Abstract
We report the crystal structure of the glycosylated ligand-binding (S1S2) domain of the kainate receptor subunit GluR6, in complex with the agonist domoate. The structure shows the expected overall homology with AMPA and NMDA receptor subunit structures but reveals an unexpected binding mode for the side chain of domoate, in which contact is made to the larger lobe only (lobe I). In common with the AMPA receptor subunit GluR2, the GluR6 S1S2 domain associates as a dimer, with many of the interdimer contacts being conserved. Subtle differences in these contacts provide a structural explanation for why GluR2 L483Y and GluR3 L507Y are nondesensitizing, but GluR6, which has a tyrosine at that site, is not. The structure incorporates native glycosylation, which has not previously been described for ionotropic glutamate receptors. The position of the sugars near the subunit interface rules out their direct involvement in subunit association but leaves open the possibility of indirect modulation. Finally, we observed several tetrameric assemblies that satisfy topological constraints with respect to connection to the receptor pore, and which are therefore candidates for the native quaternary structure.
Keywords: crystal structure, domoate, glutamate receptor
Ionotropic glutamate receptors (iGluRs) are major mediators of rapid excitatory neurotransmission in the mammalian CNS and in consequence are involved in a wide range of physiological and pathological neural processes (1). There are three main iGluR subfamilies: AMPA, NMDA, and kainate receptors. The functional importance of both AMPA and NMDA receptors in synaptic transmission and plasticity has long been recognized. It is only recently, however, that the physiological roles of kainate receptors have begun to be delineated. Studies using selective ligands and gene ablation in mice have shown that kainate receptors are involved in the regulation of synaptic transmission and plasticity in several brain regions (2).
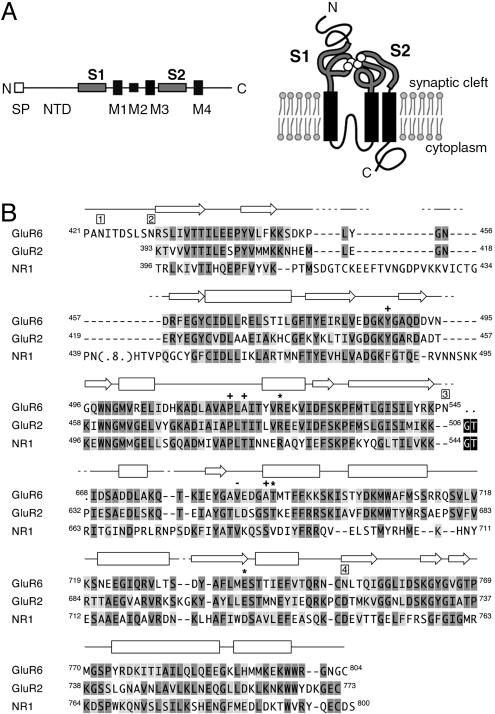
All iGluRs are thought to share a similar overall structure, with four homologous subunits arranged around a central cation-selective pore. The subunits themselves share a common membrane topology (Fig. 1A), with three transmembrane domains (termed M1, M3, and M4) and a reentrant loop similar to the P loop found in potassium channels (termed M2). This leaves two large polypeptide domains on the extracellular (synaptic) side of the membrane: the ≈530-aa N terminus and the domain between M3 and M4. Studies of chimeric receptors and recombinant constructs identified the ≈120-aa preceding M1 (termed S1) and the ≈140-aa M3–M4 loop (S2) as forming the agonist-binding domain (3, 4). The S1S2 domain can be expressed as a soluble polypeptide independently of the membrane-spanning domains, making it amenable to crystallization and structure determination by x-ray diffraction. In this way, structures for the S1S2 domains for the AMPA-selective subunit GluR2 (5–7) and the NMDA subunit NR1 (8) have been determined, with both proteins sharing a bilobate structure. Although no structure has been available for the S1S2 domain of a kainate-selective subunit, the primary sequences of the agonist-binding domains of AMPA and kainate receptor subunits are sufficiently similar (48–55%) that the GluR2 S1S2 structure can be used as a good model. This has enabled directed mutagenesis studies to probe kainate receptor function (9, 10). Nonetheless, for kainate receptors, there are limits to the inferences that can be drawn from a straight comparison with the GluR2 structure. For example, the kinetics and ion sensitivity of desensitization and deactivation appear to differ between AMPA and kainate receptors (11, 12).
Fig. 1.
