ABSTRACT
The Gram-positive actinobacteria Actinomyces spp. are key colonizers in the development of oral biofilms due to the inherent ability of Actinomyces to adhere to receptor polysaccharides on the surface of oral streptococci and host cells. This receptor-dependent bacterial interaction, or coaggregation, requires a unique sortase-catalyzed pilus consisting of the pilus shaft FimA and the coaggregation factor CafA forming the pilus tip. While the essential role of the sortase machine SrtC2 in pilus assembly, biofilm formation, and coaggregation has been established, little is known about trans-acting factors contributing to these processes. We report here a large-scale Tn5 transposon screen for mutants defective in Actinomyces oris coaggregation with Streptococcus oralis. We obtained 33 independent clones, 13 of which completely failed to aggregate with S. oralis, and the remainder of which exhibited a range of phenotypes from severely to weakly defective coaggregation. The former had Tn5 insertions in fimA, cafA, or srtC2, as expected; the latter were mapped to genes coding for uncharacterized proteins and various nuo genes encoding the NADH dehydrogenase subunits. Electron microscopy and biochemical analyses of mutants with nonpolar deletions of nuo genes and ubiE, a menaquinone C-methyltransferase-encoding gene downstream of the nuo locus, confirmed the pilus and coaggregation defects. Both nuoA and ubiE mutants were defective in oxidation of MdbA, the major oxidoreductase required for oxidative folding of pilus proteins. Furthermore, supplementation of the ubiE mutant with exogenous menaquinone-4 rescued the cell growth and pilus defects. Altogether, we propose that the A. oris electron transport chain is biochemically linked to pilus assembly via oxidative protein folding.
KEYWORDS: actinobacteria, Actinomyces, Mycobacterium, coaggregation, disulfide bond, oxidoreductases, pilus assembly, protein folding, sortase
IMPORTANCE
The Gram-positive actinobacterium A. oris expresses adhesive pili, or fimbriae, that are essential to biofilm formation and Actinomyces interactions with other bacteria, termed coaggregation. While the critical role of the conserved sortase machine in pilus assembly and the disulfide bond-forming catalyst MdbA in oxidative folding of pilins has been established, little is known about other trans-acting factors involved in these processes. Using a Tn5 transposon screen for mutants defective in coaggregation with Streptococcus oralis, we found that genetic disruption of the NADH dehydrogenase and menaquinone biosynthesis detrimentally alters pilus assembly. Further biochemical characterizations determined that menaquinone is important for reactivation of MdbA. This study supports the notion that the electron transport chain is biochemically linked to pilus assembly in A. oris via oxidative folding of pilin precursors.
INTRODUCTION
Found only in Gram-positive bacteria, such as Actinomyces spp., Corynebacterium diphtheriae, Bacillus cereus, streptococci, and enterococci, covalently linked pili, also termed fimbriae, are important virulence determinants (1, 2). These adhesive pilus structures are assembled and linked to bacterial peptidoglycan by conserved transpeptidase enzymes collectively named sortases (3). Initially reported in C. diphtheriae (1, 4), sortase enzymes that polymerize pilin subunits into covalently linked pilus polymers are often called pilus-specific sortases, or class C sortases (3, 5). Generally, the housekeeping sortase and nonpolymerizing sortase enzymes anchor pilus polymers to the bacterial cell wall (6–10) via a process similar to cell wall anchoring of surface proteins by the archetype SrtA of Staphylococcus aureus (11, 12).
Pili were discovered in many species of Actinomyces in the 1970s (13, 14), and early pilus characterizations focused on the fimbriae of Actinomyces naeslundii genospecies 2 (15), which was later renamed Actinomyces oris (16). In the biofilm-forming actinobacterium A. oris, two types of fimbriae have been identified. Type 1 fimbriae—consisting of the pilus shaft FimP and the tip pilin FimQ (17)—mediate bacterial binding to salivary proline-rich proteins on the tooth surface (18). SrtC1, the pilus-specific sortase encoded by the type 1 fimbrial gene locus, specifically catalyzes pilus polymerization of FimP and FimQ (17, 19). Type 2 fimbriae, with SrtC2 as their pilus-specific sortase (19, 20), are made of the pilus shaft FimA and the canonical tip pilin FimB (20). Biofilm formation and Actinomyces interactions with oral streptococci in biofilm, termed coaggregation (15), are well-documented phenotypes associated with the type 2 fimbriae since a fimA mutant fails to mediate biofilm formation and bacterial coaggregation (20, 21). Nonetheless, it has recently been discovered that a coaggregation factor named CafA hijacks the sortase SrtC2 machine, forming a distinct pilus tip with the pilus shaft FimA independent of FimB (22). It is now clear that CafA is the major coaggregation factor of A. oris as deletion of cafA results in the same coaggregation defect as fimA deletion; additionally, a specific antibody against recombinant CafA or addition of this protein blocks bacterial coaggregation (22). Thus, the coaggregation defect of the fimA mutant can be attributed to the loss of CafA pilus assembly on the bacterial cell wall, while the role of FimB in oral colonization and biofilm formation remains unknown.
Consistent with the above findings, in a previous small-scale Tn5 transposon screen intended to identify A. oris mutants that fail to aggregate with Streptococcus oralis, we found 3 mutants with Tn5 insertions in fimA and srtC2; the fourth mutant was mapped to vkor, which encodes a bacterial vitamin K epoxide reductase (VKOR) (23). Importantly, an in-frame, nonpolar vkor deletion mutant was shown to be severely defective in pilus assembly. Further characterizations established that VKOR is required for reoxidation of the thiol-disulfide oxidoreductase MdbA (23). It is noteworthy that a Mycobacterium tuberculosis homolog of A. oris VKOR was initially shown to replace DsbB in reoxidation of the disulfide bond forming oxidoreductase DsbA when expressed in Escherichia coli (24). Unlike E. coli dsbA, mdbA is an essential gene, and a conditional mdbA deletion mutant failed to assemble adhesive pili (23). It was shown that MdbA and VKOR together form the thiol-disulfide oxidoreductase pair that catalyzes oxidative protein folding in A. oris (25). Thus, based on available evidence centered on the type 2 fimbriae, a model of pilus assembly in A. oris has been proposed (22, 23) (Fig. 1). Synthesized in the cytoplasm, pilin precursors, such as FimA, FimB, and CafA, are transported across the cytoplasmic membrane by the Sec translocon in an unfolded state. The thiol-disulfide oxidoreductase MdbA catalyzes oxidative folding of nascent pilin precursors before they are embedded into the membrane. Membrane-bound pilin precursors are polymerized by the pilus-specific sortase SrtC2, resulting in formation of type 2 fimbriae with two distinct tip pilins, FimB and CafA. The resulting polymers are presumably anchored by the housekeeping sortase SrtA. While it is clear in this model that the sortase and oxidative folding machines are central elements of pilus assembly in A. oris, it is unknown if there are additional factors directly or indirectly involved in the pilus assembly process.
FIG 1 .
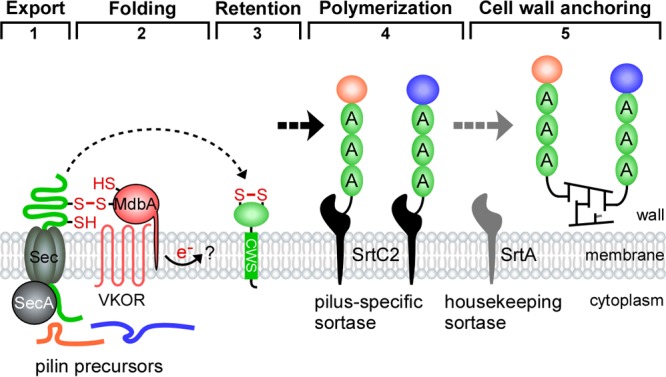
A model of pilus assembly in A. oris. Presented is a simplified model of pilus assembly in A. oris that is centered on the type 2 fimbriae (see the text for details); FimA, FimB, and CafA are colored green, orange, and blue, respectively. The thiol-disulfide oxidoreductase MdbA catalyzes oxidative folding of nascent protein precursors as they are translocated into the exoplasm by the Sec machinery. Reoxidation of MdbA requires the membrane-bound oxidoreductase VKOR. Folded pilin precursors are polymerized and anchored to the cell wall by the tandem sortase enzymes. It is not known how electrons generated from reoxidation of MdbA/VKOR are transferred (question mark). Dashed arrows denote potential multiple steps (adapted from data in references 22 and 23).
