Abstract
NADH regulates the release of calcium from the endoplasmic reticulum by modulation of inositol 1,4,5-trisphosphate receptors (IP3R), accounting for the augmented calcium release of hypoxic cells. We report selective binding of IP3R to GAPDH, whose activity leads to the local generation of NADH to regulate intracellular calcium signaling. This interaction requires cysteines 992 and 995 of IP3R and C150 of GAPDH. Addition of native GAPDH and NAD+ to WT IP3R stimulates calcium release, whereas no stimulation occurs with C992S/995S IP3R that cannot bind GAPDH. Thus, the IP3R/GAPDH interaction likely enables cellular energy dynamics to impact calcium signaling.
Keywords: calcium, hypoxia, metabolism
Inositol 1,4,5-trisphosphate (IP3) is a major second messenger molecule that binds to the IP3 receptor (IP3R), releasing intracellular endoplasmic reticulum Ca2+ stores (1, 2). Whereas the IP3R is 2,749 amino acids, the portion responsible for its Ca2+ release activity (IP3 binding domain and the calcium ion channel) comprises only ≈550 amino acids of the entire protein (1, 2). The large intervening region of IP3R contains sites for numerous binding proteins and small molecules that regulate IP3 release of Ca2+. Hence, IP3R is an integrator protein calibrating intracellular Ca2+ signaling in response to diverse stimuli.
Cells need to coordinate signaling with metabolic demands. ATP, the major component of metabolism, has already been demonstrated to regulate IP3R. At physiological levels, ATP enhances IP3-mediated calcium flux as demonstrated with purified IP3R in lipid vesicles (3), lipid bilayers (4), or permeabilized cells (5) and is thought to inhibit IP3R function when cellular ATP levels are decreased (6). In conditions where oxidative respiration is inhibited (e.g., hypoxia), cellular NADH levels are amplified (7). We showed that NADH stimulation of IP3R calcium release mediates hypoxic mobilization of calcium (8). In hypoxic PC12 cells and cerebellar Purkinje neurons, rapid increases in internal Ca2+ derived from IP3-sensitive stores were demonstrated to be directly regulated by GAPDH-derived NADH. However, it was not clear whether global increases in NADH elicited by GAPDH were responsible for this activity or whether such a signal was localized. In this study we demonstrate that GAPDH physiologically binds IP3R and discretely delivers NADH, eliciting calcium release. The GAPDH/IP3R link is a means whereby cellular energetics can regulate Ca2+ signaling.
Materials and Methods
IP3R and GAPDH Mutagenesis. Site-directed IP3R mutations were introduced by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) on a rat His-tagged WT IP3R and confirmed by sequencing. C150S GAPDH construct was generously provided by Akira Sawa and Makoto Hara (The Johns Hopkins University School of Medicine).
Culture of Cells. Human embryonic kidney 293 cells and COS-7 cells (passage numbers 5–25) were cultured as described in refs. 9 and 10.
Yeast Two-Hybrid System. Experiments were performed by using the Matchmaker 3 yeast two-hybrid system (Clontech) with all IP3R fragments cloned into the pGBKT7-binding domain vector by using EcoRI/SalI restriction sites and all GAPDH fragments cloned into the pGADT7 vector by using EcoRI/XhoI restriction sites. Expression was confirmed by Western blotting by using antibodies from Clontech. Positive transformants were selected on -Leu/-Trp/-Ade/-His plates containing 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside.
Calcium Release Measurements. Calcium release through recombinant type I (SII+)IP3R or mutant IP3R was measured as described in ref. 10. When GAPDH was added, cofactors were added according to Sigma's specifications for GAPDH activity.
Heparin Pull-Down. Purified IP3R (50 nM) and GAPDH (50 nM) were incubated in lysis buffer (150 mM NaCl/50 mM Tris, pH 7.8/1% Triton X-100/1 mM EDTA) with a 50-μl bed volume of heparin beads for 1 h at 4°C. Samples were then washed 10 times with 20 volumes of lysis buffer and prepared for Western analysis.
Coimmunoprecpitation. Coimmunoprecipitation of α-myc- and α-histidine-tagged antibodies was performed as described in ref. 9.
Lipid Vesicle Assay Containing IP3R Purified Rat Cerebellum. This assay was performed as described in ref. 6 except the uptake buffer contained 200 nM Ca2+ in addition to the 45Ca2+. When GAPDH was added to this assay, cofactors were added according to Sigma's specifications for GAPDH activity.
