Unique among animal viruses, retroviral replication proceeds through an obligate recombination step with host cell DNA. Although this step makes it extremely difficult to eradicate a retrovirus from a cell, it also confers a desirable trait to gene therapy vectors. Yet stable integration can yield unwanted side effects, as highlighted in a recent gene therapy trial in which 2 of 11 patients developed leukemia-like illness because of the integration of a murine leukemia virus (MLV)-based vector in the vicinity of the LMO2 protooncogene (1). Deciphering the mechanism(s) of integration site selection not only will shed light on a basic step(s) in retroviral biology but also holds promise for the design of more effective vectors. Although different cloning strategies have indicated an overall preference for transcription units, the mechanism of retroviral integration site selection is for the most part unknown. In this issue of PNAS, Maxfield et al. (2) assayed RAV-1 (a subgroup A Alpharetrovirus) integration into the quail metallothionein (MT) gene with and without transcriptional activation. A 100-fold increase in gene expression reduced integration 6-fold. Taken alongside a previous study that demonstrated that more modest levels of induction also inhibited integration (3), the active transcription complex appears unlikely to help guide retroviruses to their sites of integration.
Retroviruses encode their own integrase (IN) protein that is carried into the cell as part of the virus. After reverse transcription the viral cDNA, IN and other proteins comprise an integration-competent nucleoprotein complex known as the preintegration complex (PIC) (reviewed in ref. 4). Early studies indicated that transcriptionally active and/or DNase I-hypersensitive regions were preferred sites, but it was unclear to what extent relatively small sample size and the selective outgrowth of certain integrants before cloning may have influenced those results (see ref. 5 for review). With this in mind, Withers-Ward et al. (6) developed a PCR strategy to monitor relatively large numbers of integrations without the need for cloning. These results indicated a fairly global access of avian DNA by avian leukosis-sarcoma virus (ALV) as well as local hotspots that were used as much as 280-fold over random. Because integration into defined nucleoprotein complexes or DNA structures in vitro indicated preference for distorted DNA (7-9), it is possible that protein-induced DNA distortion contributed to the local hotspots observed in ALV-infected cells.
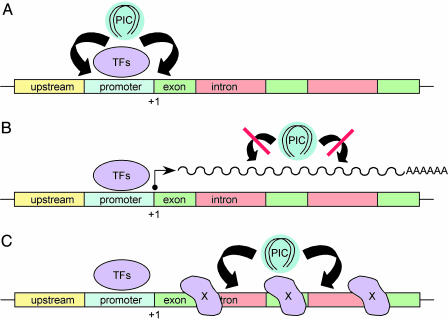
Scaled-up cloning and sequencing of HIV type 1 (HIV-1), MLV, and ALV integration sites has since afforded a human genome-wide view of target site selection. Although exact numbers have differed between studies, results can be summarized as follows. Whereas ALV displayed a slight preference for genes, transcriptional activity did not appear to play a role in site selection (10). Although MLV, like ALV, displayed a slight preference for genes, in this case regions adjacent to transcriptional start sites were highly preferred over downstream gene regions (11). Considering this preference within and nearby promoters, one can envision a connection between MLV integration and host cell transcription (Fig. 1A). In contrast to ALV and MLV, HIV-1 displayed a strong preference for integration inside genes, and more active genes were in general targeted over less active genes (12, 13). Given the propensity to integrate into active genes, one hypothesis was that PICs targeted active transcription (12) (Fig. 1B).
Fig. 1.
Models for promoter/gene targeting during retroviral integration. (A) TFs bound to the promoter region of a hypothetical gene tether PICs for integration into nearby sequences. This general scenario applies to Ty3, where TFIIIB and TFIIIC help target retrotransposition (14). Because MLV preferred promoters and 5′ ends of coding regions (11), such a scheme may apply for this virus. (B) HIV-1 preferred genes to a greater extent than ALV, but both integrated into genes more frequently than nongene regions. In contrast to MLV, integration occurred fairly evenly along genes, which might occur by means of an affinity for the transcription complex itself. The work by Maxfield et al. (2) discounts this model, at least for ALV. (C) Alternatively, PICs might target gene regions by association with an unknown chromatin factor(s) X.
