Abstract
The polygenic nature of complex psychiatric disorders suggests a common pathway that may be involved in the down-regulation of multiple genes through an epigenetic mechanism. To investigate the role of methylation in down-regulating the expression of mRNAs that may be associated with the schizophrenia phenotype, we have adopted a cell-culture model amenable to this line of investigation. We have administered methionine (2 mM) to primary cultures of cortical neurons prepared from embryonic day 16 mice and show that this treatment down-regulated reelin and glutamic acid decarboxylase 67 (GAD67) mRNA expression but not that corresponding to neuron-specific enolase mRNA. Moreover, methionine increased methylation of the reelin promoter, suggesting a possible mechanism for the observed change. These cultures contain a mixed population of neurons and glia. Approximately 83% of the neurons are GABAergic based on GAD immunoreactivity, and these neurons coexpress high levels of reelin and DNA methyltransferase (Dnmt) 1 immunoreactivity. To examine whether Dnmt1 regulates reelin gene expression, we used an antisense approach to reduce (knock down) Dnmt1 expression. The reduced Dnmt1 mRNA and protein were accompanied by increased reelin mRNA expression. More importantly, the Dnmt1 knockdown blocked the methionine-induced reelin and GAD67 mRNA down-regulation. These data support the hypothesis that the reduced amounts of reelin and GAD67 mRNAs documented in postmortem schizophrenia brain may be the consequence of a Dnmt1-mediated hypermethylation of the corresponding promoters.
Keywords: epigenetics, gene expression, methylation, schizophrenia
Diseases that show complex inheritance patterns, such as those associated with psychiatric illness, are likely to have polygenic origins. Recent research in the area of epigenetic regulation of gene expression has progressed considerably, and concepts emerging from this field provide a molecular mechanism to explain how multiple gene targets may be affected in psychiatric disease (1, 2). Here, we refer specifically to the covalent methylation of cytosine in the promoters of genes embedded in CpG islands. This modification is affected by the action of DNA methyltransferases (Dnmts) (3). We suggest that the variable symptomatology associated with the schizophrenia spectrum of mental disorders might be the consequence of inappropriate promoter methylation resulting from the dysregulation of this control mechanism operating in GABAergic neurons (2). Interestingly, sequence analyses of genomic DNA isolated from monozygotic twins discordant for schizophrenia show clear discrepancies in the methylation patterns of certain genes, suggesting the involvement of an epigenetic mechanism that may underlie the corresponding phenotypic differences (4, 5). We propose that an epigenetic origin of schizophrenia might be responsible for the differential modulation of subsets of genes expressed in GABAergic neurons in the schizophrenia brain (2, 6).
Since the initial discovery that glutamic acid decarboxylase (GAD) 67 mRNA is reduced in patients with schizophrenia (7), several reports have suggested that a GABAergic deficit is consistent with the existing data and symptomatology associated with schizophrenia (2, 8–10). That is, schizophrenia may no longer be a disease associated with a dopamine deficit but, rather, may be a defect associated with GABAergic inhibitory input onto prefrontal cortical neurons. A more recent study suggests the possibility that the GABAergic deficit may be the result of compromised glutamatergic input onto cortical GABAergic neurons (11). We (12, 13) and others (14–16) have shown that reelin and GAD67 mRNAs are reduced in the postmortem brains of patients with schizophrenia and psychotic bipolar disorder. Recent in vitro studies demonstrate a role for reelin in stimulating dendritic spine mRNA translation (17). It seems plausible that the down-regulation of reelin expression may be associated with the previously reported decrease in numbers of dendritic spines in postmortem brains of schizophrenia subjects (18). The reduced levels of GAD67 are consistent with a decreased inhibitory tone associated with prefrontal cortical dysfunction in schizophrenia (19). A recent study showing that expression of Dnmt1 mRNA is increased in prefrontal cortical GABAergic neurons of schizophrenia patients relative to nonpsychiatric subjects (6) provides a mechanism for the coordinated down-regulation of not only reelin and GAD67 but of other mRNAs coexpressed in these same GABAergic neurons. As would be predicted by this mechanism, the overexpression occurs in the same neurons that show decreased reelin and GAD67 mRNA expression (6).
Among the DNA cytosine methyltransferases, Dnmt1 is not only expressed in dividing cells but is also highly expressed in postmitotic cells such as neurons (6, 20). For example, Dnmt1 mRNA is abundant in telencephalic GABAergic neurons of the human cortex (6), suggesting the possibility that DNMT1, in addition to the maintenance of methylation patterns in mitotic cells, possesses additional functions in regulating DNA methylation patterns in nondividing neurons (2). It should be remembered that oral administration of methionine to schizophrenia patients elicits a psychotic episode in a large number of patients but not in normal subjects (21) so that it may well be that this mechanism is the Achilles' heel of psychosis associated with both schizophrenia and bipolar illness. Although the mechanism for this recrudescence of psychosis symptomatology has never been adequately accounted for, we suggest that the increased methionine results in the hypermethylation of susceptible promoters in neurons accessible to the amino acid.