GluR6 topology and sequence alignment. (A) The topology of iGluR subunits makes it possible to express the GluR6 agonist-binding domain comprising S1 and S2 as a soluble polypeptide, removing both the N-terminal domain and membrane domains M1–M4. Ligand (white) is shown bound to the S1S2 domain. SP, signal peptide. (B) The regions of the GluR6 S1S2 polypeptide modeled in the structure are shown, aligned to equivalent residues from the GluR2 and NR1 structures. The numbering for GluR6 and NR1 is for the protein including signal sequence; the numbering for GluR2 is for the mature polypeptide, to conform with common usage in the literature. Secondary structure elements in GluR6 are indicated by rectangles (α-helices) and arrows (β-strands). Gaps, introduced on the basis of structural proximity, are indicated by dashes, and dots denote residues not visible in the structures. Primary sequence homology is indicated by medium and light gray for identical and conserved residues, respectively. Linker sequences are shown in black. Positions of the four potential N-linked glycosylation sites are shown by boxed numbers above the sequence. Residues interacting with the domoate molecule via ionic interactions through the side chain (asterisks) or main chain (pluses) and via hydrophobic interactions (dashes) are indicated above the sequence (see also Fig. 3).
An atomic structure for a kainate receptor subunit agonist-binding domain is therefore crucial to complete the picture of iGluRs. Here we describe the structure of the S1S2 domain of the kainate-selective iGluR subunit GluR6 with domoic acid (domoate) bound at 3.1-Å resolution, determined for the glycosylated polypeptide. We compare the overall protein fold with those of the GluR2 and NR1 subunit S1S2 domains and describe the mode of domoate binding. Finally, we identify differences in intersubunit contacts between the GluR2 and GluR6 subunits. The relevance of these structural differences to the function of AMPA and kainate receptors is explored.
Materials and Methods
Expression, Purification, and Characterization of GluR6 S1S2His. The cDNA for rat GluR6 was used in this study. To conform with common usage in the literature, we have numbered GluR6 residues according to the full sequence (i.e., including the 31-aa signal peptide) and GluR2 according to the mature polypeptide. The GluR6 S1S2His expression construct [the GluR6 signal peptide (residues 1–31), a FLAG epitope tag (DYKDDDDK), the S1 domain residues K418 to N557, a 9-aa linker (GGSLVPRGS), and the S2 domain residues E662 to N819 and a hexa-histidine tag] was inserted into the pFASTBac vector (Invitrogen). Recombinant baculovirus was generated by using the Bac-to-Bac system protocol (Invitrogen). Virus generation, maintenance, and expression are described in the Supporting Text, which is published as supporting information on the PNAS web site. The recombinant protein was purified chromatographically (using nickel resin, Resource Q ion exchange, and gel filtration columns), including proteolytic treatment with endoproteinase Glu-C (resulting in the loss of the His tag). Details of the purification procedure are given in the Supporting Text.
Recombinant baculovirus for full-length GluR6 was generated by using the Bac-to-Bac system (data not shown). This was used to infect adherent High 5 cells in T75 flasks. Membranes were prepared from these cells 68–72 h postinfection essentially as described in ref. 13. The GluR6 S1S2 soluble polypeptide was characterized after extensive dialysis of the culture media but before purification. Sample saturation-binding assays performed on the purified polypeptide gave similar values (data not shown). [3H]Kainate equilibrium-binding assays were performed as described (13). Nonspecific binding was defined in the presence of 1 mM glutamate.
Crystallization and X-Ray Diffraction Studies. Crystals were obtained by the hanging drop vapor diffusion method in 2.4 M ammonium sulfate/0.1 M ammonium thiocyanate/0.1 M Pipes, pH 6.5/1.5 mM domoate. After 2 months at 20°C, crystals grew in plates of 100 × 150 × 40 microns in the space group C2. Data were collected on flash-frozen crystals by using 12% ethylene glycol as cryoprotectant. Oscillation data (Table 1) were collected at a wavelength of 1.08 Å on beamline 7.1 at the Stanford Synchrotron Radiation Laboratory by using a MAR Research (Hamburg) image plate detector.
Table 1. Data collection and refinement statistics.
| Space group | C2 |
| Unit cell | a = 246.4 Å; b = 106.6 Å; c = 172.7 Å; β = 133.2° |
| Resolution, Å | 30-3.1 (3.5-3.1) |
| Reflections observed, Å | 156,647 (42,357) |
| Unique reflections | 58,030 (17,019) |
| Completeness, % | 97.2 (94.2) |
| I/σ | 8.7 (2.4) |
| Rsymm* | 11.8 (42) |
| Refinement resolution | 30-3.1 (3.27-3.1) |
| Rcryst† | 27.5 (36.9) |
| Rfree‡ | 33.5 (42.2) |
| Protein residues | 1,521 |
| Solvent atoms | 78 |
| Average protein B factor, Å2; protomer a/b/c/d/e/f | 58.6/58.8/59.9/59.2/60.7/59.1 |
| Average domoate B factor, Å2; subunit a/b/c/d/e/f | 33.3/34.9/39.7/41.8/55.0/57.9 |
| rms deviation from ideality | |
| Bonds, Å | .011 |
| Angles, ° | 1.50 |
| Ramachandran core/allowed/disallowed | 88.1/11.9/0 |
Values in parentheses represent the highest-resolution shell.
