Abstract
Salicylic acid (SA) is a critical signal for the activation of plant defense responses against pathogen infections. We recently identified SA-binding protein 2 (SABP2) from tobacco as a protein that displays high affinity for SA and plays a crucial role in the activation of systemic acquired resistance to plant pathogens. Here we report the crystal structures of SABP2, alone and in complex with SA at up to 2.1-Å resolution. The structures confirm that SABP2 is a member of the α/β hydrolase superfamily of enzymes, with Ser-81, His-238, and Asp-210 as the catalytic triad. SA is bound in the active site and is completely shielded from the solvent, consistent with the high affinity of this compound for SABP2. Our biochemical studies reveal that SABP2 has strong esterase activity with methyl salicylate as the substrate, and that SA is a potent product inhibitor of this catalysis. Modeling of SABP2 with MeSA in the active site is consistent with all these biochemical observations. Our results suggest that SABP2 may be required to convert MeSA to SA as part of the signal transduction pathways that activate systemic acquired resistance and perhaps local defense responses as well.
Keywords: salicylic acid, salicylic-acid-binding protein, systemic acquired resistance, α/β hydrolase
The innate immunity system of plants shows many parallels with that of vertebrates and invertebrates (1–3). The cells at the sites of pathogen entry usually undergo apoptotic-like cell death, resulting in the formation of necrotic lesions characteristic of the hypersensitive resistance response. There is also enhanced expression of defense-associated genes, including those encoding pathogenesis-related proteins. In addition, after a delay of several hours to a few days, plants frequently develop a broad-based long-lasting resistance to secondary pathogen infection known as systemic acquired resistance (SAR).
Many studies have shown that salicylic acid (SA) is a critical signal for activation of plant defense responses both at the site of infection and systemically in distal tissues (1, 4, 5). The important role of SA has been demonstrated in a number of plant species, particularly Nicotiana tabacum and Arabidopsis thaliana. For example, plants that are SA-deficient fail to develop SAR, do not express PR genes in the uninoculated leaves, and display enhanced susceptibility to pathogens (5). Similar phenotypes were observed in pathogen-infected Arabidopsis mutants that are defective for SA accumulation (6–8). SA may also regulate cell death, possibly via a positive-feedback loop that involves reactive oxygen species (9–11), and may play a role in pathogen containment (12–14).
SA can be methylated (15–17) or conjugated to glucose (18–20) to form methyl salicylate (MeSA) and SA β-glucoside, respectively. These SA derivatives appear to be biologically inactive with respect to induction of defense responses such as PR gene expression but can be readily converted back to free active SA (15, 20) by a partially characterized SA β-glucosidase (21) and a yet-to-be-identified MeSA esterase. The gene encoding the methyl transferase that synthesizes MeSA from SA has been recently isolated (22, 23), and SA glucosyl transferase has been partially purified (24).
As part of our ongoing research to define the SA-mediated defense signaling pathway(s) and to determine the mechanism(s) of SA action, we have identified and characterized a high-affinity SA-binding protein (SABP) termed SABP2 from tobacco (25, 26). SABP2 is present in extremely low abundance and specifically binds SA with high affinity (Kd of 90 nM). It has esterase activity and SA-stimulated lipase activity. Silencing of SABP2 expression via RNA interference suppresses local resistance to tobacco mosaic virus and SA-induced PR-1 gene expression and blocks development of SAR (26).