We have shown that agents that either reduce genomic DNA methylation (5-azadeoxycytidine) or increase histone acetylation (trichostatin A, valproic acid) increase human reelin expression in vitro (22). We have also shown that when administered to mice in vivo, methionine down-regulates the expression of reelin and GAD67 mRNAs and proteins (23). Chronic methionine (Met) administration reduces reelin and GAD67 mRNA levels in the frontal cortex of wild-type and heterozygous reeler mice. The effect of l-methionine was associated with an increase in the number of methylated cytosines in the CpG island of the murine reelin promoter (23). In the present study, we examined the expression of reelin and GAD67 mRNAs in primary neuronal cultures from mouse cortex in vitro and evaluated the effects of methionine on this expression. We also determined the effects of this treatment on reelin promoter methylation for several treatment durations. Finally, we treated the cortical neuron cultures with a Dnmt1 antisense oligonucleotide to assess its involvement in the regulation of the reelin gene. After the various treatments, the expression of mRNAs encoding reelin, GAD67, neuron-specific enolase (NSE), and Dnmt1 were quantitated. Our findings are consistent with the possibility that one target of Dnmt1 action may be the reelin promoter. This regulation likely occurs either through a change in promoter methylation or as the result of the recruitment of corepressors and/or histone methyltransferases to the promoter by Dnmt1.
Methods
Cortical Cell Cultures. Cortical cell cultures were prepared as described in refs. 24 and 25. In brief, neocortices from fetal mice [embryonic day (E) 14 to E15] were dissociated and plated onto six-well plates precoated with 100 μg/ml poly(d-lysine) and 4 μg/ml laminin at a density of five hemispheres per plate for the neuron and glia cocultures, in MEM (Earle's salts) supplemented with 5% horse serum, 5% FBS, and 21 mM glucose. Proliferation of nonneuronal cells was arrested with 10 μM cytosine arabinoside at 7–8 days in vitro. Cultures were maintained as described in refs. 24 and 25.
Cultures were incubated with methionine (or vehicle) at a final concentration of 2 mM in MEM supplemented with 21 mM glucose at 37°C for 24, 36, and 72 h and cultures were harvested for either isolation of genomic DNA or total RNA. Dose–response curves were initially performed to assess methionine toxicity. The data showed little or no cell death associated with the 2 mM treatment as determined by incubating the cultures with propidium iodide/fluorescein diacetate as described in ref. 26. In contrast, the numbers of live/dead cells decreased when concentrations as high as 10 mM methionine were tested. The 2 mM methionine dose was the highest dose that failed to show increased cell death based on this assay and is ≈20 times the concentration of methionine present in the culture medium.
Immunohistochemistry. Cortical cultures were fixed with 4% paraformaldehyde in 0.1 M PBS for 30 min and rinsed three times with PBS. Nonspecific sites were blocked, and cells were then rinsed two times with 1% normal goat serum (NGS) and 1% BSA in PBS for 10 min. The following primary antibodies were used: rabbit polyclonal GAD (Chemicon); rabbit polyclonal Dnmt1 (20), mouse monoclonal NeuN (Chemicon), and mouse monoclonal reelin G10 (27). Cultures were incubated with the primary antibody (dilution of 1:500 with 1% NGS/1% BSA in PBS) overnight at 4°C. Secondary antibodies [biotinylated goat anti-mouse IgG (1:250) or Cy5-conjugated goat anti-rabbit IgG (1:500)] were incubated for 60 min at room temperature in 1% NGS/1% BSA in PBS. Cy2-conjugated streptavidin (1:500) was incubated identically. After washing, preparations were air-dried and coverslip-mounted by using an antifading agent (DABCO). Images and photomicrographs were captured by using a laser confocal microscope (Leica TCS-NT).
For counting positive neurons (Table 1), random fields were viewed by using ×40 magnification with appropriate filters. Cy2- and Cy5-positive neurons were counted independently in the same fields, and the extent of overlap was counted. From 4 to 7 fields were counted per antibody combination with each field containing anywhere from 20 to 80 immunopositive neurons.
Table 1. Colocalization of histochemical markers in E16 cortical neurons in primary culture.
| NeuN-positive neurons expressing GAD 92 ± 2.5% | GAD-positive neurons expressing NeuN 100% |
| NeuN-positive neurons expressing Dnmt1 86 ± 2.9% | Dnmt1-positive neurons expressing NeuN 97 ± 1.7% |
| Reelin-positive neurons expressing Dnmt1 92 ± 3.7% | Dnmt1-positive neurons expressing Reelin 81 ± 0.9% |
| Reelin-positive neurons expressing GAD 97 ± 1.5% | GAD-positive neurons expressing Reelin 77 ± 6.2% |
Data are reported as percentages ± SEM of at least five random fields at ×40 magnification.
Total RNA Extraction and Synthesis of Internal cRNA Standards. For each condition, two six-well plates of cultured cells were used to extract total cellular RNA by using RNAqueous-4 PCR kit (Ambion, Austin, TX). DNase I treatment was performed, and RNA amounts were determined by using the fluorescent RiboGreen RNA Quantitation kit (Molecular Probes) and a hand-held fluorometer (Turner Designs Picofluor).
Internal standard templates were generated by sited-directed mutagenesis with PCR-overlap extension (28). The reelin and GAD67 primers and standards are described in refs. 12 and 23. For DNMT1, the internal standard was generated by deleting 106 bp in the middle of the 496-bp amplicon, using overlap extension PCR. This approach allows the PCR products to be differentiated by gel electrophoresis without postamplification restriction enzyme digestion. Independently, we compared a Dnmt1 internal standard engineered with an XbaI site in the center of the amplicon and the Dnmt1 internally deleted standard cRNA. PCR reactions carried out in parallel showed that the templates were amplified with the same kinetics and efficiency.