Rsym = Σhkl Σi|I(hkl,i) - 〈I(hkl)〉|/Σhkl Σi|I(hkl,i)|.
Rcryst = Σhkl∥Fobs| - |Fcalc∥/Σhkl|Fobs|.
Rfree = Σhkl⊂T∥Fobs| - |Fcalc∥/Σhkl⊂T|Fobs| based on 5.1% of the data.
Data were processed in xds/xscale (14) and imported into MTZ format. Structure solution was performed by molecular replacement with anisotropy correction using phaser, Ver. 1.2 (15), and GluR2 S1S2 (1FTK) as a search model. Four protomers (a–d) were found initially, the fifth (e) in a repeat search, and the final protomer (f) placed by hand. Rigid body refinement on each lobe of all six protomers was performed by using refmac (16). sigmaa (17)-weighted structure factors were then used in resolve (18) in prime and switch mode, using sixfold noncrystallographic symmetry (NCS) averaging to obtain an initial electron density map. Positions and temperatures were refined in refmac, and restrained in NCS groups, as determined by escet (19). Although the free R reflection subset (5.1%) was chosen randomly, subsequent analysis using fine-resolution shells to avoid potential model overfit produced essentially the same Rfree values. Model improvement was performed by using the model-building package coot (20). The geometric parameters of the model (Table 1) were validated in procheck (21).
Analysis of the domain closure angle was performed by using the dyndom web site (www.cmp.uea.ac.uk/dyndom/main.jsp) (22). This method gave equivalent results for GluR2 S1S2 structures to the method used by Armstrong and Gouaux (6). The PDB ID codes of the GluR2 S1S2 structures used were: GluR2-kainate, 1FWO; GluR2-glutamate, 1FTJ; GluR2-AMPA, 1FTM; and GluR2-apo, 1FT0. In both dyndom and whole molecule alignments using lsqkab (14), the following residue equivalences were used: Lobe I, GluR6 residues 434–445 with GluR2 residues 396–407, 460–487 with 422–449, 501–544 with 463–506, and 751–799 with 719–767; lobe II, 670–694 with 635–659, 701–710 with 666–675, 717–731 with 682–696, and 732–747 with 699–714. Figures were generated by using macpy-mol (http://pymol.sourceforge.net).
Results and Discussion
GluR6 S1S2His Purification and Characterization. We purified the GluR6 S1S2 polypeptide after recombinant expression in insect cells (see Supporting Text and Fig. 5, which is published as supporting information on the PNAS web site), including a proteolytic digest that cleaved the C-terminal His tag (data not shown) but without any treatment to eliminate glycosylation. The binding properties of the GluR6 S1S2 polypeptide were determined by radioligand-binding assay as described in Materials and Methods. These were similar to those of the full length GluR6 subunit (Table 2, which is published as supporting information on the PNAS web site), consistent with correct folding of the soluble S1S2 domain. The purified GluR6 S1S2 polypeptide was monomeric in solution and was crystallized at neutral pH in the presence of 1.5 mM domoate (see Materials and Methods). The resulting crystals were in the C2 space-group and diffracted to a maximum resolution of 3.1 Å (Table 1). Phases were obtained by the molecular replacement method, using the GluR2 S1S2 structure (5) as described in Materials and Methods. There are six S1S2 protomers in the asymmetric unit. These are essentially identical, although there were minor differences in the visibility of the N and C termini of S1 and S2 and in the visibility of side chains. The sequence built into the density is shown aligned with GluR2 and NR1 in Fig. 1B.