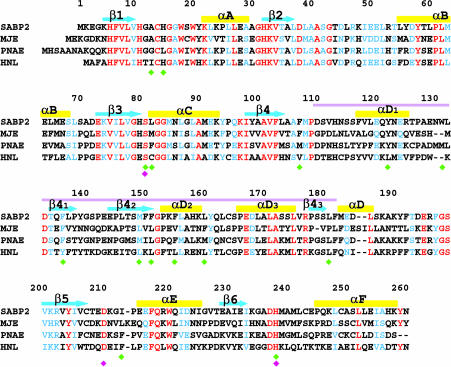
The amino acid sequence of the SABP2 protein indicates that it is a member of the α/β hydrolase superfamily (26–28). These enzymes share a conserved α/β core domain and catalyze the hydrolysis of different substrates (27, 28). Interestingly, recent studies show that several confirmed methyl esterases from plants, including methyl jasmonate (MeJA) esterase from tomato (29) and polyneuridine aldehyde esterase from the Indian medicinal plant Rauvolfia serpentina (30), also belong to this family. SABP2 shares recognizable amino acid sequence homology with these enzymes (Fig. 1 and see Fig. 5, which is published as supporting information on the PNAS web site, for a more complete sequence alignment). In addition, SABP2 shares 45% amino acid sequence identity with Hevea brasiliensis (Brazil nut) hydroxynitrile lyase (HNL) (31). It is likely that these enzymes are phylogenetically related (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 1.
Sequence comparison of tobacco SABP2 with the most similar proteins of known function. These include tomato MeJA esterase (MJE), Rauvolfia serpentina polyneuridine aldehyde esterase (PNAE), and Brazil nut HNL. Identical residues are shown in red and similar residues in blue. The cyan arrows and yellow bars identify the secondary structure elements. The purple line marks the cap domain. The catalytic triad residues are indicated with the magenta diamond, and residues that contact SA are indicated by green diamonds. See Fig. 5 for the GenBank accession nos. of these sequences and for an alignment with additional sequences.
To help in the understanding of the biochemical and biological functions of SABP2, we have determined its 3D structure in the absence and presence of SA at up to 2.1-Å resolution. Through biochemical analysis, we also demonstrate that SABP2 has esterase activity with a physiologically relevant Km value for MeSA, and that SA is a potent product inhibitor of this activity. These results, together with the genetic and physiological experiments previously reported for SABP2, suggest that MeSA may have an important role in SAR that is distinct from the role of SA.
Materials and Methods
Detailed experimental procedures can be found in the Supporting Text, which is published as supporting information on the PNAS web site.
Expression, Purification, Crystallization, and Structure Determination of SABP2. Tobacco SABP2 was overexpressed in Escherichia coli and purified by Ni-agarose affinity chromatography and gel-filtration chromatography. The free enzyme and the SA complex of SABP2 were crystallized at 4°C by the hanging-drop vapor diffusion method. x-ray diffraction data up to 2.1-Å resolution were collected at 100 K at the ×4A beamline of the National Synchrotron Light Source. The diffraction images were processed with the hkl package (32). The data processing statistics are summarized in Table 1.
Table 1. Summary of crystallographic information.
| Structure | Complex with SA | Free enzyme |
|---|---|---|
| Maximum resolution, Å | 2.1 | 2.5 |
| Number of observations | 135,796 | 614,841 |
| Rmerge, %* | 7.0 (28.1) | 12.5 (27.6) |
| Number of reflections | 34,275 | 138,444† |
| Number of unique SABP2 molecules | 2 | 8 |
| Resolution range used in refinement | 30-2.1 | 30-2.5 |
| Completeness, % | 92 (76) | 92 (67) |
| R factor, %‡ | 19.7 (21.7) | 22.7 (24.8) |
| Free R factor, % | 24.8 (28.1) | 29.7 (32.9) |
| rmsd in bond lengths, Å | 0.006 | 0.008 |
| rmsd in bond angles, ° | 1.2 | 1.2 |
rmsd, rms deviation.
Rmerge = ΣhΣi|Ihi〈Ih〉|/ΣhΣiIhi. The numbers in parentheses are for the highest-resolution shell.
The Friedel pairs are refined independently.
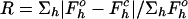 .
.
The structure of SABP2 in complex with SA was solved by the seleno-methionyl single-wavelength anomalous diffraction method (33). The structure of the free enzyme of SABP2 was determined by the molecular replacement method with the program como (34). The atomic models were built with the program xtalview (35), and the structure refinement was carried out with the program cns (36). The crystallographic information is summarized in Table 1.