RT-PCR. Increasing concentrations of individual internal standard cRNAs were added to 1 μg of total RNA isolated from cultured cells and subjected to the RT-PCR reaction as described in ref. 23. The following primers were used for PCR: DNMT1 (GenBank accession no. NM_010066) forward 5′-CCCATGCATAGGTTCACTTCCTTC-3′ and reverse 5′-TGGCTTCGTCGTAACTCTCTACCT-3′. DNMT1 PCR cycle conditions were 2 min at 94°C, followed by 28 cycles consisting of 94°C for 30 s, 61°C for 45 s, 72°C for 45 s, and 72°C for 7 min.
Images of gels were taken through a Kodak EDAS 290 as a digital image, and the signals from different samples were quantified by using Kodak 1d 3.6 image analysis software.
Bisulfite Modification and Sequencing of Genomic DNA. Genomic DNA was isolated from cortical neuronal cultures treated with either 2 mM l-methionine or vehicle for 24, 36, and 72 h (29). The DNA was bisulfite-modified and PCR-amplified as described in refs. 22 and 23, using nested primer pairs. PCR products from each group were purified and subcloned. Individual clones were sequenced at the University of Chicago Core Sequencing Facility. A minimum of 11 clones were isolated and sequenced from each treatment paradigm.
Western Blotting. Cell extracts were prepared in lysis buffer (50 mM Tris·HCl, pH 7.5/1% Nonidet P-40/0.1% sodium deoxycholate/150 mM NaCl/2 mM EDTA/10 mM EGTA) containing a mixture of protease inhibitors. Protein samples were separated on 4–20% gradient Tris-glycine gels (Invitrogen), and Western blotting was performed as described in ref. 23. Blots were incubated with a 1:4,000 dilution of antibodies against C-terminal DNMT1 and β-actin (Sigma), overnight at 4°C in 1% BSA, 2% Triton X-100, and 0.4% SDS in PBS. After primary antibody hybridization, blots were washed and incubated with the secondary anti-rabbit (DNMT1, Sigma) or anti-mouse (actin, Sigma) antibodies conjugated with horseradish peroxidase for 4 h. Enhanced chemiluminescence (ECL, Amersham Pharmacia) was used for detection with a Storm fluorescent imager (Molecular Dynamics).
Oligonucleotide Transfections. The DNMT1 antisense oligonucleotide previously was synthesized and used as described in detail in ref. 30. The four bases at the 5′ and 3′ ends are 2′-O-methylphosphorothioate, whereas the central deoxynucleotide backbone is phosphorothioate-modified [5′-(TCTA)TTTGAGTCTGCC(ATTU)-3′]. This sequence corresponds to positions –2 to +18 of the murine Dnmt1 cDNA (GenBank accession no. NM_010066). As a control, a scrambled sequence, 5′-f luorescein-tagged and 2′-O-methylphosphorothioate-modified sequence oligonucleotide [5′-(TGTG)ATTCTCCTTA(TTCG)-3′; parenthesized bases are 2′-O-methyl modified], was synthesized and used in parallel. The oligonucleotides were HPLC-purified by the manufacturer (Keystone Labs, BioSource International, Camarillo, CA).
Oligonucleotides [1 μM (final concentration) of each sequence] were transfected using Lipofectamine 2000 (8 μl/ml final concentration, Invitrogen) as described in ref. 22. The oligonucleotide transfection was repeated once after 36 h. Three days after transfection, mRNA or proteins were extracted as indicated above.
For the combined antisense transfection and methionine treatments, cultures were transfected with the antisense or scrambled oligonucleotides, and 5 h later, the medium was replaced with normal growth medium plus 2 mM methionine. Cultures were harvested 72 h later for RNA and/or protein analysis. For statistical purposes, three separate and independent transfections were carried out and subsequent analyses were performed separately at least three times.
Results
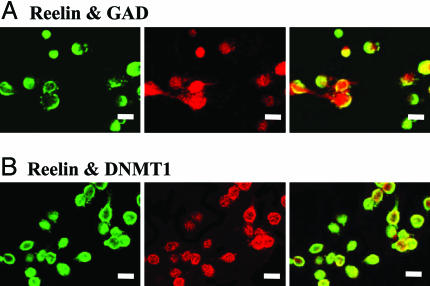
Expression Patterns of Reelin, GAD, and Dnmt1 in Cortical Cultures. Mixed cultures derived from E14–E15 mouse cortices and maintained in vitro for 8–10 days in vitro are primarily composed of postmitotic neurons with some proliferating glia. To elucidate the characteristics of these cultures, we used double-immunohistochemical staining to identify neuronal nuclear antibody (NeuN)-, reelin-, GAD-, and Dnmt1-immunopositive cells. A large number of neurons present in the cultures were GABAergic based on positive staining with a pan GAD65–67 antibody. Reelin immunoreactivity (IR) colocalized in these same cells which were neurons based on NeuN immunostaining. Dnmt1 was also expressed in predominantly NeuN-positive cells. That is, 93.5 ± 7.5% of all NeuN-positive cells were also Dnmt1-immunoreactive. Representative double-immunostaining photomicrographs are shown in Fig. 1. Fig. 1 A shows reelin IR (Upper Left), GAD IR (Upper Middle), and the corresponding colocalization (Upper Right). Fig. 1B shows Reelin IR (Lower Left), Dnmt1 IR (Lower Middle), and the colocalization (Lower Right). These two sets of panels indicate that the reelin, GAD, and Dnmt1 genes are expressed in the same neurons. Colocalization data are summarized in Table 1.