Because A. oris coaggregation and pilus assembly are tightly associated, we exploited this property in a high-throughput assay to screen more than 6,200 Tn5 transposon mutants for clones that are variably defective in coaggregation with S. oralis So34, an indicator strain expressing pilus receptor cell-surface polysaccharides (RPS) (20, 21, 26). Using a modified visual coaggregation scoring system, we were able to identify auxiliary factors contributing to pilus assembly that might have been missed in the previously reported small-scale screen, which was solely based on positive or negative coaggregation phenotype scoring (23). By characterizing a subset of these coaggregation-defective mutants, we demonstrate here that NADH dehydrogenase and menaquinone, components of the electron transport chain (ETC), are involved in reoxidation of the major disulfide bond-forming machine MdbA/VKOR in A. oris.
RESULTS
A Tn5 transposon screen revealed A. oris mutants defective in polymicrobial interactions.
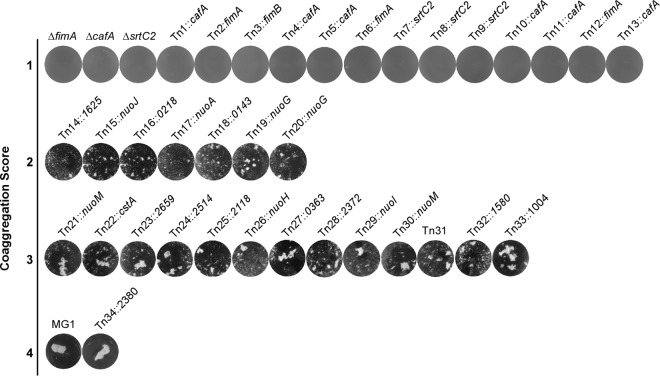
Since pilus assembly and bacterial coaggregation in A. oris are coupled (20, 22), we sought to identify trans-acting factors involved in pilus assembly by screening a large number of A. oris Tn5 mutants using a previously reported coaggregation assay in a 96-well plate format (23). Following a published protocol (23, 27), a Tn5 transposon library of roughly 6,240 A. oris mutants (>3-fold genome coverage) was constructed and screened. The coaggregation efficiency of the Tn5 mutants was scored largely based on the visual scoring system reported by Cisar et al. (28), whereby the coaggregation-positive phenotype of the parental strain MG1 was designated 4 and the coaggregation-negative phenotype of type 2 fimbria-less mutants (i.e., ΔfimA, ΔcafA, and ΔsrtC2) was set as 1; tiny clumps of aggregates were scored as 2, whereas the larger clumps were scored as 3. We obtained 13 mutants with the coaggregation score (CS) of 1 (CS-1), 7 mutants with the CS of 2 (CS-2), and 13 mutants with CS-3; of note, the Tn34 mutant exhibiting a coaggregation phenotype similar to that of MG1 was included for comparison (Fig. 2). These coaggregation-defective Tn5 mutants were then subjected to mapping by thermal asymmetric interlaced PCR (TAIL-PCR) and DNA sequencing as previously described (23).
FIG 2 .
Identification of A. oris coaggregation-defective mutants by Tn5 transposon mutagenesis. Thirty-three A. oris coaggregation-defective Tn5 mutants identified by a cell-based screen were confirmed by a standard coaggregation assay (50). Equal cell numbers of A. oris mutants and S. oralis So34 were mixed together, and coaggregation was imaged using an AlphaImager. Coaggregation scores indicate the degrees of coaggregation, with the phenotype of the parental MG1 strain scored as 4 and that of the fimA, cafA, and srtC2 deletion mutants scored as 1; scores of 2 and 3 represent small and larger clumps of bacterial aggregates. Tn5 target genes were mapped by TAIL-PCR.
As expected, 13 coaggregation-negative mutants (i.e., the CS-1 strains) were mapped to genes encoding CafA, FimA, and SrtC2, which are known fimbrial factors essential for A. oris coaggregation with oral streptococci (20–22). Intriguingly, 4 out of 7 CS-2 mutants contained Tn5 insertions in the nuoA, nuoJ, and nuoG genes; the remainder mapped to genes (i.e., ANA_1625, ANA_0218, and ANA_0143) that code for geranylgeranyl reductase (GGR), a putative metal-binding protein, and β-glucosidase, respectively. Finally, the CS-3 mutants were mapped to genes encoding many hypothetical proteins, a carbon starvation protein A (CstA), a two-component system sensor kinase, an ABC-2-type transporter, and the H, I, M subunits of NADH dehydrogenase (i.e., NuoH, NuoI, and NuoM) (Fig. 2; Table 1). Since a large number of the nuo genes were repeatedly targeted by the Tn5 transposon, they were further characterized and reported in this study; the remaining candidates will be investigated in future studies.
TABLE 1 .
Mapping of A. oris coaggregation-defective Tn5 mutants
| Tn5 mutant | Genomic Tn5 positiona | Target gene | Predicted function |
|---|---|---|---|
| 1 | 2422225 | cafA | Coaggregation factor A |
| 2 | 38189 | fimA | Type 2 fimbrial shaft pilin |
| 3 | 36541 | fimB | Type 2 fimbrial tip pilin |
| 4 | 2420317 | cafA | Coaggregation factor A |
| 5 | 2421799 | cafA | Coaggregation factor A |
| 6 | 38174 | fimA | Type 2 fimbrial shaft pilin |
| 7 | 39115 | srtC2 | Type 2 pilus-specific sortase |
| 8 | 39481 | srtC2 | Type 2 pilus-specific sortase |
| 9 | 39749 | srtC2 | Type 2 pilus-specific sortase |
| 10 | 2421087 | cafA | Coaggregation factor A |
| 11 | 2421136 | cafA | Coaggregation factor A |
| 12 | 38721 | fimA | Type 2 fimbrial tip pilin |
| 13 | 2420135 | cafA | Coaggregation factor A |
| 14 | 1755107 | ana_1625 | Geranylgeranyl reductase |
| 15 | 1744569 | nuoJ | NADH dehydrogenase I, subunit J |
| 16 | 230679 | ana_0218 | Zn2+/Mn2+ transport system substrate-binding protein |
| 17 | 1754944 | nuoA | NADH dehydrogenase I, subunit A |
| 18 | 162115 | ana_0143 | β-d-Glucoside glucohydrolase |
| 19 | 1747764 | nuoG | NADH dehydrogenase I, subunit G |
| 20 | 1748226 | nuoG | NADH dehydrogenase I, subunit G |
| 21 | 1741097 | nuoM | NADH dehydrogenase, subunit M |
| 22 | 2348384 | cstA | Carbon starvation protein A |
| 23 | 2863369 | ana_2659 | Ni/Fe-hydrogenase III large subunit |
| 24 | 2711713 | ana_2514 | Histidine kinase (2-component system) |
| 25 | 2295587 | ana_2118 | Permease component of ABC-type multidrug transport system |
| 26 | 1746526 | nuoH | NADH dehydrogenase I, subunit H |
| 27 | 382186 | ana_0363 | Conserved hypothetical protein |
| 28 | 2566963 | ana_2372 | Hypothetical protein with Ser/Arg repeats |
| 29 | 1745086 | nuoI | NADH dehydrogenase I, subunit I |
| 30 | 1739922 | nuoM | NADH dehydrogenase I, subunit M |
| 31 | 2513949 | ana_2325; ana_2326b | Ana_2325 (Cys/His-dependent amidohydrolase/peptidase); Ana_2326 (hypothetical protein) |
| 32 | 1712411 | ana_1580 | Conserved hypothetical protein |
| 33 | 1080892 | ana_1004 | Hypothetical protein |
| 34 | 2576712 | ana_2380 | Conserved hypothetical protein |
Shown is the nucleotide position of the Tn5 transposon mapped in the genome.
The Tn5 insertion was found within the intergenic region of ana_2325 and ana_2326.