Esterase Assays. The MeSA esterase activity of SABP2 was determined in two steps: incubation of SABP2 and MeSA for 30 min in the reaction buffer after which the enzyme was inactivated by boiling, and then coupling of the SA product with radioactive 14C–S-adenosylmethionine (AdoMet) by using purified SA methyltransferase. Radiolabeled products (14C-MeSA) were extracted with ethyl acetate, and radioactivity was determined in a scintillation counter. Preliminary assays verified the linearity of the reaction within the 30-min incubation times in both steps of the assay. Methylindoleacetic acid (MeIAA) and MeJA esterase activity was performed by using a similar protocol.
SA, MeSA, and MeJA Binding. MeSA and MeJA binding to SABP2 was measured by performing competition-binding assays, as described (26).
Results and Discussion
Structure Determination. The crystal structure of tobacco SABP2 in complex with SA was determined at 2.1-Å resolution by the seleno-methionyl single-wavelength anomalous diffraction method (33). The current atomic model contains residues 3–260 for the two monomers of SABP2 in the asymmetric unit. The current R factor is 19.9% (Table 1), and 87% of the residues are in the most favored regions of the Ramachandran plot. We also determined the crystal structure of free SABP2 at 2.5-Å resolution (Table 1).
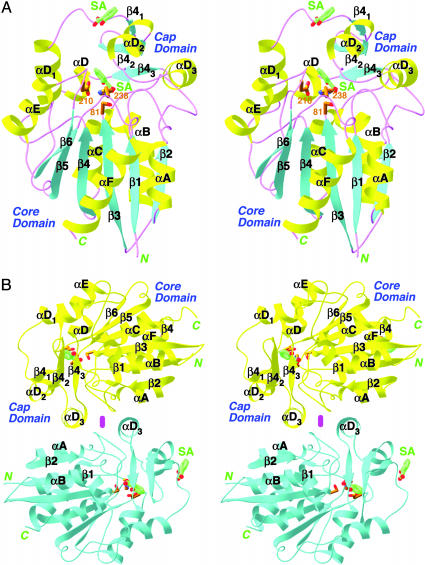
Overall Structure of SABP2. The crystal structures confirm that SABP2 is a member of the α/β hydrolase superfamily of enzymes (27, 28). The structure of SABP2 can be divided into two domains. The core domain contains a central six-stranded parallel β-sheet (named β1–β6) that is flanked on both sides by six helices (αA–αF) (Fig. 2A). The cap (or lid) domain contains a three-stranded antiparallel β-sheet (β41–β43) and three helices (αD1–αD3). The secondary structure elements are given the same names as those in the structure of HNL (31).
Fig. 2.
Structure of SABP2 in complex with SA. (A) Stereoview of the SABP2 monomer in complex with SA. The core and cap domains are labeled. The secondary structure elements, α helices, β strands, and loops are colored in yellow, cyan, and magenta, respectively. SA (in green for carbon atoms) is located in the active site as well as another site on the surface of the enzyme. This second surface-binding site may not be physiologically relevant. (B) Dimer of SABP2. The two monomers are colored in yellow and cyan, respectively. The 2-fold axis of the dimer is indicated with the magenta oval [produced with ribbons (39)].
SABP2 shares 45% amino acid sequence identity with the Brazil nut HNL, and the structures of the two enzymes are also similar to each other (Fig. 7, which is published as supporting information on the PNAS web site). The rms distance between equivalent Cα atoms of the two structures is 1.3 Å. However, despite the high degree of structural conservation, SABP2 may not possess HNL activity, because there are significant differences between the two enzymes in their active sites. In particular, a Lys residue in the active site of HNL (immediately following the second member His), which is required for HNL activity (37), is replaced by a methionine in SABP2 (Fig. 1).
