced state. QT6 cells were infected with the alpharetrovirus strain, RAV-1, for 1 h, and then grown with or without 100 μM zinc for a time period (24-48 h) sufficient to allow integration to occur. A PCR-based assay was then used to map and quantitate the integration sites within the gene by using one primer that binds to viral sequences and another that binds to sequences within the target sequence. A primer-extension reaction with an end-labeled primer was used to detect the specific integration product after analysis by electrophoresis in denaturing gels. The size of each band is a function of the position within the host gene where the provirus has integrated, and the number of bands reflects the number of integration events in the analyzed region. Given a multiplicity of infection yielding approximately two proviruses per cell (Table 1) and a genome size of 109 bp, we expected to detect, on average, approximately one integration event in a 500-bp region per 20 μg of analyzed cell DNA.
The integration site and frequency within the coding region of the MT gene was mapped by using a set of primers in which one primer spanned the transcription start site, and the other was specific for sequences within the viral U3 portion of the LTR (Fig. 3A). Fig. 3B shows the results of one representative experiment generated by this assay system. Each lane represents one PCR-extension analysis of 20 μg of genomic DNA, and eight such reactions were done in parallel. In this and other experiments, approximately one-fourth of the sites of integration were analyzed by reamplification of the DNA in each band and sequencing. In all cases (see Table 2 and Supporting Materials and Methods), the correct end of the proviral DNA was joined to the portion of the MT sequence predicted from the position of the band. As expected, no integration events were detected from uninfected QT6 DNA, and reconstruction experiments (data not shown) revealed that we could detect a single copy of integrated viral DNA.
When DNA from infected, uninduced cells was analyzed, we found an average of one band per reaction (Fig. 3B), consistent with the expected number for a random distribution of integration events in cell DNA. By contrast, many fewer bands were detectable from infected, zinc induced cells under the same conditions, despite the presence of equal or greater numbers of total integration events in these cells (Table 1). Identical results have been observed in a total of six experiments, yielding a total of 47 integration events into uninduced MT DNA, whereas only eight integration events were mapped into MT gene templates after induction by zinc (Fig. 4). Therefore, 100-fold activation of transcription of MT reduced the number of integration events into that gene region by ≈6-fold, which is a highly significant difference (P < 0.0001). In both cases, the distribution of integration events showed no preference for targeting into exonic verses intronic DNA. Although the distribution of integration sites appeared to be mostly random, the presence of several sites used twice along with one used twice in the uninduced and once in the Zn-induced samples is consistent with the existence of local hot spots, as we have described (3).
Discussion
There are several possible models for the inverse relationship between transcriptional activity observed in this study and other studies (8). First, when a DNA template is undergoing active transcription, integration might be blocked by the RNA polymerase II complex because of steric hindrance. Local inhibition by transcription factor binding of integration in vitro has been reported (15, 16), and a high concentration of transcription complexes might block access of the preintegration complex. Alternatively, productive integration is likely to require concerted reactions on both strands of the DNA target, and much of the target DNA is likely to be single-stranded during active transcription. Last, transcription might lead to remodeling of chromatin into a structure that is less favorable for integration. Indeed, it has been observed that DNA in association with nucleosomes is favored for integration, most likely reflecting increased accessibility of the major groove because of distortion of the helix (15, 17-19). It remains to be determined experimentally which of these mechanisms is responsible for our observation.
Several lines of evidence are consistent with the finding that an actively transcribing DNA template may be a disfavored target for retroviral integration. Studies in our laboratory using an ALV system have shown that essentially all of the avian genome is available for integration, and that except for specific hot-spots, integration is essentially random throughout the genome (3). Furthermore, in vitro, integration can be preferentially targeted to regions of DNA with CpG methylation (2), a state associated with gene inactivation in vivo. Also, in a previous study in our laboratory in which an artificial minigene stably transfected into QT6 cells was induced 5-fold, a 3-fold reduction in the number of integration events was detected in the upstream control region (8). This effect was especially pronounced in the TATA box and transcription-factor binding sites where there were virtually no integration events mapped during transcription activation. Also, a 2-fold reduction in integration events was mapped in the coding region of the gene after induction. In this study, we have confirmed and extended this observation by comparing integration events into a highly inducible endogenous gene under both induced and uninduced transcriptional states, and we have observed that a 100-fold induction of transcription resulted in 6-fold fewer integration events into the template DNA. Therefore, after the evaluation of integration patterns into two separate inducible gene systems (a moderately expressed gene and a very highly expressed gene) when the gene was activated, there were fewer integration events observed into the actively transcribing template. These results indicate that the inhibitory effect of active transcription on integration occurs in both highly and moderately active genes.