Precedence for a connection between retroelement integration and transcription comes from studies of close relatives of retroviruses, yeast retrotransposons. Retrotransposons share many lifestyle features with their viral cousins, including protease-dependent assembly of ribonucleoprotein complexes [termed virus-like particles (VLPs)], reverse transcription, and IN-mediated integration. The main difference is that, whereas viruses exit from cells to initiate new rounds of infection, VLPs remain intracellular and, after reverse transcription, reintegrate into the same genome. Especially considering a haploid stage during mating, yeast must carefully control retrotransposition to avoid disrupting essential genes. For Ty1 and Ty3, this control is accomplished in part by targeting integration into relatively benign regions upstream of RNA polymerase III-transcribed genes. In the case of Ty3, interactions between transcription factor (TF)IIIB and TFIIIC help mediate site-specific integration (see ref. 14 for review). Not all retrotransposons, however, use TFs for targeting. For example, an interaction between Ty5 IN and Sir4p, a host component of heterochromatin, mediates integration at telomeres and mating loci (15). Despite these mechanistic differences, prebound host factors can be thought of as attracting or tethering PICs/VLPs to specific sites for integration (Fig. 1 A and C).
Maxfield et al. (2) examined the frequency and distribution of RAV-1 integration along the quail MT gene plus/minus transcriptional activation. The avian gene was chosen for three main reasons. First, the previous study that quantified ALV integration as a function of gene expression used an artificially engineered construct introduced into cells by stable transfection (3), so it was of interest to now analyze an endogenous gene. Second, the MT gene supports a low level of constitutive expression that is rapidly induced by the addition of Zn2+ to cell culture media, and preliminary experiments revealed that Zn2+ did not influence overall integration levels (2). Third, whereas mammalian MT genes occur in multiple isoforms, the avian gene is single-copy and, thus, greatly simplifies data interpretation. Initial experiments revealed that transcriptional up-regulation persisted for at least 48 h, a length of time sufficient for integration to occur. Without induction, approximately one provirus was detected along a 500-bp stretch of the gene, a frequency corresponding to random integration. The experiment was repeated several times, yielding a total of 47 events, 5 of which were duplicate hotspots. Yet during side-by-side comparison, the same number of repetitions yielded only eight integrations in the presence of Zn2+, leading to an overall 6-fold reduction in integration (2). Because Coffin and coworkers (3) established that an ≈5- to 7-fold increase in transcription decreased integration into coding-region DNA by ≈25-40%, active transcription has suppressed gene-specific integration over a range of expression levels. Transcription-based inhibition could be due to steric hindrance via the RNA polymerase II complex, DNA duplex separation, and/or chromatin remodeling to a structure disfavoring integration (2).
Active transcription has suppressed gene-specific integration over a range of expression levels.
Given the observed inhibition, it is tempting to speculate on the mechanism of integration site selection. One possibility is that genes in general and active genes in particular are more accessible for PICs because of favorable or common nuclear environments. Yet if DNA accessibility was the only factor, one might predict different retroviruses to display near-identical patterns of gene usage in the same cell type, which has not been observed. Because MLV preferred promoter-proximal regions, TFs or other factors involved in transcription initiation might be in play (Fig. 1A). Because ALV and HIV-1 targeted genes fairly evenly along their lengths, proteins other than initiation factors are likely to be involved (Fig. 1C). Because ALV has only been profiled in human cells, where certain virus-host interactions might fail to operate, the remainder of this discussion will focus on lentiviruses. One candidate host factor for HIV-1 targeting is IN-interacting protein 1 (INI1) (16), a component of the SWI/SNF chromatin-remodeling machine (reviewed in refs. 4 and 5). A recent study, however, revealed that simian immunodeficiency virus (SIV)-based vectors favored rhesus genes similar to the preference observed for HIV-1 in human cells (17). Because INI1 is highly specific for HIV-1 and failed to interact with SIV IN (18), a role for INI1 in lentiviral targeting appears unlikely. Another particularly attractive IN-interacting protein, lens epithelium-derived growth factor/transcriptional coactivator p75 (LEDGF/p75), appears to be lentiviral-specific (19) and thus might play a role in site selection (4, 20). Additional experiments are required to address this hypothesis and to decipher the mechanism(s) at play for other retroviruses. In the long run, the results of these studies will hopefully lead to the design of safer gene therapy vectors.