A dimer of SABP2 was observed in the crystals, where residues in the cap domain of one monomer contact those in the core domain of the other (Fig. 2B). The dimer interface is rather extensive, burying ≈800 Å2 of the surface area of each monomer. Our gel filtration and light-scattering studies with the recombinant protein also showed that SABP2 is a dimer in solution at pH 7.5–8.0 (details provided in the Supporting Text). Interestingly, our earlier studies with partially purified SABP2 from natural sources suggested that it may be a monomer at physiological concentrations (25, 26). The active site of the enzyme is located far from the dimer interface (Fig. 2B), suggesting that monomers could be active catalytically.
The Active Site of SABP2. As a member of the α/β hydrolase superfamily (27, 28), the active site of SABP2 is defined by the presence of a catalytic triad, Ser-81, His-238, and Asp-210 (Fig. 2 A). These residues are strictly conserved among SABP2 and several closely related members of this superfamily (Fig. 1). The catalytic nucleophile Ser-81 is located in the sharp turn (the nucleophile elbow) between strand β3 and helix αC of the core domain, with a strained main-chain conformation. The second member of the triad, His-238, is located in the loop connecting strand β6 and helix αF, whereas the third member of the triad, Asp-210, is located in the loop connecting strand β5 and helix αE (Fig. 2 A).
The active site of SABP2 is located at the C-terminal end of the parallel β-sheet in the core domain (Fig. 2 A). The cap domain, especially strands β42, β43 and helices αD2, αD3, covers the exposed side of the active site (Fig. 2 A).
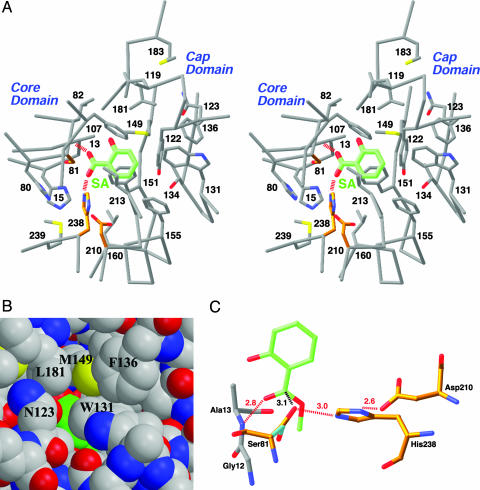
Binding Mode of SA in the Active Site. SABP2 was originally identified by its high affinity for SA, with a Kd of ≈90 nM (25). To define the binding site of SA in SABP2, we determined the crystal structure of the enzyme in complex with SA at 2.1-Å resolution (Fig. 2 A and Fig. 8, which is published as supporting information on the PNAS web site). Crystallographic analysis revealed that SA is bound in the active-site pocket of the enzyme, where it is completely shielded from the solvent and shows intimate polar and van der Waals contacts with the enzyme (Fig. 3A). This provides a molecular explanation for the high affinity of SABP2 for this compound.
Fig. 3.
The active site of SABP2 and the binding mode of SA. (A) Stereoview of the active site of SABP2 in complex with SA. The catalytic triad residues (Ser-81, His-238, and Asp-210) are shown in gold. The hydrogen bonds from the carboxylate group of SA are shown as red dashed lines. (B) Model of SABP2 in complex with SA, showing that the SA molecule (in green for carbon atoms) is shielded from the solvent in the active site. (C) Model of the binding mode of the MeSA substrate (green) to SABP2. The side chain of the catalytic Ser-81 residue assumes a different conformation for catalysis (cyan and gold in complex with SA and MeSA, respectively). The hydrogen bonds are indicated in dashed lines in red, and the distance between Ser-81, and the MeSA carboxylate carbon is indicated in black.
The carboxylate group of SA is bound deepest in the active-site pocket, and its two oxygen atoms are hydrogen-bonded to the main-chain amide of residue Ala-13 and the side chain of the His-238 (Fig. 3A), the second member of the catalytic triad. The side-chain hydroxyl of the catalytic Ser-81 is placed at an equal distance of 3 Å to the two carboxylate oxygens of SA, although the hydrogen-bonding angle is close to 90° (Fig. 3A). In addition, the Ser-81 hydroxyl is not hydrogen-bonded to the second-member His-238 residue, and the hydrogen-bonding network among the catalytic triad residues is not formed in this complex. The hydroxyl group of SA does not appear to have a hydrogen-bonding partner in the complex.