Antibodies and Reagents. Plasmids were obtained from the following sources: Matchmaker 3 yeast two-hybrid kit and MYC-tagged vector cDNA were from Clontech; anti-His antibody, anti-MYC antibody, purified GAPDH, NAD+, NADH, purified glyceraldehyde-3-phosphate and IP3 were from Sigma; polyclonal anti-IP3R was generated in our laboratory; and monoclonal GAPDH antibody was from Chemicon.
Results
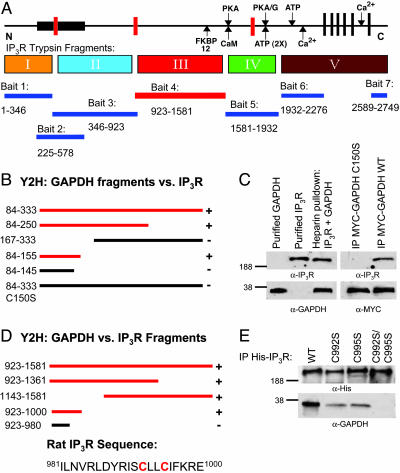
We performed yeast two-hybrid analysis for the entire rat type 1 IP3R as described in ref. 11. Using a bait comprising amino acids 923-1581, we identify numerous clones coding for GAPDH (Fig. 1A). We have mapped the portion of GAPDH that binds IP3R to 11 amino acids (145–155) including the critical catalytic cysteine-150 of GAPDH (Fig. 1B). Mutation of this cysteine to serine abolishes binding. We have confirmed the interactions of GAPDH and IP3R by demonstrating direct in vitro binding of the two purified proteins as well as coimmunoprecipitation in intact cells (Fig. 1C).
Fig. 1.
GAPDH binds to the N terminus of IP3R. (A) Schematic depiction of the functional domains of the IP3R, including the ligand-binding (bait 2), modulatory and transmembrane regions, and trypsin digest (I–V) domains. The seven regions used as bait in the yeast two-hybrid (Y2H) screen also are indicated below the protein and the corresponding amino acids (rat type 1 sequence). When GAPDH was screened against all IP3R baits, only bait 4 supported growth on selective media. (B) Y2H for GAPDH-binding sites. Red bars depict IP3R baits capable of interacting with GAPDH. (C Left) Purified full-length WT IP3R and GAPDH in vitro binding demonstrated by Western analysis. (C Right) Coimmunoprecipitation experiments from human embryonic kidney 293 lysates of endogenous IP3R and exogenously expressed myc-tagged GAPDH WT and C150S visualized by Western blot analysis. (D) Y2H for IP3R binding sites. Red bars depict IP3R baits capable of interacting with GAPDH. (E) Western blot analysis with anti-GAPDH antibody demonstrates IP3R mutants with impaired or abolished binding to GAPDH. Full-length His-tagged WT and mutant IP3Rs were expressed in COS-7 cells and purified on nickel bead columns.
Conversely, we mapped the sequence of IP3R that binds GAPDH to amino acids 981–1000 (Fig. 1D). This sequence contains two cysteines (C992 and C995) which could confer a disulfide linkage with GAPDH C150. To test this possibility, we mutated these two cysteines in full length IP3R (Fig. 1E). Mutation of either C992 or C995 to serine diminishes binding, whereas mutation of both abolishes binding.
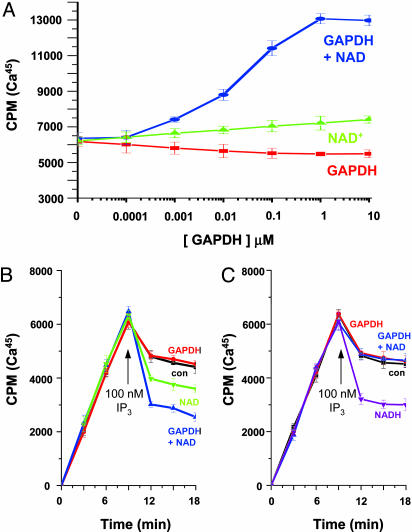
The binding of GAPDH to IP3R is precise and physiological, suggesting that GAPDH could synthesize NADH close to the channel, thus regulating discrete Ca2+ signaling. To examine this possibility, we monitored calcium flux of purified IP3R protein reconstituted into lipid vesicles as described in ref. 6. Although GAPDH alone is ineffective, the combination of NAD+ and GAPDH doubles the activity of IP3R in response to IP3 (Fig. 2A). Half-maximal augmentation of Ca2+ flux occurs at ≈10 nM GAPDH, which may reflect its binding affinity for IP3R. NAD+ (100 μM) alone produces only a small increase in Ca2+ release. In our earlier study, 50 μM NADH produced half-maximal increases in calcium flux, whereas NAD+ had negligible effects at 100 μM (8). The ability of 10 nM GAPDH with 100 μM NAD+ to elicit the same increase in calcium flux as 50 μM NADH is a 5,000-fold amplification and likely reflects the catalytic activity of the enzyme.