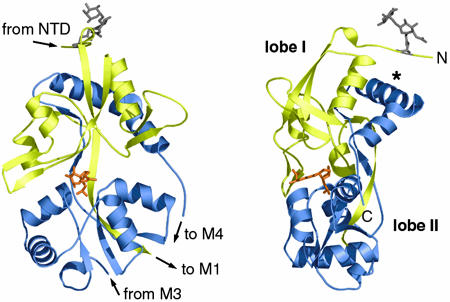
GluR6, GluR2, and NR1 S1S2 Domains Share a Common Fold. The overall fold of the GluR6 S1S2 polypeptide (Fig. 2) is essentially the same as that seen previously for the S1S2 domains of AMPA receptor subunit GluR2 and the NMDA receptor subunit NR1 (Fig. 6 A and B, which is published as supporting information on the PNAS web site). We do, however, see clear density for an additional 10 aa of the N terminus of S1 compared with these structures, providing further constraints on the relative positioning of the S1S2 domain and the remainder of the N terminus. The GluR6 S1S2 domain forms a bilobed α/β structure, with the ligand bound in the cleft between the lobes. Defining the N terminus as the “top” of the protomer, the S1 domain comprises the bulk of the upper lobe (lobe I) with seven β-strands, three α-helices, and two large loops, before crossing to the lower lobe (lobe II) via an 11-residue β-strand. At this point, there is a break in the electron density, corresponding approximately to the M1–M3 pore-forming membrane domain. The S2 domain resumes close to the end of the S1 domain and forms the lower lobe with five α-helices and three β-strands before returning to the upper lobe via another extended β-strand. As expected from homology with GluR2 and NR1 (5, 8), we observed a disulphide link between cysteines 750 and 804 in protomers a, b, and d. Protomers c, e, and f do not appear to contain a disulphide, with no peaks >3.5 σ in Fo–Fc maps near C750. Indeed, C750 appears to be oxidized in these protomers. However, because C804 is at the very C terminus of our structure and exhibits large temperature factors, we cannot rule out the presence of a disulphide in all protomers.
Fig. 2.
The structure determined for GluR6 S1S2 (protomer a) is shown in two views ≈90° apart. S1 is colored yellow, and S2 is blue. The N-acetylglucosamine–fucose sugar moieties modeled at N423 (gray) and domoate (orange) are shown in stick representation. Labels indicate the relative location of the chains within the full-length subunit, lobes I and II, and the construct N and C termini. *, helix 774–788.
N-Linked Glycosylation. The iGluR S1S2 domain structures described to date have been determined for unglycosylated proteins, whereas our structure is of the glycosylated polypeptide. There are nine consensus glycosylation sites [NX(S/T)] in the extracellular domains of GluR6, four of which are in the S1S2 domain expressed in this study (termed here sites 1, N423; 2, N430; 3, N546, and 4, N751; indicated by boxed numerals in Fig. 1B). Two sites appear to be glycosylated and two not. Site 1 is glycosylated in all protomers and was modeled as an unbranched N-acetylglucosamine (NAG) adduct. A fucose ring was modeled linked to the NAG in protomer a (Fig. 2). This corresponds to part of a paucimannosidic N-glycan, which has been described as the dominant glycosylation in insect cells (23). The asparagine side chain for site 2 is visible in protomers b, c, e, and f and is apparently unglycosylated in all of these protomers, whereas no significant difference density is observed within 8 Å of site 2 in protomers a and d. Site 3 is in a disordered region in all protomers except b, where it does not appear to be glycosylated. Finally, site 4 is glycosylated in all protomers except a and was modeled with single NAG moieties.
Lobe Closure and Agonist Potency. Structures of the GluR2 and NR1 S1S2 domains have been determined in their unliganded (apo) forms and several ligand-bound states (e.g., see refs. 5–8, 24–26). These structures have varied in two main respects: the angle of closure of the binding cleft and the intersubunit contacts observed, both of which have functional implications. In GluR2 S1S2, the degree of domain closure correlates closely with ligand efficacy (6, 25, 26). The full agonists glutamate and AMPA close the cleft to the greatest extent (19–21°) relative to the unliganded (apo) form. The partial agonist kainate, in contrast, partly closes the cleft (12°), whereas with the antagonist DNQX bound domain closure is prevented, the lobes being only ≈2.5° closer relative to the apo form (6).
We compared the GluR6 S1S2 structure with GluR2 S1S2 in complex with glutamate (GluR2-glu), AMPA (GluR2-AMPA), kainate (GluR2-KA), and no ligand (GluR2-apo), as described in Materials and Methods. The lowest rms deviation was for the GluR2-KA structure (0.87 Å), followed by glu (1.21 Å), AMPA (1.24 Å), and apo (1.75 Å) structures. When lobes I and II were aligned independently, all rms values were <1 Å (range 0.80–0.96 Å), as expected if the differences were due to hinge motions. We therefore calculated the degree of rotation around the putative interlobe hinge (see Materials and Methods). Independent alignment of lobes I and II required no hinge closure when GluR6 was compared with GluR2-KA. Alignment with both the GluR2-glu and GluR2-AMPA structures required a rotation of 10° to a more “closed” conformation, whereas alignment with GluR2-apo required a rotation of 15° to a more “open” conformation. These observations are consistent with the notion that domoate acts as a partial agonist at GluR6.