Genetic disruption of the A. oris NADH dehydrogenase (complex I) subunits caused significant defects in CafA-mediated coaggregation and CafA pilus assembly.
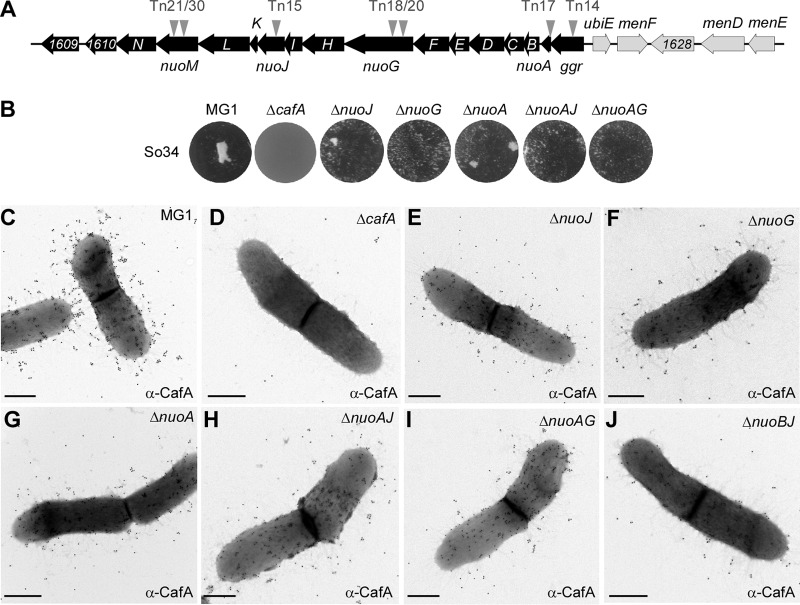
The ggr, nuoA, nuoG, nuoH, nuoJ, and nuoM genes revealed from the Tn5 screen presented above are part of the nuo gene locus in A. oris MG1 (http://genome.brop.org/), which are predicted to encode the NADH dehydrogenase enzyme—often referred to as the NADH:ubiquinone oxidoreductase or respiratory complex I of the ETC (29) (Fig. 3A). To confirm that the coaggregation defects of the Tn5::nuo mutants are not due to polar effects, we generated individual mutants and mutants with combinations of in-frame, nonpolar deletions of nuoJ, nuoG, and nuoA. The generated mutants were confirmed for their inability to aggregate with S. oralis via CafA-mediated coaggregation using a standard coaggregation assay (22), whereby A. oris cells were mixed in equal volume with S. oralis So34 cells and coaggregation was determined after a few minutes of mixing. As shown in Fig. 3B, deletion of nuoJ, nuoG, or nuoA or a combination of nuoA and nuoJ or nuoA and nuoG caused significant coaggregation defects compared to the wild-type level.
FIG 3 .
Involvement of nuo genes in CafA-mediated coaggregation and pilus assembly. (A) The nuo operon and adjacent genes are shown in black, and genes involved in ubiquinone/menaquinone biosynthesis are shown in gray. Arrowheads indicate the locations of the Tn5 transposon. (B) Coaggregation of A. oris strains and S. oralis So34 was performed as described in the legend to Fig. 2. (C to J) A. oris cells were immobilized in nickel-carbon grids and labeled with anti-CafA antibodies, followed by labeling with anti-rabbit IgG antibodies conjugated to 18-nm gold particles. Samples were stained with 1% uranyl acetate and viewed by a transmission electron microscope. Scale bars indicate 0.5 µm.
Since CafA pilus assembly is essential for A. oris coaggregation (22), we next examined whether the observed coaggregation defects are due to pilus assembly defects of CafA by immunoelectron microscopy (IEM). In this assay, A. oris cells were stained with antibodies against CafA (anti-CafA), followed by staining with gold particles conjugated with IgG, and the samples were viewed by transmission electron microscopy (TEM) after staining with 1% uranyl acetate. In the parental strain MG1, CafA signal was found abundantly on the bacterial surface and at the distal end of pili; as expected, the CafA signal was absent in the cafA deletion mutant (22). Importantly, CafA labeling was significantly reduced in the nuoJ, nuoG, nuoA, nuoAJ, nuoAG, and nuoBJ mutants (Fig. 3C to J). Since the CafA assembly defects among these mutants were apparently equal, we chose to focus on the nuoA mutant for further characterizations (see below) as nuoA is typically the first gene in bacterial nuo operons (30). To support that the generated nuoA mutant is nonpolar, we introduced into this mutant a plasmid constitutively expressing nuoA; as shown in Fig. S1 in the supplemental material, ectopic expression of nuoA rescued the assembly defects of both type 1 and type 2 pili of the nuoA mutant. Of note, the defects in pilus assembly and coaggregation might not be due to the cell growth defect as the growth rate of the nuoA mutant was not significantly different from that of the parental strain (see Fig. S3 in the supplemental material). Together, the results confirm the requirement of the NADH dehydrogenase for optimal bacterial coaggregation and CafA pilus assembly.
Requirement of nuoA for pilus assembly. A. oris cells of the indicated strains were immobilized on carbon-coated nickel grids and stained with anti-FimA (A to D), anti-CafA (E to H), or anti-type 1 (I to L) antibodies, followed by staining with IgG conjugated to 18-nm gold particles. Samples were stained with 1% uranyl acetate prior to being analyzed by electron microscopy. Scale bars indicate 0.5 µm. Download FIG S1, PDF file, 2.7 MB (2.8MB, pdf) .
Copyright © 2017 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The menaquinone C-methyltransferase UbiE is involved in A. oris coaggregation, biofilm formation, and pilus assembly.
In E. coli, the NADH dehydrogenase, encoded by 14 nuo genes (nuoA to -N), generates energy by coupling the transfer of electrons from NADH to ubiquinone with the proton translocation across the membrane (29). We observed that upstream of the A. oris nuo locus in the MG1 genome are genes encoding part of the ubiquinone/menaquinone biosynthesis pathway, with the C-methyltransferase encoding-gene ubiE adjacent to ggr (Fig. 3A). In E. coli, UbiE catalyzes the carbon methylation reaction in the biosynthesis of ubiquinone and menaquinone, which are essential components of the ETC (31). This information prompted us to examine whether the ubiquinone/menaquinone biosynthesis pathway is also linked to A. oris coaggregation and pilus assembly. Since ubiE is not an essential gene in E. coli (31), we decided to generate an in-frame, nonpolar deletion mutant of A. oris ubiE. Using published assays, the generated mutant was examined for its ability to aggregate with S. oralis, to mediate biofilm formation, and to produce pili (22).
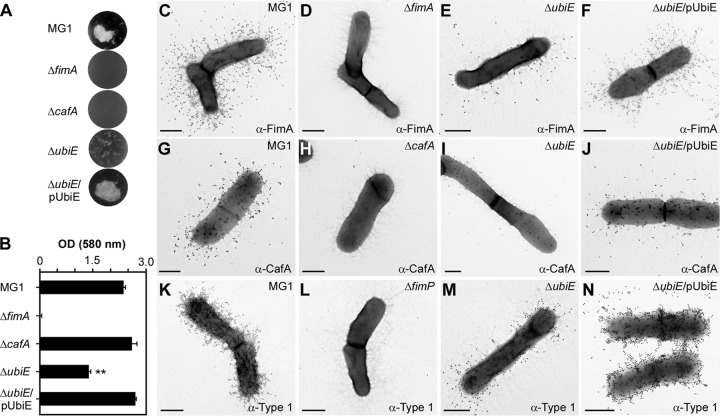
In the coaggregation assay, the ubiE mutant exhibited a severe defect in bacterial coaggregation with S. oralis compared to the parental strain, MG1, and this defect was rescued by ectopic expression of UbiE (Fig. 4A). The same set of strains was then examined in biofilm formation assays, in which A. oris biofilms were cultivated in heart infusion broth (HIB) supplemented with 1% sucrose, followed by biofilm staining with 1% crystal violet and quantification by the optical density at 580 nm (OD580). Consistent with the results described above, deletion of ubiE significantly reduced biofilm formation compared to the wild-type level, and expression of ubiE in trans restored biofilm formation to the wild-type level (Fig. 4B).