Fig. 1.
Immunohistochemistry of the mouse cortical cultures. The photomicrographs show the colocalization of reelin with additional histochemical markers in the primary cortical neuronal cultures. (A) Reelin IR, color-coded green (Left); GAD IR, color-coded red (Center); overlay of reelin and GAD (Right). (Scale bar: 10 mm.) (B) Reelin IR, color-coded green (Left); DNMT1 IR, color-coded red (Center); overlay of Reelin and DNMT1 (Right). (Scale bar: 10 mm.) The figure shows representative images taken from random fields. A more complete statistical analysis of these data are presented in Table 1.
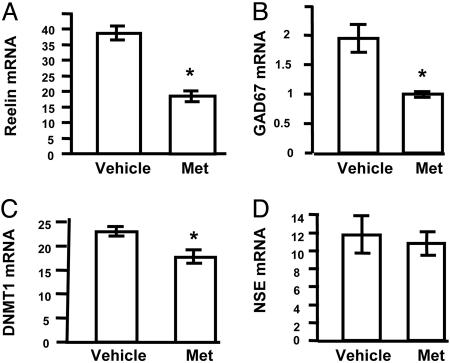
l-Methionine-Induced Down-Regulation of mRNA Expression. We treated primary neuronal cultures with 2 mM methionine treatment, which is 20 times higher than the culture media concentration, and examined the expression of reelin, GAD67, and Dnmt1 mRNAs as compared with the vehicle-treated group. As shown in Fig. 2 A–C, methionine treatment resulted in reduced mRNA levels for reelin, GAD67, and also Dnmt1. The down-regulation mediated by methionine was most pronounced with respect to the reelin and GAD67 mRNAs, although the change in Dnmt1 mRNA was significant. In contrast, this treatment had no effect on the expression of the neuronal marker mRNA, NSE (Fig. 2D). Experiments performed in parallel showed that treatment of these cultures with 2 mM glycine had no effect on the expression of either reelin or GAD67 mRNAs (data not shown).
Fig. 2.
Treatment of mouse cortical cultures with methionine down-regulates mRNA expression. Mouse primary cortical cultures were prepared and treated with 2 mM methionine as outlined in Methods. After 72 h, RNA was harvested, and the amounts of specific transcripts were quantified by using competitive RT-PCR with internal standards. These included reelin (A), GAD67 (B), Dnmt1 (C), and NSE (D) mRNAs. mRNA units are pg/μg total RNA. The data represent mean ± SE of three independent RNA measurements from three separate treatments. The vehicle-treated group was time-matched and performed in parallel. *, significance at P < 0.001.
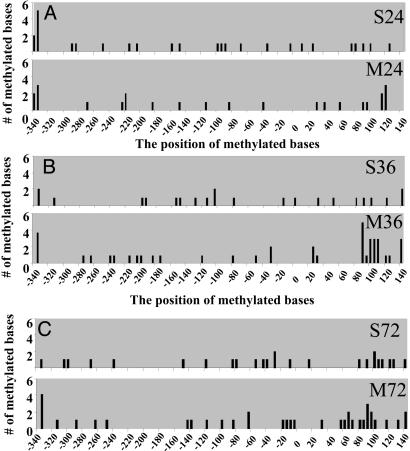
l-Methionine-Induced Hypermethylation of the Reelin Promoter. Primary neuronal cultures were treated with 2 mM methionine for 24, 36, and 72 h, and genomic DNA was extracted. The control set from each group was the time-matched/vehicle-treated group that served to confirm that methylation did not vary during the treatment. DNA was modified using the sodium metabisulfite method, and the reaction products were amplified. Individual clones were obtained (at least 11 from each group) and sequenced. Methylation was assessed by examining both the total number of methylated sites at each time point and by examining the methylation pattern obtained from individual clones that were summed to generate the methylation maps presented in Fig. 3. Inspection of the data show that the total number of methylated sites did not vary significantly over the duration for the saline (S)-treated group (S24, 3.4 ± 0.87; S36, 2.8 ± 1.0; S72, 3.1 ± 0.44). The amount of methylation did not increase after 24 h of methionine (M24, 3.0 ± 0.5). However, the change was significant after 36 h (M36, 5.1 ± 1.0), and this increase remained elevated after 36 h (M72, 4.6 ± 1.0). The extent of the methylation then remained at the elevated levels and did not increase further after the 72-h treatment. Inspection of the time-dependent methylation profiles presented in Fig. 3 does not indicate a readily discernable sequence specific pattern to the increased methylation.
Fig. 3.