The phenyl ring of SA is located in a highly hydrophobic environment, surrounded by side chains from the core and the cap domains (Fig. 3A). In particular, the side chains of Asn-123, Trp-131, Phe-136, Met-149, and Leu-181 in the cap domain help shield the SA molecule from the solvent (Fig. 3B).
There are only minor structural differences between the free enzyme and the SA–SABP2 complex, and the rms distance between equivalent Cα atoms of the two structures is only 0.45 Å. Our studies of SABP2 by solution NMR spectroscopy showed chemical-shift changes for only a few residues in the presence of SA (Fig. 9, which is published as supporting information on the PNAS web site), indicating that the overall structure of the free and SA-bound forms of SABP2 are quite similar and consistent with observations from crystallographic analysis. However, because the active site is completely shielded from the solvent, the enzyme is expected to undergo an open–closed transition to allow substrate binding and product release. These NMR data indicate that no more than a small fraction of the enzyme is present in the open form in solution, even in the absence of SA.
Electron density for a second SA molecule was also observed in the crystal structure (Fig. 2 A). However, this molecule is located on the surface of the enzyme, at a crystal packing interface. This binding site is unlikely to possess high affinity for SA. Therefore, this second binding site may be a crystallographic artifact.
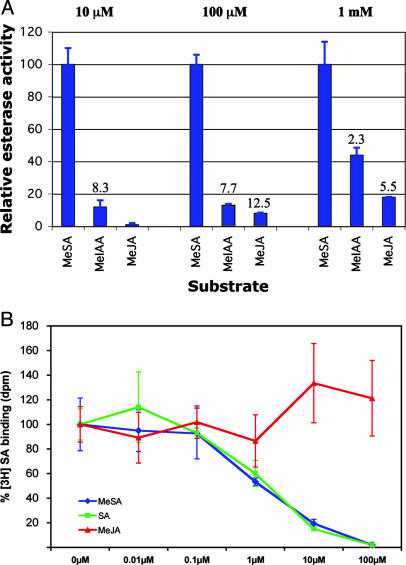
Esterase Activity of SABP2 on Natural Substrates. Because SABP2 is a member of the α/β hydrolase family, its activity was initially investigated by using enzymatic assays with various substrates. SABP2 displayed lipase activity on artificial substrates para-nitrophenyl (pNP) palmitate and pNP myristate, as well as esterase activity on pNP butyrate and 4-methylumbelliferone butyrate (26). Recent observations that the α/β hydrolase family also includes two plant methyl esterases (29, 30) (Fig. 1), as well as structural data showing binding of SA in the active site, prompted us to examine whether SABP2 could use as substrates physiologically important compounds such as methylated derivatives of plant hormones. MeSA, MeJA, and MeIAA were first tested at 1 mM concentration. SABP2 had esterase activity with all three substrates, but its activity with MeSA was the highest (Fig. 4A). The apparent Km value for MeSA is 8.6 μM, and the kcat value is 0.45 s–1.
Fig. 4.
Comparisons of methyl esterase activities and binding affinities of SABP2. (A) Relative methyl esterase activity of SABP2 with MeSA, MeIAA, and MeJA substrates at three different concentrations (10 μM, 100 μM, and 1 mM). The activity with MeSA at each of the substrate concentrations was set at 100%. The activity ratios between MeSA and the other substrates are indicated. All results are the average of three independent experiments. (B) SABP2 binds MeSA but not MeJA. Binding of [3H]SA by SABP2 in the absence of any competitor (SA, MeSA, or MeJA) was set to 100%. MeSA (blue) competes with [3H]SA for binding to SABP2 with the same potency as SA (green), whereas MeJA (red) does not compete for binding. Results (±SD) from competition-binding assays are presented as an average of three replicate binding reactions. The binding experiments were repeated three times with similar results.