FIG 4 .
Requirement of ubiE for bacterial coaggregation, biofilm formation, and pilus assembly. (A) Coaggregation of A. oris strains and S. oralis So34 was determined as described in the legend to Fig. 2. (B) Biofilms were obtained by growing the indicated strains in HIB containing 1% sucrose for 48 h. Harvested biofilms were subjected to crystal violet staining and optical density measurement at 580 nm using a microplate reader. The results are shown as representatives of 3 independent experiments performed in triplicate. **, P < 0.0025, calculated using a one-way ANOVA (Duncan’s method, nonparametric) with GraphPad Prism. (C to N) A. oris cells were immobilized in nickel grids and stained with anti-FimA (C to F), anti-CafA (G to J), or anti-type 1 (K to N) antibodies, followed by staining with IgG conjugated to 18-nm gold particles. Samples were stained with 1% uranyl acetate prior to being analyzed by electron microscopy. Scale bars indicate 0.5 µm.
To examine if the coaggregation and biofilm defects are associated with pilus assembly, we employed IEM using anti-CafA and antibodies against the type 2 fimbrial shaft FimA (anti-FimA). Compared to the MG1 strain, which produced abundant signals of FimA and CafA, FimA and CafA detection was drastically reduced in the ubiE mutant; these defects were rescued in the ubiE mutant expressing UbiE from a plasmid (Fig. 4C to J; see Fig. S2 in the supplemental material). Finally, to determine if the role of UbiE also extends to the type 1 fimbriae, we used antibodies against type 1 fimbriae (32) (anti-type 1) in IEM experiments. Consistently, the type 1 fimbrial signal was significantly reduced in the ubiE mutant compared to the parental MG1 and UbiE complementing strains (Fig. 4K to N). Altogether, these results suggest that the defects of ubiE deletion in bacterial coaggregation and biofilm formation are directly linked to assembly deficiencies of adhesive fimbriae, which are the major factors required for the aforementioned processes in A. oris (20, 22).
Requirement of nuoA and ubiE for surface expression of CafA. The expression of CafA on the cell surface was determined by whole-cell ELISA using polyclonal anti-CafA antibodies. The absorbance measurements at 450 nm, compared to the cafA mutant as background, were determined from three independent experiments performed in triplicate. Error bars represent standard deviations. * and *** indicate P < 0.05 and P < 0.001, respectively, determined using the unpaired, two-tailed t test with GraphPad Prism. Download FIG S2, PDF file, 0.1 MB (80KB, pdf) .
Copyright © 2017 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Generation times of the A. oris MG1 and ΔnuoA mutant strains. Growth of the wild-type (WT) MG1 strain and ΔnuoA and ΔnuoA/pNuoA mutant strains was measured by optical density (OD600). Generation times were calculated as described in Materials and Methods. The results are representative of three independent experiments performed in triplicate. Error bars represent standard deviations. ns, not significant. Download FIG S3, PDF file, 0.1 MB (60KB, pdf) .
Copyright © 2017 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Requirement of the menaquinone C-methyltransferase UbiE and NADH dehydrogenase subunit NuoA in reoxidation of the major thiol-disulfide oxidoreductase MdbA.
As mentioned above, disulfide bond formation is required for correct folding of pilin precursors, and this process is catalyzed by the major thiol-disulfide oxidoreductase MdbA in A. oris (23). Given that UbiE and several subunits of the NADH dehydrogenase are associated with pilus assembly, as presented above, we hypothesized that the pilus assembly defects in the ubiE and nuo mutants might be due to aberrant oxidative folding of pilus proteins. Reactivation of MdbA normally requires the membrane-bound oxidoreductase VKOR, but overexpression of MdbA in a vkor mutant can compensate for the loss of VKOR (23). Here, we examined if ectopic expression of MdbA in the absence of UbiE or Nuo subunits would rescue the pilus assembly defects by introducing a recombinant plasmid that constitutively expresses MdbA into the ubiE and nuoA mutants. The resulting strains, along with the mutants, were examined for their ability to assemble pili by IEM with anti-FimA. Compared to the parental MG1 strain, the ubiE mutant produced significantly less FimA (compare Fig. 4C and Fig. 5A), whereas overexpression of MdbA in this mutant resulted in abundant production of FimA pili (Fig. 5B). While the assembly defect of FimA in the nuoA mutant was not as severe as that of the ubiE mutant, MdbA overexpression in this nuoA mutant also increased pilus production (Fig. 5C and D; Fig. S2). These results suggest that MdbA was not sufficiently reoxidized by VKOR in the absence of UbiE or NuoA.
FIG 5 .
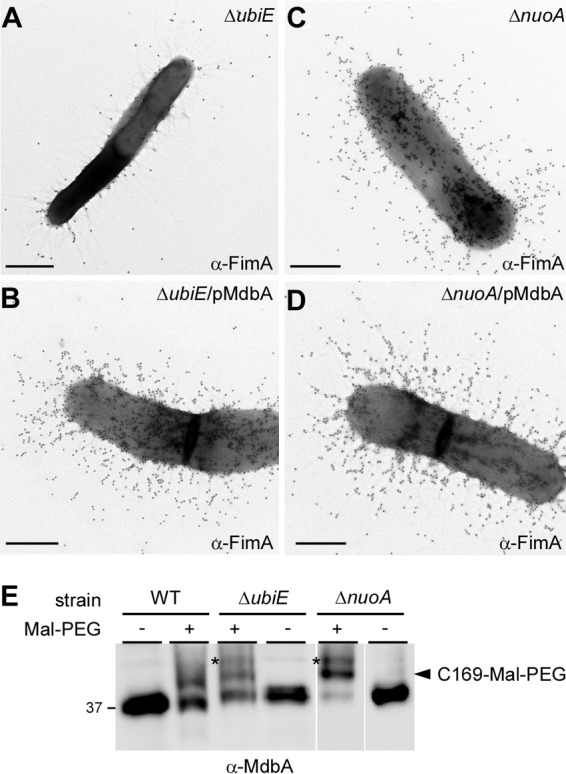
Requirement for NuoA and UbiE in oxidation of the thiol-disulfide oxidoreductase MdbA (A to D). Cells of the indicated A. oris strains were subjected to immunogold labeling with anti-FimA as described in the legend to Fig. 4. Scale bars indicate 0.5 µm. (E) Whole-cell lysates of A. oris strains were prepared by mechanical disruption and treated (+) or not treated (−) with Mal-PEG. Protein samples were immunoblotted with antibodies against the thiol-disulfide oxidoreductase MdbA (α-MdbA). A reduced form of MdbA is shown by an asterisk, whereas the MdbA species labeled by Mal-PEG at C169 are indicated by a black arrowhead.
To confirm this, we examined the oxidation status of MdbA by alkylation with methoxypolyethylene glycol-maleimide (Mal-PEG), as previously reported (23, 33). In this assay, protein samples acid trapped by trichloroacetic acid (TCA) precipitation from cell lysates of the MG1, ΔubiE, and ΔnuoA strains were treated with 10 mM Mal-PEG or mock treated prior to immunoblotting with antibodies against MdbA (anti-MdbA). As the wild-type MG1 strain contains MdbA with the catalytic CXXC motif C139/142 and a noncatalytic cysteine residue, C169 (23), Mal-PEG treatment caused an upshift in MdbA mobility that was due to Mal-PEG modification of the free sulfhydryl group of C169, as previously reported (23) (Fig. 5E; lanes WT, black arrowhead). Significantly, Mal-PEG treatment of the ΔubiE, and ΔnuoA mutants resulted in a higher upshift than that of the MG1 strain under the same condition, consistent with Mal-PEG modification of additional reduced sulfhydryl groups at the active site of MdbA (Fig. 5E, last 4 lanes, asterisks). These results support that the catalytic cysteine residues C139/142 in the CXXC motif are in a reduced form when ubiE and nuoA are genetically disrupted, very much like the phenotype of MdbA when VKOR is absent (23).
Exogenous menaquinone rescues the pilus assembly and cell growth defects of the ubiE mutant.