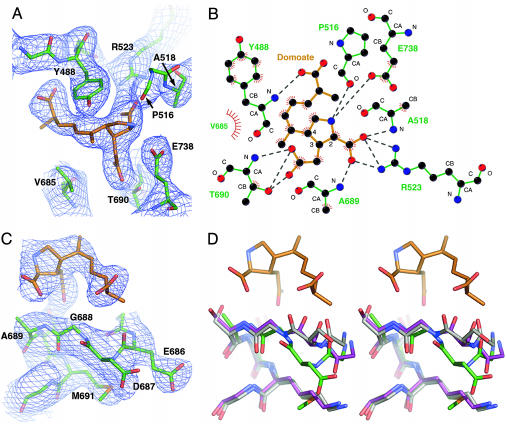
Agonist-Binding Specificity and Selectivity. The similarity to the GluR2–kainate complex extends to the ligand-binding mode. The ring and carboxylate groups are shared between domoate and kainate, and this moiety binds to GluR6 in an essentially identical manner to kainate in GluR2 (Fig. 3 A and B). All residues directly interacting with the ligand were clearly visible in initial, refined, and composite omit maps. The most significant difference between domoate and previously characterized ligands in complex with iGluRs is the large side chain that extends from position 4 on the pyrrolidine ring. In our model, this side chain correlates well with the electron density from the initial maps (Fig. 3 A and C), giving us good confidence in the position of the ligand. Interestingly, despite its size, the domoate side chain interacts only with lobe I (via the NH of Y488) and makes no apparent contact with lobe II. This differs from the predictions of modeling studies of domoate binding to GluR2 (28). The region of lobe II closest to the domoate side chain is bent away from the ligand (Fig. 3C). This apparent displacement is more obvious when this region is compared with the equivalent residues in GluR2 (Fig. 3D) and is possibly due to a polar interaction we observe between the backbone amide of N687 and the sulfur of M691. Mobility in this region seems to be important for ligand specificity, because the same region of GluR2 (D651 and S652) has been shown to undergo a 180° peptide bond flip when complexed with full agonists (6).
Fig. 3.
Views of the domoate-binding pocket; domoate carbon atoms are colored orange and GluR6 carbons, green. (A) Electron density for domoate and the main interacting residues in protomer a is shown with a stick representation of the final model structure. The fourfold NCS averaged density (contouredat1σ) was calculated with sigmaa-weighted coefficients and phases from the initial molecular replacement solution (four promoters and no ligands). Only the main-chain carboxyl of P516 is shown for clarity. (B) The observed ligand–receptor interactions in protomer a are shown in schematic form (generated by using ligplot; see ref. 37). Dotted gray lines indicate polar interactions and red hatching hydrophobic contacts. Main-chain atoms are labeled, as are substituted pyrrolidine ring carbons. (C) Electron density and model structure are shown for residues E686 to T692 (main chain only for T692) and domoate. (D) The same view is shown in stereo for an alignment of the GluR6 (green carbons), GluR2-AMPA (purple carbons), and GluR2-KA (gray carbons) structures (for clarity, the side chains of residue GluR6 E686 and its GluR2 equivalent are omitted).
We also looked at the positioning of residue N721 within the binding pocket. This site has been shown to be involved in ligand selectivity in kainate receptors (13, 29). For instance, whereas GluR6 is not activated by the ligand AMPA, the GluR6 N721S mutant both binds to and is activated by AMPA (13). This same mutation also increases the rate of channel deactivation after application of domoate. Curiously, however, residue N721 is not close to any part of the ligand in our structure. It appears to instead form a polar interaction with E440 on the other side of the cleft, as the GluR2 homologues T686 and E402 do in ligand-bound structures. It is possible that this interaction restricts domain closure, thereby preventing GluR6 subunits from adopting the degree of closure necessary for AMPA binding. However, the exact mechanism by which the N721S mutation in GluR6 converts the subunit into one that responds to AMPA remains to be determined.