See companion article on page 1436.
References
- 1.Hacein-Bey-Abina, S., Von Kalle, C., Schmidt, M., McCormack, M. P., Wulffraat, N., Leboulch, P., Lim, A., Osborne, C. S., Pawliuk, R., Morillon, E., et al. (2003) Science 302, 415-419. [DOI] [PubMed] [Google Scholar]
- 2.Maxfield, L. F., Fraize, C. D. & Coffin, J. M. (2005) Proc. Natl. Acad. Sci. USA 102, 1436-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weidhaas, J. B., Angelichio, E. L., Fenner, S. & Coffin, J. M. (2000) J. Virol. 74, 8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turlure, F., Devroe, E., Silver, P. A. & Engelman, A. (2004) Front. Biosci. 9, 3187-3208. [DOI] [PubMed] [Google Scholar]
- 5.Engelman, A. (2003) Curr. Top. Microbiol. Immunol. 281, 209-238. [DOI] [PubMed] [Google Scholar]
- 6.Withers-Ward, E. S., Kitamura, Y., Barnes, J. P. & Coffin, J. M. (1994) Genes Dev. 8, 1473-1487. [DOI] [PubMed] [Google Scholar]
- 7.Pryciak, P. M. & Varmus, H. E. (1992) Cell 69, 769-780. [DOI] [PubMed] [Google Scholar]
- 8.Pruss, D., Bushman, F. & Wolffe, A. (1994) Proc. Natl. Acad. Sci. USA 91, 5913-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katz, R. A., Gravuer, K. & Skalka, A. M. (1998) J. Biol. Chem. 273, 24190-24195. [DOI] [PubMed] [Google Scholar]
- 10.Narezkina, A., Taganov, K. D., Litwin, S., Stoyanova, R., Hayashi, J., Seeger, C., Skalka, A. M. & Katz, R. A. (2004) J. Virol. 78, 11656-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu, X., Li, Y., Crise, B. & Burgess, S. M. (2003) Science 300, 1749-1751. [DOI] [PubMed] [Google Scholar]
- 12.Schroder, A. R. W., Shinn, P., Chen, H., Berry, C., Ecker, J. R. & Bushman, F. (2002) Cell 110, 521-529. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell, R. S., Beitzel, B. F., Schroder, A. R. W., Shinn, P., Chen, H., Berry, C. C., Ecker, J. R. & Bushman, F. D. (2004) PLoS Biology 2, e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandmeyer, S. (2003) Proc. Natl. Acad. Sci. USA 100, 5586-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie, W., Gai, X., Zhu, Y., Zappulla, D. C., Sternglanz, R. & Voytas, D. F. (2001) Mol. Cell. Biol. 21, 6606-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalpana, G. V., Marmon, S., Wang, W., Crabtree, G. R. & Goff, S. P. (1994) Science 266, 2002-2006. [DOI] [PubMed] [Google Scholar]
- 17.Hematti, P., Hong, B.-K., Ferguson, C., Adler, R., Hanawa, H., Sellers, S., Holt, I. E., Eckfeldt, C. E., Sharma, Y., Schmidt, M., et al. (2004) PLoS Biology 2, e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yung, E., Sorin, M., Wang, E.-J., Perumal, S., Ott, D. & Kalpana, G. V. (2004) J. Virol. 78, 2222-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llano, M., Vanegas, M., Fregoso, O., Saenz, D., Chung, S., Peretz, M. & Poeschla, E. M. (2004) J. Virol. 78, 9524-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cherepanov, P., Maertens, G., Proost, P., Devreese, B., Van Beeumen, J., Engelborghs, Y., De Clercq, E. & Debyser, Z. (2003) J. Biol. Chem. 278, 372-381. [DOI] [PubMed] [Google Scholar]