In the majority of Gram-positive bacteria, menaquinone plays a central role in the ETC, functioning as a conduit to receive electrons from electron donors (e.g., NADH dehydrogenase) and transfer them to an electron acceptor (e.g., cytochrome c reductase) (34, 35). As UbiE catalyzes the conversion of dimethyl-menaquinone to menaquinone (31, 36), the lack of ubiE potentially reduces the quinone pool in the mutant cells, leading to the pleiotropic effects discussed above. To examine this, we monitored the growth rates of A. oris strains grown in HIB by optical density (OD600) over time. Indeed, the ubiE mutant displayed a slow-growth defect compared to the parental strain, and complementation of this strain with the pUbiE plasmid rescued this growth defect (Fig. 6A). Importantly, addition of 0.1 mM menaquinone-4 (MK-4) to the culture medium increased the growth rate to a level comparable to that of the complementing strain after 16 h of growth (Fig. 6A, black and gray triangles).
FIG 6 .
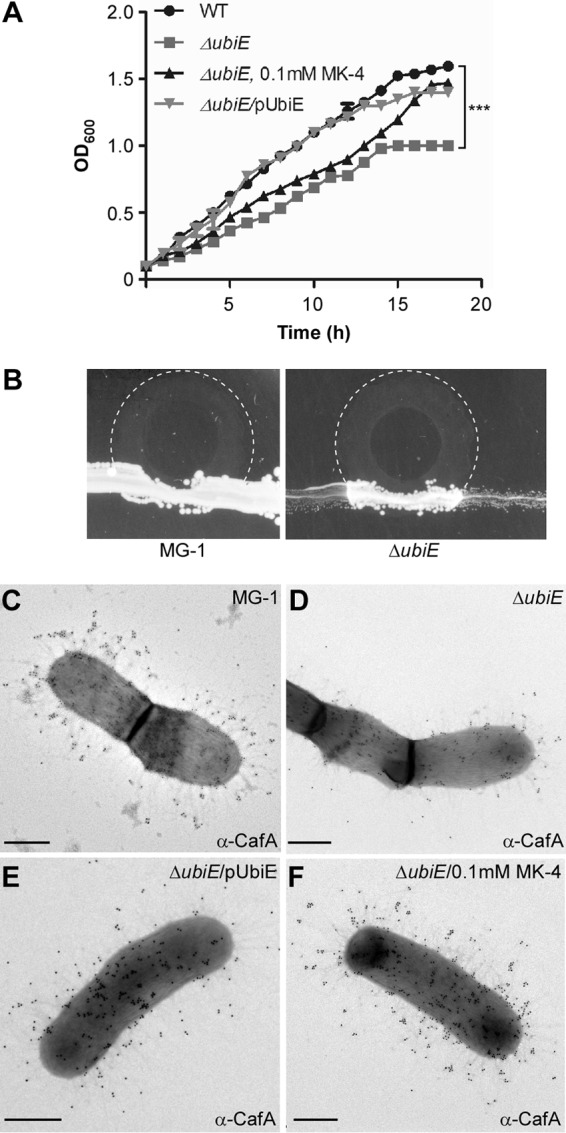
Exogenous menaquinone rescues the growth and pilus assembly defects of the ΔubiE mutant. (A) Growth of the wild-type (WT) strain (solid circles), ΔubiE (gray squares), ΔubiE/pUbiE (gray inverted triangles), and ΔubiE strains in the presence of 0.1 mM menaquinone-4 (MK-4 [triangles]) was measured by optical density (OD600). The results are representative of three independent experiments performed in duplicate. ***, P ≤ 0.0001, as determined by paired two-tailed t test with GraphPad Prism. (B) A. oris strains were streaked as a broad band on HIA plates. A 3-μl drop of 50 mM MK-4 was placed on the border of the streaks, and the growth of the strains at 37°C was recorded after 48 h. Areas of MK-4 diffusion are marked with dashed lines. (C to F) A. oris cells of the WT and ΔubiE, ΔubiE/pUbiE, and ΔubiE mutants grown in the presence of 0.1 mM MK-4 were subjected to immunogold labeling with anti-CafA as described in the legend to Fig. 4. Scale bars indicate 0.5 µm.
To further confirm this phenotype, we adapted a streaking assay with menaquinone (37), whereby the parent MG1 strain and its ubiE isogenic mutant were streaked as a broad band on the heart infusion agar plate, and a 3-μl drop of 50 mM MK-4 was placed on the border of the streak. Cell growth at 37°C was recorded after 48 h. As shown in Fig. 6B, the MG1 strain exhibited abundant growth inside and outside the MK-4 diffusing zone (dashed lines), whereas the ubiE mutant cells inside the MK-4 diffusing zone grew significantly better than those found outside.
Finally, to determine whether menaquinone can also rescue pilus assembly defects, we collected the ubiE mutant cells grown in the presence or absence of MK-4 for IEM using anti-CafA. Indeed, the mutant cells grown in the presence of MK-4 produced abundant CafA signal at a level comparable to that of strain MG1 and the complementing strains (Fig. 6C to F). Altogether, the results support that the defects of the ΔubiE mutant in pilus assembly, cell growth, and MdbA reoxidation are mainly due to the reduced level of endogenous menaquinone.
DISCUSSION
Because pilus assembly and bacterial coaggregation are mutually inclusive in A. oris, this inherent property was used in our Tn5 transposon screen intended to identify factors involved in pilus assembly. This screen revealed numerous mutants that display various defects in Actinomyces coaggregation with S. oralis. As expected, roughly one-third of the coaggregation-defective mutants were mapped to genes encoding the major pilus shaft FimA, the coaggregation factor CafA, and the pilus sortase machine SrtC2 (Fig. 2). Mapping of the remainder surprisingly uncovered the association of coaggregation with the NADH dehydrogenase (complex I) (Table 1), whose genes are adjacent to the ubiquinone/menaquinone biosynthesis gene locus. By genetically and biochemically characterizing two representative mutants with in-frame, nonpolar mutations ΔnuoA and ΔubiE, we have established that the two ETC components, the NADH dehydrogenase and menaquinone, are linked to pilus assembly via reoxidation of the major disulfide bond-forming machine MdbA, which is essential for oxidative folding of pilus proteins in A. oris (23, 25).
By electron microscopy, both the ΔnuoA and ΔubiE mutants are shown to be defective in pilus assembly, although the latter displays the most striking defects (Fig. 3 and 4). Since overexpression of MdbA in the ΔubiE mutant rescues its pilus defects (Fig. 5), we argued that in the absence of UbiE, the disulfide bond-forming machine MdbA is not fully reoxidized, given that reoxidation of MdbA is required for its activity and oxidative folding of A. oris pilus proteins (23). Indeed, by alkylation with Mal-PEG we detected a reduced form of MdbA, although oxidized MdbA was also observed (see Fig. 5E). The latter might be due to inefficient alkylation by Mal-PEG and/or spontaneous oxidation of MdbA during the sampling process. Since menaquinone, 2-methyl-3-polyprenyl-1,4-naphthoquinone, is the main quinone utilized in the ETC of Gram-positive actinobacteria (38), the lack of UbiE certainly reduces the menaquinone pool in the cells, as previously demonstrated in the E. coli ubiE mutant (31). In fact, by adding MK-4, a menaquinone with 4 isoprene units, to the ΔubiE cell cultures, we were able to rescue not only the cell growth deficiency but also the pilus assembly defect of the ΔubiE mutant (Fig. 6). The results strongly support that menaquinone is the major quinone source capable of reoxidizing the disulfide bond machine MdbA/VKOR in A. oris.