Methylation patterns of genomic DNA isolated from saline- and methionine-treated primary mouse cortical cultures. After the saline/methionine treatment, genomic DNA was isolated and treated with sodium bisulfite. PCR primers were used to amplify the reelin promoter region. The amplified material was subcloned, and individual clones were sequenced to determine the pattern and total number of methylated bases. A minimum of 11 clones from each treatment set were sequenced. The saline-treated controls were time-matched for each group and performed in parallel. (A) Twenty-four-hour treatment of saline (S24) and methionine (M24). The total number of methylated cytosines for S24 is 27, and for M24 is 24. (B) Thirty-six-hour treatment of saline (S36) and methionine (M36). Total number of methylated cytosines for S36 is 22, and for M36 is 41. (C) Seventy-two-hour treatment of saline (S72) and methionine (M72). The total number of methylated cytosines for S72 is 25, and for M72 is 37.
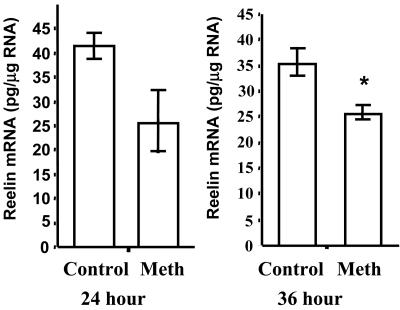
We also examined the time-dependence of the methionine-induced down-regulation of reelin to determine whether the changes in methylation correlated with the observed mRNA changes. After a 24-h treatment paradigm, the levels of reelin mRNA were decreased (Fig. 4 Left), although this decrease was not quite significant (P < 0.08). In contrast, the 36-h methionine treatment decreased reelin mRNA levels by 30%, and this change was significant (Fig. 4 Right). The increase in promoter methylation was evident after 36 h of methionine and remained increased after another 36 h of treatment.
Fig. 4.
Time dependence of the methionine-induced reelin mRNA down-regulation. Data presented in Fig. 2 show the effects after a 72-h methionine treatment on reelin mRNA levels. Fig. 3 shows that changes in reelin promoter methylation begin to be significant somewhat earlier. In this experiment, we treated mouse primary cortical cultures for 24 and 36 h with 2 mM methionine and harvested RNA for analysis. The data show that whereas there appears to be a trend toward down-regulation at 24 h (P < 0.08), this becomes significant after 36 h of treatment (P < 0.001). RNA levels were quantified by using competitive RT-PCR with internal standards.
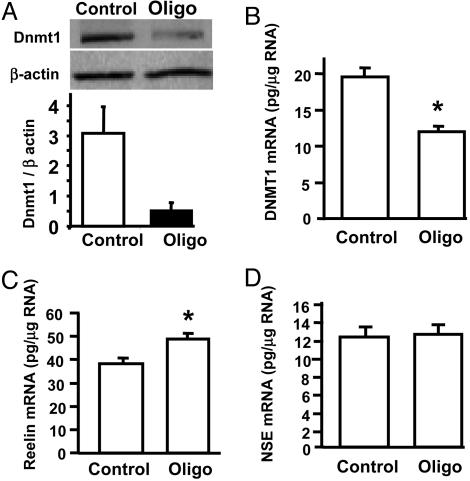
Dnmt1 Antisense Decreases Dnmt1 Expression and Attenuates the Methionine-Induced Reelin mRNA Down-Regulation. Antisense Dnmt1 and scrambled oligos were transfected in parallel, and both mRNA and protein were harvested for analysis. To confirm the effects of antisense oligonucleotide on this culture, we measured the Dnmt1 mRNA as well as Dnmt1 protein after the treatments. In contrast to the scrambled oligonucleotide-treated cells, the antisense oligonucleotide-treated samples showed a decrease in Dnmt1 protein. Fig. 5A Upper shows a representative Western blot and the corresponding measure of the ratio of the optical density relative to the β-actin optical density is shown in Fig. 5A Bottom. Dnmt1 mRNA was reduced as determined by competitive RT-PCR (Fig. 5B). In addition, treatment of the cortical cultures with the Dnmt1 antisense oligonucleotide significantly increased reelin mRNA levels (Fig. 5C). In contrast, there was no effect on the expression of NSE mRNA (Fig. 5D).
Fig. 5.
Dnmt1 antisense oligonucleotide increases reelin mRNA expression. The Dnmt1 antisense oligonucleotide was incubated with the primary cortical cultures to down-regulate the corresponding mRNA and protein (see Methods). A shows a representative Western blot (Upper) demonstrating a down-regulation of Dnmt1 protein (Oligo, antisense treated; Control, scrambled oligo). The amount of residual Dnmt1 after antisense treatment was measured to be ≈18%; t test, P < 0.02 (Lower). B shows the results of a competitive RT-PCR assay showing that the Dnmt1 mRNA is also reduced by the antisense treatment. C demonstrates that the Dnmt1 antisense increases reelin mRNA levels (P < 0.001) but does not affect the levels of the NSE mRNA (D).
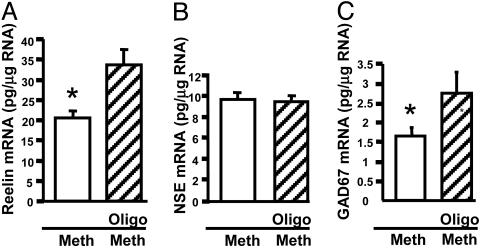
The effect of Dnmt1 knockdown-mediated increase in reelin mRNA was small but significant. We tested whether an antisense-mediated down-regulation of Dnmt1 affected the methionine-induced down-regulation of reelin and GAD67 mRNAs. The combined treatment involved the simultaneous administration of antisense (or scrambled) oligonucleotide and 2.0 mM methionine, and after 72 h, RNA was harvested and analyzed. As shown, methionine down-regulated reelin (Fig. 6A) and GAD67 (Fig. 6C) mRNAs. In the presence of the Dnmt1 antisense, the reelin and GAD67 mRNA down-regulation was blocked. In contrast, the antisense treatment did not affect the levels of the NSE mRNA either in the absence or presence of methionine (Fig. 6B).