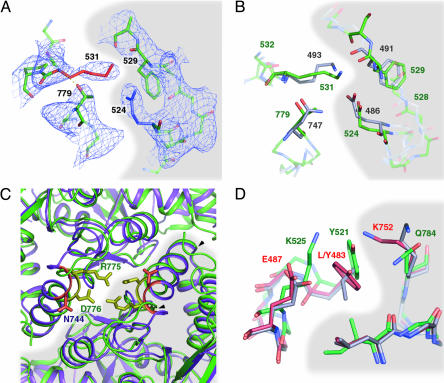
GluR6 S1S2 Forms a Twofold Symmetric Dimer. Many of the GluR2 S1S2 structures have contained a common twofold symmetric dimer (termed here the “AMPA-like” dimer). The crystal structure of GluR6 S1S2 reveals three AMPA-like dimers (b:d, c:e, and a:a). The surface area buried per monomer is ≈1,600 Å2. This is more extensive than in GluR2 (1,150 Å2; ref. 6), partially due to a shift in the 774–788 helix and also to the N terminus, which makes additional contacts with the adjacent subunit. The overall orientation of the dimers is similar in the GluR6 and GluR2 structures, and we observe some of the same interprotomer interactions. For example, there is a salt bridge formed across the dimer in GluR2 (between E486 and K493). The equivalent residues in GluR6, E524, and K531 are in a similar conformation (Fig. 4 A and B), although our electron density maps do not allow us to say definitively whether they form a salt bridge. Similarly, in the same region, both F529 and T779 occupy equivalent positions to their GluR2 counterparts (F491 and N747, respectively). Interestingly, however, unlike the bivalent N747 in GluR2, which interacts with both K493 and E486 across the interface, the monovalent T779 in GluR6 makes no intersubunit contacts but instead forms a polar interaction with the backbone NH of K531. We see a further potential crossdimer salt bridge not seen in GluR2, between R775 and D776 (Fig. 4C). This interaction requires an inward shift and rotation of the helix comprising residues 774–788 compared with the equivalent helix in GluR2. Intriguingly, both of these residues are variable in GluR2, with residue 743 subject to RNA editing (from R to G; ref. 30) and 744 the first residue in the flip-and-flop splice variants (T and N, respectively). It is therefore possible that a different combination of these residues in GluR2 might also form a similar contact.
Fig. 4.
Views of the AMPA-like dimer interface in GluR6 and GluR2. The interface between dimers is highlighted by shading. Structures were aligned over lobe I only. (A) Density is shown around residues K531 and T779 (protomer c on the left) and residues E524 F529 (protomer e on the right). A polar interaction is observed between the T779 side chain and the K531 main chain nitrogen. (B) The same view of GluR6 (green residue labels) is shown aligned with GluR2 protomers a and c (AMPA complex, carbons, and residue labels in gray). GluR2 N747 forms hydrogen bonds with both the K493 main chain and the E486 side chain. (C) An interaction observed only in GluR6 is shown for the same protomer pairs (GluR6, green cartoon; GluR2, purple cartoon), looking down the twofold axis from lobe I. GluR6 residues R775 and D776 (yellow; green labels) and GluR2 residues G743 and G743 and N744 (orange; purple label) are highlighted. There is a clear movement of helix 774–788 (arrowheads) compared with the equivalent helix in GluR2 (742–755). (D) The environment around GluR6 residue Y521 in the dimer formed by protomer a with itself (green carbons) is shown compared with the equivalent residues in wild-type GluR2 (gray carbons) and GluR2 L483Y (1LB8; red carbons). Residues Y521 to K525 are shown on the left (residues L/Y483 to E487 in GluR2) and I780 to Q784 (L748 to K752 in GluR2) on the right. For clarity, side chains are shown only for the first and last residues in each chain.
The AMPA-Like Dimer and Receptor Desensitization. In GluR2, there is a good correlation between stability of the dimer (measured by equilibrium centrifugation) and the degree of desensitization, leading Sun et al. (31) to propose a model where rearrangement of the dimer is required for the receptor to desensitize (31). They showed that L483Y, the GluR2 homologue of the nondesensitizing GluR3 mutant L507Y (32), increased the stability of the dimer through an intersubunit contact (31). Further, they showed that cyclothiazide, which blocks desensitization in AMPA receptors, binds to a site in the dimer interface. Their model of receptor activation predicted that mutations that stabilize the dimer will reduce or eliminate desensitization, whereas those that destabilize it will accelerate desensitization. This has been found largely to be the case in GluR2 (31, 32).
We therefore identified the interactions of Y521, the GluR6 homologue of GluR2 residue 483. The residues that interact with the tyrosine in the GluR2 L483Y mutant (L748 and K752) are partly conserved in GluR6 (as I780 and Q784). Despite this, GluR6 desensitizes strongly in response to agonist. The most obvious explanation would be that in GluR6, Q784 and Y521 do not interact as strongly as their homologues in GluR2, but GluR6 Q784K desensitizes only marginally slower than wild type (9). The explanation for this striking functional divergence appears to be due instead to structural differences between GluR2 and GluR6 in the surrounding region (Fig. 4D). Although GluR2 Y483 interacts with K752 across the dimer interface, an equivalent interaction seems to occur within the GluR6 protomer between Y521 and K525. We predict that GluR6 desensitizes, because Y521 is held in a strong interaction with K525. In support of this hypothesis, mutation of the GluR3 homologue of GluR6 K525 (E511) to lysine increases desensitization of the GluR3 L507Y mutant (32).