In E. coli, DsbA is the primary disulfide bond-forming catalyst, which catalyzes disulfide bond formation of nascent polypeptides transported to the periplasm by the Sec apparatus (39–41). DsbA becomes reduced after catalysis, and reoxidation of DsbA requires DsbB (42, 43). Our results presented here are in line with previous studies in E. coli that demonstrate the participation of the respiratory ETC in the reoxidation of the DsbA/DsbB system (44, 45). Kobayashi and colleagues showed that E. coli mutants lacking hemA—coding for glutamyl-tRNA reductase involved in protoheme and siroheme biosynthesis—or ubiA and menA, which encode products involved in menaquinone biosynthesis, are defective in reoxidation of DsbA when cells are grown under conditions deficient in protoheme or ubiquinone/menaquinone, respectively (44). By reconstituting the E. coli disulfide bond-forming machine DsbA/DsbB with purified components, Bader and colleagues demonstrated that under anaerobic conditions, menaquinone serves as an electron acceptor during DsbA/DsbB reoxidation; under aerobic growth ubiquinone acts as electron acceptor that reoxidizes DsbB, which in turn reoxidizes DsbA (45). It is interesting to note that structural studies of a VKOR homolog of Synechococcus sp. reveal the protein is complexed with ubiquinone (46). Thus, it is plausible that a similar mechanism of MdbA/VKOR reoxidation occurs in A. oris, with menaquinone presumably acting as an electron acceptor for VKOR during this process; how menaquinone mechanistically reoxidizes MdbA/VKOR remains to be investigated in future studies.
Of note, the effect of the NADH dehydrogenase in MdbA/VKOR reoxidation, which has not been reported before with regard to DsbA/DsbB reoxidation, is somewhat puzzling. In many eukaryotic and prokaryotic systems, complex I (NADH quinone:dehydrogenase) serves as an entry point for electrons to enter the respiratory chain, transferring 2 electrons from NADH to ubiquinone (47). In E. coli, the NuoB, NuoD, NuoH, and NuoM subunits form a ubiquinone binding pocket (48). Thus, it is possible that genetic disruption of the Nuo subunits in A. oris might disturb electron transfer and/or the quinone pool in the cells. However, the effects seen in the nuo deletion mutants in pilus assembly are not as striking as those observed in the ubiE mutant, although their defects in coaggregation are obvious (Fig. 3). Thus, it is possible that genetic disruption of the Nuo subunits potentially causes pleiotropic effects. It is likely that many other dehydrogenases, such as the malate and succinate (49), may compensate for the loss of the NADH dehydrogenase.
While the association of the NADH dehydrogenase and menaquinone in pilus assembly is clear, the role of other factors identified from the aforementioned coaggregation-defective screen with Tn5 mutagenesis is not obvious as many of these factors are hypothetical proteins (Table 1). Except for ANA_0218 and ANA_0143, which are predicted to be a Zn2+/Mn2+ transport system substrate-binding protein and β-d-glucoside glucohydrolase, respectively, the coaggregation defect of the remainder is not glaring. Given the nature of the two proteins mentioned above, it is more likely they may participate in receptor-related binding than their involvement in pilus assembly. Finally, it is noteworthy that the Tn5 screen in this study is not saturated, which may explain why a ubiE mutant was not detected in this screen. Future Tn5 screens with multifold coverage designed to directly target pilus assembly will be necessary to reveal any additional novel pilus factors.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and cell growth.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. A. oris cells were grown in heart infusion broth (HIB) or on heart infusion agar (HIA) plates at 37°C with 5% CO2. S. oralis cells were grown in HIB containing 1% glucose in a Coy anaerobic chamber. E. coli DH5α cells were grown in Luria broth (LB) at 37°C. When required, kanamycin was added to the bacterial cultures at a final concentration of 35 or 50 µg ml−1. Reagents were purchased from Sigma unless indicated otherwise.
Strains and plasmids used in this study. Download TABLE S1, PDF file, 0.1 MB (92.3KB, pdf) .
Copyright © 2017 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of recombinant plasmids.
The nuo promoter and the nuoA coding sequence were PCR amplified with A. oris MG1 genomic DNA as a template using the primers Pnuo-F-KpnI/PnuoA-R and nuoA-F/ nuoA-R-HindIII (see Table S2 in the supplemental material), respectively, and Phusion DNA polymerase (New England Biolabs). Overlapping PCR was employed to fuse the two sequences accordingly (50). The fused fragment was cloned into the pCW10 vector (50), and the generated plasmid was electroporated into an A. oris nuoA deletion strain. Similarly, a ubiE complementing plasmid was generated and electroporated into a ubiE deletion strain (Table S1).
Primers used in this study. Download TABLE S2, PDF file, 0.1 MB (83.9KB, pdf) .
Copyright © 2017 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene deletion in A. oris.
All A. oris nonpolar, in-frame deletion mutants were generated using a galK counterselection method described previously (51). In this method, 1-kb fragments up- and downstream of a targeted gene were amplified using appropriate primers (Table S2) and fused using overlapping PCR. The 2-kb fragment was then cloned into the integrative plasmid pCWU2 (Table S1), and the resulting plasmid was electroporated into the A. oris strain CW1, which lacks the galK gene (27). Cointegrants resulting from a single crossover event were selected on kanamycin-containing HIA plates. Loss of the recombinant plasmid by a second crossover event resulting in wild-type and mutant alleles was selected using medium containing 0.2% 2-deoxygalactose (2-DG). Generated mutants were identified by PCR. For double mutants, such as the ΔnuoAJ and ΔnuoAG strains, single mutants were used as a starting strain.
Identification of A. oris coaggregation-defective mutants by Tn5 transposon mutagenesis.
Following a published protocol (51), a library of approximately 6,200 kanamycin-resistant Tn5 mutants was generated from the parental MG1 strain. This library was used to screen for A. oris mutants defective in coaggregation with S. oralis using a cell-based screen in 96-well plates as previously reported (23). Coaggregation was ranked from 1 to 4, largely based on the scoring system described by Cisar and colleagues (28), with the coaggregation phenotypes of A. oris MG1 and S. oralis So34 scored as 4 and the ΔfimA mutant and S. oralis So34 scored as 1; the coaggregation scores 2 and 3 were designated for small and larger clumps of aggregates, respectively.
Consequently, 33 coaggregation-defective mutants were obtained, and TAIL-PCR was employed to map Tn5 insertion sites in these mutants, detailed in our published procedures (27, 51). In brief, two sequential PCRs were performed; the first reaction started with a colony of Tn5 mutants suspended in reaction buffer containing primers Tn5-1 and AD-1 (Table S2) and Apex Taq DNA polymerase (Genesee Scientific). The product of this PCR was used for the next one with primers Tn5-2 (Table S2) and AD-1. Finally, the obtained product of this reaction was gel purified and submitted for DNA sequencing using primer Tn5-3 (Table S2). The resulting DNA sequences were subjected to a BLAST search against the MG1 genome (http://www.homd.org/) to identify the Tn5 insertion sites.
Immunoelectron microscopy.
Immunogold labeling of A. oris cells was performed as previously described, with some modifications (52). Cells were washed once and suspended in 0.1 M NaCl. Seven microliters of bacterial cell suspensions was placed on carbon-coated nickel grids, and then samples were washed with phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA), followed by blocking with 0.1% gelatin in PBS–1% BSA. Adhered cells were stained with primary antibodies diluted in PBS–1% BSA (1:100 for anti-FimA, 1:50 for anti-CafA, and 1:1,000 for anti-type 1), followed by staining with IgG antibodies conjugated to 18-nm gold particles (Jackson ImmunoResearch Laboratories, Inc.). Finally, samples were washed with water, stained with 1% uranyl acetate, and analyzed using a JEOL JEM-1400 electron microscope. The results are representative of three independent experiments in which the reported phenotypes were observed at a level of at least 95% in the fields of view.
Coaggregation assays.
Coaggregation assays were performed as previously described, with some modifications (20, 52). Briefly, stationary-phase cultures of A. oris and S. oralis strain So34 cells were harvested by centrifugation and suspended in TBS buffer (200 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1 mM CaCl2). Equivalent cell numbers of A. oris and S. oralis strains, based on OD600, were mixed for a few minutes, and bacterial aggregates were imaged using an AlphaImager.
Biofilm assays.
In vitro biofilm formation assays were performed according to a published protocol (50). Briefly, overnight A. oris cell cultures were used to inoculate fresh cultures (1:100 dilution) in 1.5 ml of HIB containing 1% sucrose and kanamycin in 24-well plates. After incubation in a CO2 incubator at 37°C for 48 h, the biofilms were gently washed with PBS and dried before staining with 1% crystal violet. After washing the unbound dye, the stained biofilms were subject to ethanol treatment before being quantified by absorbance measurement at 580 nm with a Tecan M1000 microplate reader.