Fig. 6.
Dnmt1 antisense blocks the methionine-mediated reelin and GAD67 mRNA down-regulation. Primary cortical cultures were treated with the Dnmt1 antisense oligonucleotide before the methionine treatment to test whether Dnmt1 is a component of the pathway from methionine to the reelin promoter. See Methods for specific details of the treatment paradigm. In brief, cultures were transfected with the Dnmt1 antisense (Oligo, hatched bars) and in parallel the control oligo (open bars) and were subsequently treated with methionine (Meth) for 72 h. RNA was harvested, and the levels of reelin (A), NSE (B), and GAD67 (C) mRNAs were quantified by using competitive RT-PCR with internal standards (28). The data show that whereas the treatment did not affect NSE mRNA expression, the methionine-mediated down-regulation was attenuated in the presence of the Dnmt1 antisense knockdown. *, P < 0.001.
Discussion
Data presented herein confirm and extend observations made in vivo regarding chronic methionine administration and the down-regulation of selected mRNAs expressed in GABAergic neurons of the frontal cortex of treated mice (23). Protracted methionine treatment of primary mixed mouse cortical cultures in vitro also resulted in an ≈50% reduction in the reelin and GAD67 mRNA content, a somewhat smaller decrease in Dnmt1 mRNA, and no change in NSE mRNA. Although these neuronal cultures were treated with cytosine arabinoside, the treatment was not started until the fifth day in vitro, resulting in a mixed glia/neuronal population. Double immunostaining with selected neuronal markers showed that the majority of NeuN-immunopositive neurons were GABAergic, largely expressing both GAD and reelin. Interestingly, many of these same neurons also expressed Dnmt1 IR. This latter finding is consistent with another study performed in postmortem human brain that shows that Dnmt1 mRNA is preferentially expressed in human cortical GABAergic neurons and that it is increased in these same neurons in the prefrontal cortex of patients diagnosed with schizophrenia (6).
Because of the increased Dnmt1 mRNA in schizophrenia patients and reduced reelin mRNA in these same neurons, we used the cortical culture system as a model to more directly investigate the mechanism through which Dnmt1 regulates reelin mRNA expression. We established first that methionine down-regulates the expression of reelin and GAD67 mRNAs in our model and furthermore demonstrated that the treatment increased the methylation of the reelin promoter. Moreover, the methionine treatment was time dependent in terms of both reelin mRNA down-regulation and promoter methylation. The effects of the antisense treatment of the cultures on reelin mRNA levels was only modest. However, the antisense treatment attenuated the methionine-induced down-regulation of reelin mRNA expression. This result links, for the first time, the expression of Dnmt1 to the regulation of specific genes (reelin and GAD67) relevant to the biology of synaptic transmission in a defined neuronal phenotype (GABAergic).
Dnmt1 is commonly thought to be responsible for maintaining methylation patterns in the daughter strands of dividing cells. The observation that Dnmt1 IR is abundant in postmitotic neurons recently was reported (20), although the function of the protein in neurons was not defined. In fact, Dnmt1 is well studied in the context of gene regulation and methylation, but the vast majority of work has been in cells that lie outside the realm of the nervous system (31). Our group recently expanded on this observation (20) by examining the in situ hybridization patterns of Dnmt1 mRNA in postmortem human brain (6). This study demonstrated that the expression of Dnmt1 mRNA is present in neurons and that these neurons also express markers identifying them as GABAergic. The signal observed in non-GABAergic neurons is considerably less pronounced. Moreover, the abundance of Dnmt1 mRNA is higher in GABAergic neurons of schizophrenia patients as compared to nonpsychiatric subjects. Interestingly, the higher expression of Dnmt1 mRNA is coincident with those neurons in which reelin and GAD67 mRNAs are down-regulated. Whereas these observations are largely correlational, the present study was designed to strengthen a link between Dnmt1 and the coordinate regulation of multiple genes that are expressed in GABAergic neurons.
Although the localization of Dnmt1 to GABAergic neurons is clear (6), considerably less is currently known regarding its function in these neurons. The methyl cytosine-binding protein MeCP2, which also copurifies with histone deacetylase complexes, is believed to be associated with transcriptional repression. Humans lacking functional MeCP2 develop Rett syndrome, which shows a predominantly neuronal phenotype (32). This observation is somewhat surprising considering the ubiquitous localization of the protein. As would be predicted, mouse Dnmt1 mutants show gross hypomethylation (33). In mosaic Dnmt mutant mice whose brains contain 30% hypomethylated CNS cells, those mutant cells that were hypomethylated are quickly eliminated from the brain after birth. This finding suggests that the hypomethylated neurons are functionally compromised and selected against at certain postnatal stages. In a second approach designed to gain insight into Dnmt1 function in brain, this same group restricted the Dnmt1 knockout to postmitotic neurons (34). When these mice were examined for their response to ischemic brain injury, they found that reduced levels of Dnmt1 correlated with neuroprotection. Our in vitro data support the hypothesis that the observed up-regulation of Dnmt1 in GABAergic neurons (6) may be responsible for the observed decrease in reelin and GAD67 mRNA levels observed in the schizophrenia brain (13).