Because of implications for understanding retrovirus replication, particularly in the context of side effects of genetherapy vectors (20), there has been much recent interest in understanding retroviral-integration targeting. By mapping a large set of HIV integration sites on the human genome, Schröder et al. (4) found a strong (but not absolute) bias toward active genes. By using the same approach, Wu et al. (5) reported that a murine leukemia virus-based vector used for current gene therapy studies targets the promoter-proximal regions of genes, both up and downstream from the transcription initiation site. Similar studies with ALV in human cells also seem to show a small preference for active genes (21, 22). Although our analysis implied that the uninduced MT gene was used at a frequency that was not greatly different from that expected if integration sites were distributed randomly over the genome, we would not have been able to detect a small difference. An important issue in considering these apparently disparate results is whether the observed preference for integration into genes implied by the site-distribution experiments is a consequence of transcriptional activity of the sequence at the time of integration or its propensity to be activated in the specific cell type studied. Our results seem more consistent with the latter possibility. It is likely that a property of frequently expressed genes other than transcription (such as their location within the nucleus or the euchromatic nature of their chromatin structure) may make these sequences a preferred target for retroviral integration. All cellular factors that have been identified through an association with the integration process (HMGa1, BAF, LEDGF/p75, and Ini1) (7) have normal cellular functions in chromatin remodeling and organization. It is the role that these processes play in gene expression that may help determine how retroviral DNA is targeted to host sequences.
Supplementary Material
Acknowledgments
We thank Glen Andrews and Stephen Hughes for the generous gift of plasmids. We also thank Mike Berne and the Tufts University Core Facility for sequencing and the production of oligonucleotides and the Tufts Gastroenterology Research on Absorbtive and Secretory Processes (GRASP) Center for technical support. This work was supported by National Cancer Institute Grant R01 CA 92192. J.M.C. is a research professor of the American Cancer Society, with support from the F. M. Kirby Foundation. L.F.M. was a Fellow of the Leukemia and Lymphoma Society.
Abbreviations: MT, metallothionein; qRT-PCR, quantitative RT-PCR; ALV, avian leukosis virus.
Data deposition: The quail metallothionein sequence reported in this paper has been deposited in the GenBank database (accession no. AY866409).
See Commentary on page 1275.
References
- 1.Coffin, J. M., Hughes, S. H. & Varmus, H. E. (1997) (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 2.Kitamura, Y., Lee, Y. M. H. & Coffin, J. M. (1992) Proc. Natl. Acad. Sci. USA 89, 5532-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Withers-Ward, E. S., Kitamura, Y., Barnes, J. P. & Coffin, J. M. (1994) Genes Dev. 8, 1473-1487. [DOI] [PubMed] [Google Scholar]
- 4.Schröder, A. R., Shinn, P., Chen, H., Berry, C., Ecker, J. R. & Bushman, F. (2002) Cell 110, 521-529. [DOI] [PubMed] [Google Scholar]
- 5.Wu, X., Li, Y., Crise, B. & Burgess, S. M. (2003) Science 300, 1749-1751. [DOI] [PubMed] [Google Scholar]
- 6.Bushman, F. D. (2002) Curr. Top. Microbiol. Immunol. 261, 165-177. [DOI] [PubMed] [Google Scholar]
- 7.Engelman, A. (2003) Curr. Top. Microbiol. Immunol. 281, 209-238. [DOI] [PubMed] [Google Scholar]
- 8.Weidhaas, J. B., Angelichio, E. L., Fenner, S. & Coffin, J. M. (2000) J. Virol. 74, 8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei, D. Y. & Andrews, G. K. (1988) Nucleic Acids Res. 16, 537-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernando, L. P., Wei, D. Y. & Andrews, G. K. (1989) J. Nutr. 119, 309-318. [DOI] [PubMed] [Google Scholar]
- 11.Sealy, L., Privalsky, M. L., Moscovici, G., Moscovici, C. & Bishop, J. M. (1983) Virology 130, 155-178. [DOI] [PubMed] [Google Scholar]
- 12.Moscovici, C., Moscovici, M. G., Jimenez, H., Lai, M. M. C., Hayman, M. J. & Vogt, P. K. (1977) Cell 11, 95-103. [DOI] [PubMed] [Google Scholar]
- 13.Tereba, A. & Astrin, S. M. (1980) J. Virol. 35, 888-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) (Cold Spring Harbor Lab. Press, Woodbury, NY), pp. 11.23-11.30.
- 15.Pryciak, P. M. & Varmus, H. E. (1992) Cell 69, 769-780. [DOI] [PubMed] [Google Scholar]
- 16.Bor, Y. C., Bushman, F. D. & Orgel, L. E. (1995) Proc. Natl. Acad. Sci. USA 92, 10334-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pryciak, P. M., Sil, A. & Varmus, H. E. (1992) EMBO J. 11, 291-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller, H. & Varmus, H. E. (1994) EMBO J. 13, 4704-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruss, D., Bushman, F. D. & Wolffe, A. P. (1994) Proc. Natl. Acad. Sci. USA 91, 5913-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hacein-Bey-Abina, S., Von Kalle, C., Schmidt, M., McCormack, M. P., Wulffraat, N., Leboulch, P., Lim, A., Osborne, C. S., Pawliuk, R., Morillon, E., et al. (2003) Science 302, 415-419. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell, R. S., Beitzel, B. F., Schröder, A. R., Shinn, P., Chen, H., Berry, C. C., Ecker, J. R. & Bushman, F. D. (2004) PLoS Biol. 2, E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narezkina, A., Taganov, K. D., Litwin, S., Stoyanova, R., Hayashi, J., Seeger, C., Skalka, A. M. & Katz, R. A. (2004) J. Virol. 78, 11656-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.