Cell growth assays.
Cell growth of A. oris strains was monitored by a plate assay and optical density (OD600) in HIB cultures as previously described (50). For the plate assay, the MG1 strain and the ΔubiE mutant were streaked as a broad band on HIA plates. A 3-ml drop of 50 mM MK-4 in ethanol was placed on the border of the streaks. Cell growth at 37°C was recorded after 2 days. For growth in HIB, overnight cultures were used to inoculate fresh cultures in HIB supplemented with 35 μg ml−1 of kanamycin with starting OD600 of 0.1. The OD600 reading was taken every hour, and the OD values were presented as averages of three independent experiments performed in duplicate. Calculation of generation time was performed using the formulas k = (log Nt − log N0)/0.301 t and g = 1/k, where N0 and Nt are OD600 values at times 0 and t, respectively, where t is the time elapsed between the Nt and N0 recordings. k corresponds to growth rate, and g corresponds to generation time expressed in hours (53). Generation times were determined from at least two independent experiments performed in triplicate. Note that the MG1, ΔnuoA, and ΔubiE strains contain an empty vector conferring kanamycin resistance.
Determination of MdbA redox status by alkylation with Mal-PEG.
Mid-logarithmic cultures of A. oris were harvested and suspended in SMM buffer (0.5 M sucrose, 10 mM MgCl2, 10 mM maleate [pH 6.8]). Bacterial cell suspensions were treated with mutanolysin at 37°C for 2 h. Protoplasts were collected by centrifugation at 1,500 × g for 10 min, suspended in alkylation buffer (100 mM Tris-HCl [pH 6.8], 1% SDS, 1× protease inhibitor) plus 10% TCA, and lysed by mechanical disruption using a microtube homogenizer (BeadBug) with 0.1-mm glass beads (MP Biomedical). The resulting cell lysates were incubated in ice for 30 min prior to acetone wash and air drying. For alkylation, obtained protein samples were suspended in alkylation buffer containing 10 mM Mal-PEG and incubated at 37°C for 1 h, followed by TCA precipitation and acetone wash. All protein samples were suspended in SDS sample buffer, separated by SDS-PAGE, and immunoblotted with anti-MdbA (1:2,000 dilution).
Statistical analysis.
Statistical analysis in this study was performed using GraphPad Prism 5, with significant differences determined by one-way analysis of variance (ANOVA) (biofilm assays) or the paired two-tailed t test (growth curves, generation time, and enzyme-linked immunosorbent assay [ELISA]). The results are presented as the average values from at least two independent experiments performed in triplicate ± standard deviations (SD).
Supplemental materials and methods. Download TEXT S1, PDF file, 0.1 MB (135.9KB, pdf) .
Copyright © 2017 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank Bethany Tiner and Sara Siegel (UTHealth), Melissa Reardon-Robinson (Harvard Medical School), and other lab members for critical review and discussion of the manuscript.
Research reported in this publication was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under award no. DE025015 and DE017382 to H.T.-T.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors have no conflicts of interest to declare.
Footnotes
Citation Sanchez BC, Chang C, Wu C, Tran B, Ton-That H. 2017. Electron transport chain is biochemically linked to pilus assembly required for polymicrobial interactions and biofilm formation in the Gram-positive actinobacterium Actinomyces oris. mBio 8:e00399-17. https://doi.org/10.1128/mBio.00399-17.
REFERENCES
- 1.Mandlik A, Swierczynski A, Das A, Ton-That H. 2008. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol 16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proft T, Baker EN. 2009. Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell Mol Life Sci 66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spirig T, Weiner EM, Clubb RT. 2011. Sortase enzymes in Gram-positive bacteria. Mol Microbiol 82:1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ton-That H, Schneewind O. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol 50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 5.Dramsi S, Trieu-Cuot P, Bierne H. 2005. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol 156:289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Swaminathan A, Mandlik A, Swierczynski A, Gaspar A, Das A, Ton-That H. 2007. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol 66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budzik JM, Oh SY, Schneewind O. 2008. Cell wall anchor structure of BcpA pili in Bacillus anthracis. J Biol Chem 283:36676–36686. doi: 10.1074/jbc.M806796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobbs AH, Rosini R, Rinaudo CD, Maione D, Grandi G, Telford JL. 2008. Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect Immun 76:3550–3560. doi: 10.1128/IAI.01613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kline KA, Kau AL, Chen SL, Lim A, Pinkner JS, Rosch J, Nallapareddy SR, Murray BE, Henriques-Normark B, Beatty W, Caparon MG, Hultgren SJ. 2009. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol 191:3237–3247. doi: 10.1128/JB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sillanpää J, Chang C, Singh KV, Montealegre MC, Nallapareddy SR, Harvey BR, Ton-That H, Murray BE. 2013. Contribution of individual Ebp pilus subunits of Enterococcus faecalis OG1RF to pilus biogenesis, biofilm formation and urinary tract infection. PLoS One 8:e68813. doi: 10.1371/journal.pone.0068813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazmanian SK, Liu G, Ton-That H, Schneewind O. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 12.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci U S A 96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellen RP, Walker DL, Chan KH. 1978. Association of long surface appendages with adherence-related functions of the Gram-positive species Actinomyces naeslundii. J Bacteriol 134:1171–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cisar JO, Vatter AE. 1979. Surface fibrils (fimbriae) of Actinomyces viscosus T14V. Infect Immun 24:523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeung MK. 1999. Molecular and genetic analyses of Actinomyces spp. Crit Rev Oral Biol Med 10:120–138. doi: 10.1177/10454411990100020101. [DOI] [PubMed] [Google Scholar]
- 16.Henssge U, Do T, Radford DR, Gilbert SC, Clark D, Beighton D. 2009. Emended description of Actinomyces naeslundii and descriptions of Actinomyces oris sp. nov. and Actinomyces johnsonii sp. nov., previously identified as Actinomyces naeslundii genospecies 1, 2 and WVA 963. Int J Syst Evol Microbiol 59:509–516. doi: 10.1099/ijs.0.000950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra A, Das A, Cisar JO, Ton-That H. 2007. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J Bacteriol 189:3156–3165. doi: 10.1128/JB.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbons RJ, Hay DI, Cisar JO, Clark WB. 1988. Adsorbed salivary proline-rich protein 1 and statherin: receptors for type 1 fimbriae of Actinomyces viscosus T14V-J1 on apatitic surfaces. Infect Immun 56:2990–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, Mishra A, Yang J, Cisar JO, Das A, Ton-That H. 2011. Dual function of a tip fimbrillin of Actinomyces in fimbrial assembly and receptor binding. J Bacteriol 193:3197–3206. doi: 10.1128/JB.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra A, Wu C, Yang J, Cisar JO, Das A, Ton-That H. 2010. The Actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol Microbiol 77:841–854. doi: 10.1111/j.1365-2958.2010.07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishra A, Devarajan B, Reardon ME, Dwivedi P, Krishnan V, Cisar JO, Das A, Narayana SV, Ton-That H. 2011. Two autonomous structural modules in the fimbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral biofilm development. Mol Microbiol 81:1205–1220. doi: 10.1111/j.1365-2958.2011.07745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reardon-Robinson ME, Wu C, Mishra A, Chang C, Bier N, Das A, Ton-That H. 2014. Pilus hijacking by a bacterial coaggregation factor critical for oral biofilm development. Proc Natl Acad Sci U S A 111:3835–3840. doi: 10.1073/pnas.1321417111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reardon-Robinson ME, Osipiuk J, Chang C, Wu C, Jooya N, Joachimiak A, Das A, Ton-That H. 2015. A disulfide bond-forming machine is linked to the sortase-mediated pilus assembly pathway in the Gram-positive bacterium Actinomyces oris. J Biol Chem 290:21393–21405. doi: 10.1074/jbc.M115.672253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Dutton RJ, Beckwith J, Boyd D. 2011. Membrane topology and mutational analysis of Mycobacterium tuberculosis VKOR, a protein involved in disulfide bond formation and a homologue of human vitamin K epoxide reductase. Antioxid Redox Signal 14:1413–1420. doi: 10.1089/ars.2010.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reardon-Robinson ME, Ton-That H. 2015. Disulfide-bond-forming pathways in Gram-positive bacteria. J Bacteriol 198:746–754. doi: 10.1128/JB.00769-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cisar JO, Sandberg AL, Abeygunawardana C, Reddy GP, Bush CA. 1995. Lectin recognition of host-like saccharide motifs in streptococcal cell wall polysaccharides. Glycobiology 5:655–662. doi: 10.1093/glycob/5.7.655. [DOI] [PubMed] [Google Scholar]