Is Dnmt1 an active regulator of gene expression in postmitotic neurons? In other words, is methylation a dynamic switch that can be activated or reversed once neurons receive the requisite signals? For this to be the case, active demethylation would have to occur as well. One potential candidate for the demethylase reaction is MBD2 (35). MBD2 binds to methylated DNA with high affinity and has been shown to copurify to an extent with the macromolecular histone deacetylase NuRD complex (36). MBD2 imparts novel properties to the NuRD complex that allow it to interact with methylated DNA (35). If MBD2 demethylates methylated cytosines, one might speculate that the association of the NuRD complex, which facilitates transcriptional repression, with MBD2 would subsequently mediate dissociation, hence allowing transcription to proceed. Consistent with this speculation is a report, in which recombinant MBD2 and the histone deacetylase inhibitor valproic acid were introduced into nondividing cells to show that an artificially methylated template becomes demethylated (37). We have recently shown that valproic acid reverses the methylation pattern of the reelin promoter in cortices of mice treated simultaneously with methionine (38). Whether the beneficial effects of valproic acid in the treatment of mood disorders are related to its histone deacetylase activity is still unclear. Nevertheless, this mechanism is in line with the findings presented herein. Our studies raise the interesting possibility that active demethylation occurs in neurons, consistent with the concept that methylation may be reversible. Moreover, our data suggest the possibility that agents that actively promote demethylation may prove beneficial in the context of psychiatric disorders.
Acknowledgments
We thank Dr. Shoji Tajima (Osaka University, Osaka) for the generous gift of Dnmt1 antibody; Andre Goffinet (University Louvain Medical School, Brussels) for the generous gift of mouse monoclonal reelin G10; and Dr. Francine M. Benes (Laboratories for Structural Neuroscience, McLean Hospital, Belmony, MA, and Program in Neuroscience, Harvard Medical School, Boston), Dr. Edward G. Jones (Center for Neuroscience, University of California, Davis), and Dr. Bryan L. Roth (Department of Biochemistry, Case Western Reserve University Medical School, Cleveland) for constructive criticism and suggestions in the preparation of the manuscript. This work was supported by National Institutes of Mental Health Grants MH62682 (to D.R.G.), MH062188 (to A.G.), and MH062090 (to E.C.).
Abbreviations: Dnmt, DNA methyltransferase; En, embryonic day n; GAD, glutamic acid decarboxylase; IR, immunoreactivity; NeuN, neuronal nuclear antibody; NSE, neuron-specific enolase.
References
- 1.Petronis, A. (2001) Trends Genet. 17, 142–146. [DOI] [PubMed] [Google Scholar]
- 2.Costa, E., Grayson, D. R., Mitchell, C. P., Tremolizzo, L., Veldic, M. & Guidotti, A. (2003) Crit. Rev. Neurobiol. 15, 121–142. [DOI] [PubMed] [Google Scholar]
- 3.Robertson, K. D. (2001) Nat. Rev. Genet. 1, 11–19. [DOI] [PubMed] [Google Scholar]
- 4.Tsujita, T., Niikawa, N., Yamashita, H., Imamura, A., Hamada, A., Nakane, Y. & Okazaki, Y. (1998) Am. J. Psychiatry 155, 422–424. [DOI] [PubMed] [Google Scholar]
- 5.Petronis, A., Gottesman, I. I., Kan, P., Kennedy, J. L., Basile, V. S., Paterson, A. D. & Popendikyte, V. (2003) Schizophr. Bull. 29, 169–178. [DOI] [PubMed] [Google Scholar]
- 6.Veldic, M., Caruncho, H. J., Liu, W. S., Davis, J., Satta, R., Grayson, D. R., Guidotti, A. & Costa, E. (2004) Proc. Natl. Acad. Sci. USA 101, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akbarian, S., Kim, J. J., Potkin, S. G., Hagman, J. O., Tafazzoli, A., Bunney, W. E., Jr., & Jones, E. G. (1995) Arch. Gen. Psychiatry 52, 258–266. [DOI] [PubMed] [Google Scholar]
- 8.Benes, F. M. & Berretta, S. (2001) Neuropsychopharmacology 25, 1–27. [DOI] [PubMed] [Google Scholar]
- 9.Lewis, D. A., Volk, D. W. & Hashimoto, T. (2004) Psychopharmacology (Berlin) 174, 143–150. [DOI] [PubMed] [Google Scholar]
- 10.Wassef, A., Baker, J. & Kochan, L. D. (2003) J. Clin. Psychopharmacol. 23, 601–640. [DOI] [PubMed] [Google Scholar]