The Km value for MeSA is within the range of reported in vivo MeSA concentrations during tobacco mosaic virus infection (16), but 1 mM is a concentration that is unlikely to be encountered in vivo for any of the above substrates. We therefore retested the relative esterase activity of SABP2 in the presence of lower concentrations of MeSA, MeIAA, and MeJA. At both 10 and 100 μM substrate concentrations, esterase activity with MeIAA and MeJA was <15% of that with MeSA, and no MeJA esterase activity was observed at 10 μM concentration (Fig. 4A). Thus, although SABP2 displays esterase activity with all of these substrates when they are present at high concentrations, it is highly specific for MeSA (among the substrates tested) at more physiologically relevant concentrations (16).
To help understand how SABP2 can catalyze the hydrolysis of MeSA, we modeled this substrate into the active site based on the structure of the complex obtained with SA (Fig. 3C), which is the product of the reaction. MeSA can be readily accommodated in the active site and requires little conformational changes in the enzyme. The only residue that does change is the catalytic Ser-81. By assuming a different torsion angle, the side chain of this residue can become hydrogen bonded to that of His-238, completing the catalytic triad (Fig. 3C). In addition, the side-chain hydroxyl is placed directly over the carboxyl carbon of MeSA, in a perfect position for initiating the nucleophilic attack. The methyl group in the model is pointed toward a small cavity at the bottom of the active-site pocket. The model, together with our structural information on the SA–SABP2 complex (Fig. 3A), also shows that the active site is only large enough to accommodate the phenyl ring of MeSA, therefore explaining the poor activity of SABP2 with the MeIAA and MeJA substrates (Fig. 4A).
SA Is a Potent Inhibitor of the Esterase Activity of SABP2. These studies demonstrate that SABP2 possesses specific esterase activity toward MeSA (Fig. 4A) and that SA, the product of this reaction, is bound in the active site (Fig. 2 A). These findings, combined with SABP2's high binding affinity for SA (25, 26), suggest that SA is a potent product inhibitor of SABP2's MeSA esterase activity. Indeed, at a substrate concentration of 1 μM, MeSA esterase activity was inhibited by 40% with 25 and 100 nM SA, by 60% with 1 μM SA, and by 95% with 2 μM SA. This potent inhibitory activity is consistent with the 90 nM Kd value of SA for SABP2 (25).
The kinetic and structural data also suggest that MeSA should compete with SA for binding to SABP2. Our data show that MeSA competed with [3H]SA for binding to SABP2 as effectively as unlabeled SA (Fig. 4B). However, additional analyses indicated that >90% of MeSA was hydrolyzed to SA in the binding assay, and at least part of the observed competition was due to the SA product. As a control, MeJA failed to compete with [3H]SA for binding, even when present in a 1,000-fold molar excess (Fig. 4B).
Characterization of the Active-Site Ser81Ala Mutation. To confirm that Ser-81 of the catalytic triad is essential for SABP2's esterase activity, we created the Ser81Ala mutant. This protein has little or no esterase activity on MeSA, MeJA, and MeIAA, as well as with three artificial substrates, pNP acetate, pNP butyrate, and pNP caproate (data not shown). However, the mutant protein maintains similar binding affinity for SA (data not shown). This is consistent with our structural observation that the side chain hydroxyl of Ser-81 has only weak interactions with SA in the complex (Fig. 3A).
Conclusion
We have determined the crystal structures of tobacco SABP2, alone and in complex with SA, at up to 2.1-Å resolution. The structures confirm that SABP2 is a member of the α/β hydrolase superfamily of enzymes, with Ser-81, His-238, and Asp-210 as the catalytic triad. SA is bound in the active site and is completely shielded from the solvent, consistent with the high affinity of this compound for SABP2. Our biochemical studies reveal for the first time that SABP2 has strong esterase activity with MeSA, and that SA is a potent product inhibitor of this catalysis.