- 27.Wu C, Huang IH, Chang C, Reardon-Robinson ME, Das A, Ton-That H. 2014. Lethality of sortase depletion in Actinomyces oris caused by excessive membrane accumulation of a surface glycoprotein. Mol Microbiol 94:1227–1241. doi: 10.1111/mmi.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cisar JO, Kolenbrander PE, McIntire FC. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect Immun 24:742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedrich T, Dekovic DK, Burschel S. 2016. Assembly of the Escherichia coli NADH:ubiquinone oxidoreductase (respiratory complex I). Biochim Biophys Acta 1857:214–223. doi: 10.1016/j.bbabio.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Virzintiene E, Trane M, Hägerhäll C. 2011. Revised transmembrane orientation of the NADH:quinone oxidoreductase subunit NuoA. FEBS Lett 585:3277–3283. doi: 10.1016/j.febslet.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Lee PT, Hsu AY, Ha HT, Clarke CF. 1997. A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J Bacteriol 179:1748–1754. doi: 10.1128/jb.179.5.1748-1754.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark WB, Wheeler TT, Cisar JO. 1984. Specific inhibition of adsorption of Actinomyces viscosus T14V to saliva-treated hydroxyapatite by antibody against type 1 fimbriae. Infect Immun 43:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reardon-Robinson ME, Osipiuk J, Jooya N, Chang C, Joachimiak A, Das A, Ton-That H. 2015. A thiol-disulfide oxidoreductase of the Gram-positive pathogen Corynebacterium diphtheriae is essential for viability, pilus assembly, toxin production and virulence. Mol Microbiol 98:1037–1050. doi: 10.1111/mmi.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurosu M, Begari E. 2010. Vitamin K2 in electron transport system: are enzymes involved in vitamin K2 biosynthesis promising drug targets? Molecules 15:1531–1553. doi: 10.3390/molecules15031531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhiman RK, Mahapatra S, Slayden RA, Boyne ME, Lenaerts A, Hinshaw JC, Angala SK, Chatterjee D, Biswas K, Narayanasamy P, Kurosu M, Crick DC. 2009. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol Microbiol 72:85–97. doi: 10.1111/j.1365-2958.2009.06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koike-Takeshita A, Koyama T, Ogura K. 1997. Identification of a novel gene cluster participating in menaquinone (vitamin K2) biosynthesis. Cloning and sequence determination of the 2-heptaprenyl-1,4-naphthoquinone methyltransferase gene of Bacillus stearothermophilus. J Biol Chem 272:12380–12383. doi: 10.1074/jbc.272.19.12380. [DOI] [PubMed] [Google Scholar]
- 37.Franza T, Delavenne E, Derré-Bobillot A, Juillard V, Boulay M, Demey E, Vinh J, Lamberet G, Gaudu P. 2016. A partial metabolic pathway enables group B streptococcus to overcome quinone deficiency in a host bacterial community. Mol Microbiol 102:81–91. doi: 10.1111/mmi.13447. [DOI] [PubMed] [Google Scholar]
- 38.Collins MD, Jones D. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev 45:316–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bardwell JC, McGovern K, Beckwith J. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 40.Kamitani S, Akiyama Y, Ito K. 1992. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J 11:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kadokura H, Beckwith J. 2009. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell 138:1164–1173. doi: 10.1016/j.cell.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bardwell JC, Lee JO, Jander G, Martin N, Belin D, Beckwith J. 1993. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci U S A 90:1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Missiakas D, Georgopoulos C, Raina S. 1993. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci U S A 90:7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobayashi T, Kishigami S, Sone M, Inokuchi H, Mogi T, Ito K. 1997. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc Natl Acad Sci U S A 94:11857–11862. doi: 10.1073/pnas.94.22.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC. 1999. Oxidative protein folding is driven by the electron transport system. Cell 98:217–227. doi: 10.1016/S0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 46.Li W, Schulman S, Dutton RJ, Boyd D, Beckwith J, Rapoport TA. 2010. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature 463:507–512. doi: 10.1038/nature08720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamaru-Ogiso E, Narayanan M, Sakyiama JA. 2014. Roles of semiquinone species in proton pumping mechanism by complex I. J Bioenerg Biomembr 46:269–277. doi: 10.1007/s10863-014-9557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong X, Xie T, Yu L, Hesterberg M, Scheide D, Friedrich T, Yu CA. 2003. The ubiquinone-binding site in NADH:ubiquinone oxidoreductase from Escherichia coli. J Biol Chem 278:25731–25737. doi: 10.1074/jbc.M302361200. [DOI] [PubMed] [Google Scholar]
- 49.Bott M, Niebisch A. 2003. The respiratory chain of Corynebacterium glutamicum. J Biotechnol 104:129–153. doi: 10.1016/S0168-1656(03)00144-5. [DOI] [PubMed] [Google Scholar]
- 50.Siegel SD, Wu C, Ton-That H. 2016. A type I signal peptidase is required for pilus assembly in the Gram-positive, biofilm-forming bacterium Actinomyces oris. J Bacteriol 198:2064–2073. doi: 10.1128/JB.00353-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu C, Reardon-Robinson ME, Ton-That H. 2016. Genetics and cell morphology analyses of the Actinomyces oris srtA mutant. Methods Mol Biol 1440:109–122. doi: 10.1007/978-1-4939-3676-2_9. [DOI] [PubMed] [Google Scholar]
- 52.Chang C, Huang IH, Hendrickx AP, Ton-That H. 2013. Visualization of Gram-positive bacterial pili. Methods Mol Biol 966:77–95. doi: 10.1007/978-1-62703-245-2_5. [DOI] [PubMed] [Google Scholar]
- 53.Willey JM, Sherwood LM, Woolverton CJ. 2008. Prescott, Harley, and Klein’s microbiology, 7th ed. McGraw-Hill, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Requirement of nuoA for pilus assembly. A. oris cells of the indicated strains were immobilized on carbon-coated nickel grids and stained with anti-FimA (A to D), anti-CafA (E to H), or anti-type 1 (I to L) antibodies, followed by staining with IgG conjugated to 18-nm gold particles. Samples were stained with 1% uranyl acetate prior to being analyzed by electron microscopy. Scale bars indicate 0.5 µm. Download FIG S1, PDF file, 2.7 MB (2.8MB, pdf) .
Copyright © 2017 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Requirement of nuoA and ubiE for surface expression of CafA. The expression of CafA on the cell surface was determined by whole-cell ELISA using polyclonal anti-CafA antibodies. The absorbance measurements at 450 nm, compared to the cafA mutant as background, were determined from three independent experiments performed in triplicate. Error bars represent standard deviations. * and *** indicate P < 0.05 and P < 0.001, respectively, determined using the unpaired, two-tailed t test with GraphPad Prism. Download FIG S2, PDF file, 0.1 MB (80KB, pdf) .
Copyright © 2017 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Generation times of the A. oris MG1 and ΔnuoA mutant strains. Growth of the wild-type (WT) MG1 strain and ΔnuoA and ΔnuoA/pNuoA mutant strains was measured by optical density (OD600). Generation times were calculated as described in Materials and Methods. The results are representative of three independent experiments performed in triplicate. Error bars represent standard deviations. ns, not significant. Download FIG S3, PDF file, 0.1 MB (60KB, pdf) .
Copyright © 2017 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and plasmids used in this study. Download TABLE S1, PDF file, 0.1 MB (92.3KB, pdf) .
Copyright © 2017 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S2, PDF file, 0.1 MB (83.9KB, pdf) .
Copyright © 2017 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental materials and methods. Download TEXT S1, PDF file, 0.1 MB (135.9KB, pdf) .
Copyright © 2017 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.