The interactions we observe around Y521 fit in well with a link between dimer stability and desensitization, but other reported GluR6 mutations appear to contradict the model's predictions. In particular, desensitization is slowed in the GluR6 K531G mutant (9), despite its involvement in a dimer contact (Fig. 4A). Mutation of T779 to glycine also had unexpected effects, with desensitization faster in response to glutamate but slower in response to kainate (9). It is therefore extremely surprising that we do not observe any significant structural differences between GluR6 and GluR2 around these residues. It has been proposed that K531G slows desensitization by enabling a closer association with the opposing protomer (33), but on examination of our structure, we do not believe this to be a plausible explanation. We believe that the most likely reason why certain disruptive changes made within the dimer interface of GluR6 slow desensitization is that GluR6 and GluR2 differ in the contacts they make, either in the transition to the desensitized state or in the desensitized state itself. A subunit conformation that can be ascribed to the desensitized state has not yet been identified in any iGluR ligand-binding domain structure, but further investigation of these observed differences between GluR2 and GluR6 may well shed light on this important question.
Receptor Assembly and Domain Organization. iGluRs are thought to be tetramers. We therefore looked for tetramers in the GluR6 S1S2 unit cell, identifying four tetrameric assemblies that bury between 1,250 and 1,850 Å per protomer. All four of these tetramers are parallel, so the membrane domains would be on the same plane. These assemblies have not, to our knowledge, been observed in GluR2 S1S2 crystal forms (Fig. 7, which is published as supporting information on the PNAS web site). Two of these were formed from dimers of AMPA-like dimers. A dimer of dimers assembly (27) for iGluRs is now supported both by electrophysiological data identifying functional 2-fold symmetry (11, 34) and a recently described structure for GluR2 determined by electron microscopy (35). Despite this, it is still generally thought that, from analogy with potassium channels, the pore will be ≈4-fold, distorted to a greater or lesser extent. A mechanism of ligand-mediated regulation by a conversion between dimer to tetramer has been proposed for a KTN (K transporter's nucleotide-binding) domain-containing potassium transporter (36). However, the physiological relevance of the tetramers we have described here, as well as the exact topological basis for the potential symmetry mismatch between the ligand-binding and pore domains of intact channels in the cell membrane, awaits further structural, physiological, and biochemical delineation.
Supplementary Material
Acknowledgments
We are grateful for the help and advice received from the members of the Heinemann and Choe laboratories during the course of this project and to the staff at the Stanford Linear Accelerator Center. We also thank Geoff Swanson for critical reading of the manuscript. This work was supported in part by funding from American Heart Association and National Institutes of Health graduate student fellowships (to M.H.N.), a Klingenstein Foundation Fellowship, The Royal Society, and the U.K. Medical Research Council (to T.G.), the U.S.–Israel Binational Science Foundation and the Israel Science Foundation (to Y.S.-B.), and the National Institutes of Health (to S.F.H. and S.C.).
Abbreviation: iGluR, ionotropic glutamate receptor.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1YAE).