- 11.Woo, T.-U. W., Walsh, J. P. & Benes, F. M. (2004) Arch. Gen. Psychol. 61, 649–657. [DOI] [PubMed] [Google Scholar]
- 12.Impagnatiello, F., Guidotti, A. R., Pesold, C., Dwivedi, Y., Caruncho, H., Pisu, M. G., Uzunov, D. P., Smalheiser, N. R., Davis, J. M., Pandey, G. N., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guidotti, A., Auta, J., Davis, J. M., DiGiorgi-Gerenini, V., Dwivedi, J., Grayson, D. R., Impagnatiello, F., Pandey, G. N., Pesold, C., Sharma, R. F., et al. (2000) Arch. Gen. Psychiatry 57, 1061–1069. [DOI] [PubMed] [Google Scholar]
- 14.Fatemi, S. H., Kroll, J. L. & Stary, J. M. (2001) NeuroReport 12, 3209–3215. [DOI] [PubMed] [Google Scholar]
- 15.Eastwood, S. L. & Harrison, P. J. (2003) Mol. Psychiatry 8, 821–831. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto, T., Volk, D. W., Eggan, S. M., Mirnics, K., Pierri, J. N., Sun, Z., Sampson, A. R. & Lewis, D. A. (2003) J. Neurosci. 23, 6315–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong, E., Caruncho, H., Liu, W. S., Smalheiser, N. R., Grayson, D. R., Costa, E. & Guidotti, A. (2003) Proc. Natl. Acad. Sci. USA 18, 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glantz, L. A. & Lewis, D. A. (2000) Arch. Gen. Psychiatry 57, 65–73. [DOI] [PubMed] [Google Scholar]
- 19.Lewis, D. A., Pierri, J. N., Volk, D. W., Melchitzky, D. S. & Woo, T.-U. W. (1999) Biol. Psychiatry 46, 616–626. [DOI] [PubMed] [Google Scholar]
- 20.Inano, K., Suetake, I., Ueda, T., Miyake, Y., Nakamura, M., Okada, M. & Tajima, S. (2000) J. Biochem. (Tokyo) 128, 315–321. [DOI] [PubMed] [Google Scholar]
- 21.Costa, E., Chen, Y., Davis, J., Dong, E., Noh, J. S., Tremolizzo, L., Veldic, M., Grayson, D. R. & Guidotti, A. (2002) Mol. Interv. 2, 47–57. [DOI] [PubMed] [Google Scholar]
- 22.Chen, Y., Sharma, R., Costa, R. H., Costa, E. & Grayson, D. R. (2002) Nucleic Acids Res. 3, 2930–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tremolizzo, L., Carboni, G., Ruzicka, W. B., Mitchell, C. P., Sugaya, I., Tueting, P., Sharma, R., Grayson, D. R., Costa, E. & Guidotti, A. (2002) Proc. Natl. Acad. Sci. USA 99, 10795–17100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noh, J. S. & Gwag, B. J. (1997) Exp. Neurol. 146, 604–608. [DOI] [PubMed] [Google Scholar]
- 25.Noh, J. S., Kim, E. Y., Kang, J. S., Kim, H. R., Oh, Y. J. & Kwag, B. J. (1999) Exp. Neurol. 159, 217–224. [DOI] [PubMed] [Google Scholar]
- 26.Manev, H., Caredda, S. & Grayson, D. R. (1991) NeuroReport 2, 589–592. [DOI] [PubMed] [Google Scholar]
- 27.DeBergeyck, V., Nakajima, K., Lambert de Rouvroit, C., Naerhuyzen, B., Goffinet, A. M., Miyata, T., Ogawa, M. & Mikoshiba, K. (1997) Mol. Brain Res. 50, 85–90. [DOI] [PubMed] [Google Scholar]
- 28.Grayson, D. R. & Ikonomovic, S. (1998) in Neuromethods, eds. Boulton, A. A., Baker, G. B. & Bateson, A. (Humana, Clifton, NJ), Vol. 33, pp. 127–151. [Google Scholar]
- 29.Zuccotti, M. & Monk, M. (1995) Nat. Genet. 9, 316–320. [DOI] [PubMed] [Google Scholar]
- 30.Szyf, M. (2002) Methods 27, 184–191. [DOI] [PubMed] [Google Scholar]
- 31.El-Osta, A. (2003) BioEssays 25, 1071–1084. [DOI] [PubMed] [Google Scholar]
- 32.Tucker, K. L. (2001) Methylated Neuron 30, 649–652. [DOI] [PubMed] [Google Scholar]
- 33.Fan, G., Beard, C., Chen, R. Z., Csankovszki, G., Sun, Y., Siniaia, M., Biniszkiewicz, D., Bates, B., Lee, P. P., Kuhn, R., et al. (2001) J. Neurosci. 21, 788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Endres, M., Fan, G., Meisel, A., Dirnagl, U. & Jaenisch, R. (2001) NeuroReport 12, 3763–3766. [DOI] [PubMed] [Google Scholar]
- 35.Bowen, N. J., Fujita, N., Kajita, M. & Wade, P. A. (2004) Biochim. Biophys. Acta 1677, 52–57. [DOI] [PubMed] [Google Scholar]
- 36.Feng, Q. & Zhang, Y. (2001) Genes Dev. 15, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Detich, N., Bovenzi, V. & Szyf, M. (2003) J. Biol. Chem. 278, 27586–27592. [DOI] [PubMed] [Google Scholar]
- 38.Tremolizzo, L., Doueiri, M. S., Dong, E., Grayson, D. R., Davis, J., Pinna, G., Tueting, P., Rodriguez-Menendez, V., Costa, E. & Guidotti, A., Biol. Psychiatry, in press. [DOI] [PubMed]