Fig. 2.
Local NADH production through GAPDH regulates IP3R calcium release. (A) Ca45 uptake into lipid vesicles containing purified IP3R. Vesicle uptake in the presence of 200 nM Ca2+ and 100 nM IP3 at varying concentrations of either GAPDH alone (red), NAD+ alone (green), or GAPDH + NAD+ (blue). (B and C) Ca45 uptake and release from COS-7 crude WT (B) or C992S/C995S (C)IP3R-containing microsomes in the presence of oxalate. IP3 (100 nM) was added (arrow) to either control microsomes (black), microsomes with 1 μM GAPDH (red), 1 mM NAD+ (green), 1 μM GAPDH and 1 mM NAD+ (blue), or 500 μM NADH (purple).
We next examined crude microsomal membrane preparations from COS-7 cells transfected with either WT IP3R or C992S/C995S IP3R that cannot bind GAPDH (Fig. 2 B and C). The addition of GAPDH and NAD+ to WT IP3R causes maximal vesicular Ca2+ release at submaximal IP3 concentrations, whereas GAPDH alone has no effect. In contrast with the purified vesicle data, NAD+ alone causes significant calcium release. This result could reflect endogenous GAPDH associated with the IP3R. Consistent with this hypothesis, C992S/C995S IP3R does not respond to the addition of NAD+/GAPDH but remains able to release calcium in the presence of NADH. This finding establishes that binding of GAPDH to IP3R is required for the enzyme to synthesize NADH to alter local IP3R calcium flux.
Discussion
The main finding of this study is that GAPDH physiologically binds to IP3R-delivering NADH in close proximity to the channel, thus regulating intracellular Ca2+ signaling. This conclusion is supported by several key findings, including abolition of the GAPDH augmentation of Ca2+ release by selective blockade of IP3R/GAPDH binding. Locally generated NADH regulating IP3R is reminiscent of the scaffolding protein PSD95 linking neuronal nitric oxide synthase to the glutamate NMDA receptor for its nitrosylation and activation (12). Perhaps other small signaling molecules, created by spatially targeted and effector-coupled enzymes, provide analogous local regulation.
Our initial study characterizing NADH stimulation of IP3R-mediated Ca2+ release (8) focused on the pathophysiological augmentation of Ca2+ release by hypoxia. The GAPDH/IP3R complex appears perfectly positioned to facilitate cellular death in response to inhibition of oxidative respiration in the mitochondria. Increases in glycolysis due to inhibition of oxidative respiration (13) could augment GAPDH activity, sensitizing the IP3R for activation. Inhibition of oxidative respiration leads to mitochondrial depolarization and release of cytochrome c (11), which also sensitizes the IP3R for activation. These two processes may work coordinately in close proximity to mitochondria to increase cytosolic Ca2+ during programmed cell death.
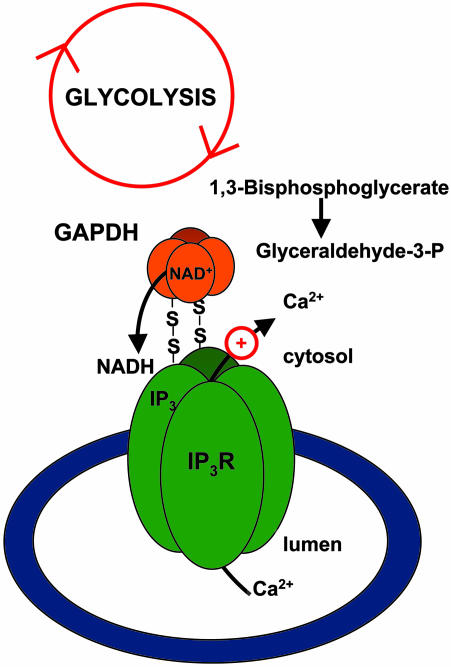
Our finding that GAPDH is physiologically bound to IP3R suggests that changes in the formation of NADH during normal alterations in cellular physiology may also regulate Ca2+ signaling through IP3R (Fig. 3). This concept is supported by numerous studies showing that alterations of glycolytic enzymes can impact cytosolic Ca2+ levels (14–16). Physiologic intracellular NADH levels are ≈1–10 μM (7), whereas half-maximal augmentation of IP3R calcium release occurs at 50 μM NADH. This result suggests that changes in GAPDH activity can modify local NADH levels within physiologically relevant ranges and likely regulate IP3R activity in response to the myriad of signals that occur during respiratory metabolism.