In tobacco mosaic virus-infected resistant tobacco plants, which later develop SAR, MeSA has been shown to accumulate to high intracellular concentrations (16). However, MeSA generally is undetected, because it is a volatile liquid at room temperature, and most procedures to quantify SA in tissue extracts include a drying step. Recently, the SAMT gene, which encodes a methyltransferase that synthesizes MeSA from SA using AdoMet as the methyl donor, was isolated (22, 23) and shown to be induced locally at the site of damage on a leaf (23). It has been shown that some MeSA produced in vegetative tissue after infection and during development of SAR is emitted into the atmosphere, where it helps attract enemies of insect herbivores (38). However, the physiological role of the relatively high intracellular MeSA concentration is not clear, nor is the function of any MeSA synthesized when SAR is triggered by microbial attack. Because MeSA is more hydrophobic than SA and can therefore cross membranes more readily than SA, it is possible that both short- and long-distance transmission of SA synthesized at the site of infection requires converting it first to MeSA. Additionally, MeSA may be an inactive form of SA that is used for storage. Our studies suggest that the role of SABP2 in plant host defense may be not as a receptor for SA (26) but rather in the hydrolysis of biologically inactive MeSA into active SA in the target cells. This is consistent with observations that SABP2-silenced plants fail to develop SAR and have suppressed local defense responses (26). The potent inhibition of SABP2 by the product of the reaction, SA, may further help fine-tune intracellular SA levels. The presence of homologous proteins with MeSA esterase activity in other plant species (Fig. 6) suggests that MeSA is a general component of SA-dependent plant innate immune response.
Supplementary Material
Acknowledgments
We thank Randy Abramowitz and Xiaochun Yang for setting up the X4A beamline. This work was supported in part by a grant from the Protein Structure Initiative of the National Institutes of Health [Grant P50 GM62413 (to G.T.M. and L.T.)], by a R01 grant from the National Institutes of Health [AI49475 (to L.T.)], and by National Science Foundation Grants MCB-0312466 (to E.P.) and IBN-0241531 (to D.F.K.).
Abbreviations: SAR, systemic acquired resistance; SA, salicylic acid; MeSA, methyl salicylate; SABP, SA-binding protein; HNL, hydroxynitrile lyase; pNP, para-nitrophenyl; MeJA, methyl jasmonate; MeIAA, methylindoleacetic acid.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1Y7H, 1Y7I, and IXKL).
References
- 1.Dangl, J. L. & Jones, J. D. G. (2001) Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- 2.Holt, B. F., III, Hubert, D. A. & Dangl, J. L. (2003) Curr. Opin. Immunol. 15, 20–25. [DOI] [PubMed] [Google Scholar]
- 3.Cohn, J., Sessa, G. & Martin, G. B. (2001) Curr. Opin. Immunol. 13, 55–62. [DOI] [PubMed] [Google Scholar]
- 4.Hammond-Kosack, K. E. & Jones, J. D. G. (1996) Plant Cell 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dempsey, D., Shah, J. & Klessig, D. F. (1999) Crit. Rev. Plant Sci. 18, 547–575. [Google Scholar]
- 6.Dong, X. (2001) Curr. Opin. Plant Biol. 4, 309–314. [DOI] [PubMed] [Google Scholar]
- 7.Glazebrook, J. (2001) Curr. Opin. Plant Biol. 4, 301–308. [DOI] [PubMed] [Google Scholar]
- 8.Kunkel, B. N. & Brooks, D. M. (2002) Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- 9.van Camp, W., van Montagu, M. & Inze, D. (1998) Trends Plant Sci. 3, 330–334. [Google Scholar]