References
- 1.Dingledine, R., Borges, K., Bowie, D. & Traynelis, S. F. (1999) Pharmacol. Rev. 51, 7–61. [PubMed] [Google Scholar]
- 2.Huettner, J. E. (2003) Prog. Neurobiol. 70, 387–407. [DOI] [PubMed] [Google Scholar]
- 3.Stern-Bach, Y., Bettler, B., Hartley, M., Sheppard, P. O., O'Hara, P. J. & Heinemann, S. F. (1994) Neuron 13, 1345–1357. [DOI] [PubMed] [Google Scholar]
- 4.Kuusinen, A., Arvola, M. & Keinänen, K. (1995) EMBO J. 14, 6327–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong, N., Sun, Y., Chen, G.-Q. & Gouaux, E. (1998) Nature 395, 913–917. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong, N. & Gouaux, E. (2000) Neuron 28, 165–181. [DOI] [PubMed] [Google Scholar]
- 7.Hogner, A., Kastrup, J. S., Jin, R., Liljefors, T., Mayer, M. L., Egebjerg, J., Larsen, I. K. & Gouaux, E. (2002) J. Mol. Biol. 322, 93–109. [DOI] [PubMed] [Google Scholar]
- 8.Furukawa, H. & Gouaux, E. (2003) EMBO J. 22, 2873–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleck, M. W., Cornell, E. & Mah, S. J. (2003) J. Neurosci. 23, 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paternain, A. V., Cohen, A., Stern-Bach, Y. & Lerma, J. (2003) J. Neurosci. 23, 8641–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowie, D. & Lange, G. D. (2002) J. Neurosci. 22, 3392–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowie, D. (2002) J. Physiol. 539, 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swanson, G. T., Gereau IV, R. W., Green, T. & Heinemann, S. F. (1997) Neuron 19, 913–926. [DOI] [PubMed] [Google Scholar]
- 14.Kabsch, W. (1976) Acta Crystallogr. A 32, 922–923. [Google Scholar]
- 15.Storoni, L. C., McCoy, A. J. & Read, R. J. (2004) Acta Crystallogr. D 60, 432–438. [DOI] [PubMed] [Google Scholar]
- 16.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 17.Reid, R. J. (1986) Acta Crystallogr. A 42, 140–149. [Google Scholar]
- 18.Terwilliger, T. C. (2002) Acta Crystallogr. D 58, 2082–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider, T. R. (2000) Acta Crystallogr. D 56 (Pt 6), 714–721. [DOI] [PubMed] [Google Scholar]
- 20.Emsley, P. & Cowtan, K. (2004) Acta Crystallogr. D 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 21.Akabas, M. H., Stauffer, D. A., Xu, M. & Karlin, A. (1992) Science 258, 307–310. [DOI] [PubMed] [Google Scholar]
- 22.Hayward, S. & Lee, R. A. (2002) J. Mol. Graphics Model 21, 181–183. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe, S., Kokuho, T., Takahashi, H., Takahashi, M., Kubota, T. & Inumaru, S. (2002) J. Biol. Chem. 277, 5090–5093. [DOI] [PubMed] [Google Scholar]
- 24.Jin, R. & Gouaux, E. (2003) Biochemistry 42, 5201–5213. [DOI] [PubMed] [Google Scholar]
- 25.Jin, R., Banke, T. G., Mayer, M. L., Traynelis, S. F. & Gouaux, E. (2003) Nat. Neurosci. 6, 803–810. [DOI] [PubMed] [Google Scholar]
- 26.Lunn, M. L., Hogner, A., Stensbol, T. B., Gouaux, E., Egebjerg, J. & Kastrup, J. S. (2003) J. Med. Chem. 46, 872–875. [DOI] [PubMed] [Google Scholar]
- 27.Madden, D. R. (2002) Nat. Rev. Neurosci. 3, 91–101. [DOI] [PubMed] [Google Scholar]
- 28.Carcache, L. M., Rodriguez, J. & Rein, K. S. (2003) Bioorg. Med. Chem. 11, 551–559. [DOI] [PubMed] [Google Scholar]
- 29.Swanson, G. T., Green, T., Sakai, R., Contractor, A., Che, W., Kamiya, H. & Heinemann, S. F. (2002) Neuron 34, 589–598. [DOI] [PubMed] [Google Scholar]
- 30.Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J. R., Kuner, T., Monyer, H., Higuchi, M., Bach, A. & Seeburg, P. H. (1994) Science 266, 1709–1713. [DOI] [PubMed] [Google Scholar]
- 31.Sun, Y., Olson, R., Horning, M., Armstrong, N., Mayer, M. & Gouaux, E. (2002) Nature 417, 245–253. [DOI] [PubMed] [Google Scholar]
- 32.Stern-Bach, Y., Russo, S., Neuman, M. & Rosenmund, C. (1998) Neuron 21, 907–918. [DOI] [PubMed] [Google Scholar]
- 33.Horning, M. S. & Mayer, M. L. (2004) Neuron 41, 379–388. [DOI] [PubMed] [Google Scholar]
- 34.Sobolevsky, A. I., Yelshansky, M. V. & Wollmuth, L. P. (2004) Neuron 41, 367–378. [DOI] [PubMed] [Google Scholar]
- 35.Tichelaar, W., Safferling, M., Keinanen, K., Stark, H. & Madden, D. R. (2004) J. Mol. Biol. 344, 435–442. [DOI] [PubMed] [Google Scholar]
- 36.Roosild, T. P., Miller, S., Booth, I. R. & Choe, S. (2002) Cell 109, 781–791. [DOI] [PubMed] [Google Scholar]
- 37.Wallace, A. C., Laskowski, R. A. & Thornton, J. M. (1995) Protein Eng. 8, 127–134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.