Fig. 3.
GAPDH creates local NADH for IP3R regulation. This cartoon depicts a mechanism whereby GAPDH may regulate IP3R Ca2+ function. Because both the IP3R and GAPDH both work as tetramers, we hypothesize that two noncatalytic GAPDH subunits bind through C150 to individual IP3R subunits through C992 or C995. This complex leaves two catalytic subunits of GAPDH positioned in direct proximity with the IP3R. The glycolysis-mediated activation of GAPDH causes increased local NADH, which, in the presence of IP3, stimulates IP3R activity. Released Ca2+ may enter mitochondria to enhance oxidative respiration (physiological) or release cytochrome c (pathophysiological) (data not shown).
Acknowledgments
We thank Louis Dang and Sungjin Park for experimental support and Makoto Hara and Dr. Akira Sawa for unpublished reagents and helpful discussions. This work was supported by the U.S. Public Health Service Grant MH-18501 and Center Grant MH68830 (to S.H.S.) and the National Research Service Award NH65090 (to R.L.P.).
Abbreviations: IP3, inositol 1,4,5-trisphosphate; IP3R, inositol 1,4,5-trisphosphate receptor.
References
- 1.Patterson, R. L., Boehning, D. & Snyder, S. H. (2004) Annu. Rev. Biochem. 73, 437-465. [DOI] [PubMed] [Google Scholar]
- 2.Berridge, M. J., Lipp, P. & Bootman, M. D. (2000) Nat. Rev. Mol. Cell Biol. 1, 11-21. [DOI] [PubMed] [Google Scholar]
- 3.Bezprozvanny, I. & Ehrlich, B. E. (1993) Neuron 10, 1175-1184. [DOI] [PubMed] [Google Scholar]
- 4.Kaznacheyeva, E., Lupu, V. D. & Bezprozvanny, I. (1998) J. Gen. Physiol. 111, 847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maes, K., Missiaen, L., Parys, J. B., De Smet, P., Sienaert, I., Waelkens, E., Callewaert, G. & De Smedt, H. (2001) J. Biol. Chem. 276, 3492-3497. [DOI] [PubMed] [Google Scholar]
- 6.Ferris, C. D., Huganir, R. L. & Snyder, S. H. (1990) Proc. Natl. Acad. Sci. USA 87, 2147-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veech, R. L., Lawson, J. W., Cornell, N. W. & Krebs, H. A. (1979) J. Biol. Chem. 254, 6538-6547. [PubMed] [Google Scholar]
- 8.Kaplin, A. I., Snyder, S. H. & Linden, D. J. (1996) J. Neurosci. 16, 2002-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson, R. L., van Rossum, D. B., Barrow, R. K. & Snyder, S. H. (2004) Proc. Natl. Acad. Sci. USA 101, 2328-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boehning, D. & Joseph, S. K. (2000) J. Biol. Chem. 275, 21492-21499. [DOI] [PubMed] [Google Scholar]
- 11.Boehning, D., Patterson, R. L., Sedaghat, L., Glebova, N. O., Kurosaki, T. & Snyder, S. H. (2003) Nat. Cell Biol. 5, 1051-1061, and erratum (2004) 6, 77. [DOI] [PubMed] [Google Scholar]
- 12.Christopherson, K. S., Hillier, B. J., Lim, W. A. & Bredt, D. S. (1999) J. Biol. Chem. 274, 27467-27473. [DOI] [PubMed] [Google Scholar]
- 13.Duffy, T. E., Nelson, S. R. & Lowry, O. H. (1972) J. Neurochem. 19, 959-977. [DOI] [PubMed] [Google Scholar]
- 14.Bertram, R., Satin, L., Zhang, M., Smolen, P. & Sherman, A. (2004) Biophys. J. 87, 3074-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh, P., Salih, M., Leddy, J. J. & Tuana, B. S. (2004) J. Biol. Chem. 279, 35176-35182. [DOI] [PubMed] [Google Scholar]
- 16.Juntti-Berggren, L., Webb, D. L., Arkhammar, P. O., Schultz, V., Schweda, E. K., Tornheim, K. & Berggren, P. O. (2003) J. Biol. Chem. 278, 40710-40716. [DOI] [PubMed] [Google Scholar]