- 10.Draper, J. (1997) Trends Plant Sci. 2, 162–165. [Google Scholar]
- 11.Shirasu, K., Nakajima, H., Rajasekhar, V. K., Dixon, R. A. & Lamb, C. (1997) Plant Cell 9, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mur, L. A., Bi, Y. M., Darby, R. M., Firek, S. & Draper, J. (1997) Plant J. 12, 1113–1126. [DOI] [PubMed] [Google Scholar]
- 13.Delaney, T. P., Uknes, S., Vernooij, B., Friedrich, L. B., Weymann, K. B., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., et al. (1994) Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- 14.Gaffney, T., Friedrich, L. B., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H. & Ryals, J. (1993) Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- 15.Shulaev, V., Silverman, P. & Raskin, I. (1997) Nature 385, 718–721. [Google Scholar]
- 16.Seskar, M., Shulaev, V. & Raskin, I. (1998) Plant Physiol. 116, 387–392. [PMC free article] [Google Scholar]
- 17.Dudareva, N., Raguso, R. A., Wang, J., Ross, J. R. & Pichersky, E. (1998) Plant Physiol. 116, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enyedi, A. J., Yalpani, N., Silverman, P. & Raskin, I. (1992) Proc. Natl. Acad. Sci. USA 85, 2480–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malamy, J., Hennig, J. & Klessig, D. F. (1992) Plant Cell 4, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hennig, J., Malamy, J., Grynkiewicz, G., Indulski, J. & Klessig, D. F. (1993) Plant J. 4, 593–600. [DOI] [PubMed] [Google Scholar]
- 21.Seo, S., Ishizuka, K. & Ohashi, Y. (1995) Plant Cell Physiol. 36, 447–453. [Google Scholar]
- 22.Ross, J. R., Nam, K. H., D'Auria, J. C. & Pichersky, E. (1999) Arch. Biochem. Biophys. 367, 9–16. [DOI] [PubMed] [Google Scholar]
- 23.Chen, F., D'Auria, J. C., Tholl, D., Ross, J. R., Gershenzon, J., Noel, J. P. & Pichersky, E. (2003) Plant J. 36, 577–588. [DOI] [PubMed] [Google Scholar]
- 24.Yalpani, N., Schulz, M., Davies, M. P. & Balke, N. E. (1992) Plant Physiol. 100, 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du, H. & Klessig, D. F. (1997) Plant Physiol. 113, 1319–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, D. & Klessig, D. F. (2003) Proc. Natl. Acad. Sci. USA 100, 16101–16106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., Harel, M., Remington, S. J., Silman, I., Schrag, J., et al. (1992) Protein Eng. 5, 197–211. [DOI] [PubMed] [Google Scholar]
- 28.Nardini, M. & Dijkstra, B. W. (1999) Curr. Opin. Struct. Biol. 9, 732–737. [DOI] [PubMed] [Google Scholar]
- 29.Stuhlfelder, C., Mueller, M. J. & Warzecha, H. (2004) Eur. J. Biochem. 271, 2976–2983. [DOI] [PubMed] [Google Scholar]
- 30.Dogru, E., Warzecha, H., Seibel, F., Haebel, S., Lottspeich, F. & Stockigt, J. (2000) Eur. J. Biochem. 267, 1397–1406. [DOI] [PubMed] [Google Scholar]
- 31.Wagner, U. G., Hasslacher, M., Griengl, H., Schwab, H. & Kratky, C. (1996) Structure (London) 4, 811–822. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 33.Hendrickson, W. A. (1991) Science 254, 51–58. [DOI] [PubMed] [Google Scholar]
- 34.Jogl, G., Tao, X., Xu, Y. & Tong, L. (2001) Acta Crystallogr. D 57, 1127–1134. [DOI] [PubMed] [Google Scholar]
- 35.McRee, D. E. (1999) J. Struct. Biol. 125, 156–165. [DOI] [PubMed] [Google Scholar]
- 36.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 37.Gruber, K., Gartler, G., Krammer, B., Schwab, H. & Kratky, C. (2004) J. Biol. Chem. 279, 20501–20510. [DOI] [PubMed] [Google Scholar]
- 38.Pichersky, E. & Gershenzon, J. (2002) Curr. Opin. Plant Biol. 5, 237–243. [DOI] [PubMed] [Google Scholar]
- 39.Carson, M. (1987) J. Mol. Graphics 5, 103–106. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.