ABSTRACT
Coxiella burnetii is the causative agent of Q fever, a zoonotic disease that threatens both human and animal health. Due to the paucity of experimental animal models, little is known about how host factors interface with bacterial components and affect pathogenesis. Here, we used Drosophila melanogaster, in conjunction with the biosafety level 2 (BSL2) Nine Mile phase II (NMII) clone 4 strain of C. burnetii, as a model to investigate host and bacterial components implicated in infection. We demonstrate that adult Drosophila flies are susceptible to C. burnetii NMII infection and that this bacterial strain, which activates the immune deficiency (IMD) pathway, is able to replicate and cause mortality in the animals. We show that in the absence of Eiger, the only known tumor necrosis factor (TNF) superfamily homolog in Drosophila, Coxiella-infected flies exhibit reduced mortality from infection. We also demonstrate that the Coxiella type 4 secretion system (T4SS) is critical for the formation of the Coxiella-containing vacuole and establishment of infection in Drosophila. Altogether, our data reveal that the Drosophila TNF homolog Eiger and the Coxiella T4SS are implicated in the pathogenesis of C. burnetii in flies. The Drosophila/NMII model mimics relevant aspects of the infection in mammals, such as a critical role of host TNF and the bacterial T4SS in pathogenesis. Our work also demonstrates the usefulness of this BSL2 model to investigate both host and Coxiella components implicated in infection.
KEYWORDS: Q fever, innate immunity, IMD, TNF, Eiger, NMII, pathogenesis, T4SS, tumor necrosis factor
INTRODUCTION
Coxiella burnetii is an obligate intracellular Gram-negative bacterium and the causative agent of the zoonosis Q fever (1). Acute C. burnetii infection in humans is characterized primarily by influenza-like symptoms and pneumonia. Domestic ruminants act as reservoir hosts of C. burnetii and have been implicated in several outbreaks of Q fever worldwide (1–3). Based on morbidity, low infectious dose, and the environmental stability of the organism, the U.S. Centers for Disease Control and Prevention (CDC) has designated C. burnetii a category B biological weapon agent (4). C. burnetii presents two antigenic forms: a pathogenic phase I variant and an attenuated phase II variant that has a truncated O chain in its lipopolysaccharide (5, 6). C. burnetii phase I is associated with Q fever, whereas phase II does not cause disease in immunocompetent hosts (7–9). The Nine Mile phase II (NMII) clone 4/RSA439 is an attenuated strain of C. burnetii derived from the virulent Nine Mile phase I (NMI) strain through repeated passages in embryonated eggs (5). Although attenuated in immunocompetent hosts, the NMII strain has been shown to be virulent to SCID mice (10) and to cause fever in gamma interferon knockout (IFN-γ−/−) and Toll-like receptor 2 knockout (TLR2−/−) mice (11). Because C. burnetii NMI and NMII strains present similar replication kinetics in tissue culture models, the NMII strain has been used as a safer option for investigating Coxiella pathogenesis in vitro (12–17). Recent studies using C. burnetii NMII have revealed that the bacterial type 4 secretion system (T4SS) and its secreted components are Coxiella virulence factors (13, 15, 17, 18).
In order to better address Coxiella-host interactions, a reliable immunocompetent-host model suitable for biosafety level 2 (BSL2) is needed as an alternative, since animal models that utilize virulent phase I strains require BSL3 facilities. Despite recent progress in understanding Coxiella pathogenesis, host mechanisms associated with the control of infection and bacterial factors implicated in replication and establishment of infection remain largely unknown. Although C. burnetii has been detected in tick populations worldwide (19–21), the role of ticks in the epidemiology of Q fever remains unclear (22). A recent study demonstrated that C. burnetii has emerged from Coxiella-like endosymbiont organisms found in ticks, revealing evidence of how the bacterium evolved from arthropods to infect mammalian cells by the acquisition of virulence factors (23). Another study used larvae of the greater wax moth, Galleria mellonella, to investigate antibiotic efficacy following Coxiella infection and the role of dotA/dotB, two components of the Coxiella T4SS, in establishing infection (24). The wax moth model revealed relevant information on antimicrobials and Coxiella biology in arthropods; however, this host system lacks the genetic malleability found in other models, such the recently described Caenorhabditis elegans nematode model (25) or the arthropod Drosophila melanogaster. Thus, a genetically tractable arthropod model that supports Coxiella replication would be useful in addressing the host immune response induced by Coxiella infection and the bacterial factors implicated in the formation of the Coxiella-containing vacuole (CCV) leading to the establishment of infection.
The fruit fly D. melanogaster is a powerful, genetically malleable model for studying host-pathogen interactions and innate immunity (26–29), bolstered by the fact that nearly 75% of human genes implicated in disease have a functional homolog in flies (30). Secretion of antimicrobial peptides (AMPs), melanization, and the phagocytic activity of hemocytes are the primary innate immune mechanisms that the flies use to combat infection (27, 31). Activation of intracellular immune pathways following infection leads to the expression of AMPs, which are small cationic molecules that disrupt pathogen homeostasis. Activation of the immune deficiency (IMD) pathway, primarily by Gram-negative bacterial infection, leads to the expression of the AMPs Drosocin, Diptericin, Cecropin, and Attacin. The Toll pathway is primarily activated by fungi and Gram-positive bacteria, resulting in the expression of the AMPs Drosomycin and Defensin (32–35). Immune signaling pathways are evolutionarily conserved among species, and the Drosophila IMD and Toll pathways show similarities to the mammalian tumor necrosis factor (TNF) and Toll-like receptor pathways, respectively (31). In addition to the IMD and Toll pathways, Drosophila Eiger, the only known TNF homolog in flies, is also activated during bacterial infection and influences host pathology and susceptibility to infection (36–40). It has been shown that Eiger contributes to the pathology induced by infection with Salmonella enterica serovar Typhimurium (37). Brandt et al. proposed that Salmonella secreted factors stimulated an Eiger-mediated immune response that is detrimental to both the bacterium and host. Interestingly, Drosophila Eiger mutants were significantly more susceptible to extracellular pathogens than wild-type flies (38). This study suggested that the Eiger-mediated immune response aided in the clearance of extracellular pathogens; however, mortality from intracellular-pathogen challenge was unchanged or reduced in Eiger mutants. Drosophila has also been used to reveal virulence factors associated with Francisella tularensis pathogenesis (34), gut immunocompetence during Pseudomonas entomophila infection (41), and phagocytic activity during Mycobacterium marinum infection (32). Taking the data together, the use of Drosophila to investigate bacterial pathogenesis and host immune responses identifies key signaling mechanisms that may lead to the development of novel therapeutics designed to control infection in natural hosts.
In this study, we used D. melanogaster as a model to reveal both host and bacterial factors implicated in the pathogenesis of C. burnetii. We demonstrate that adult Drosophila flies are susceptible to the NMII clone 4 strain of C. burnetii and that the strain is able to replicate in adult flies. While the IMD pathway was activated following infection, bacterial growth was affected only by the loss of the IMD transcription factor Relish. We also show that Eiger mutant flies display reduced mortality to C. burnetii, correlated with increased levels of the antimicrobial peptide Drosocin. Finally, our results show that the T4SS is an essential factor for the establishment of Coxiella infection in the animals. Altogether, we demonstrate that Drosophila is a novel animal model to investigate Coxiella infection and the host immune response.
RESULTS
C. burnetii replicates in Drosophila hemocyte-derived S2 cells.
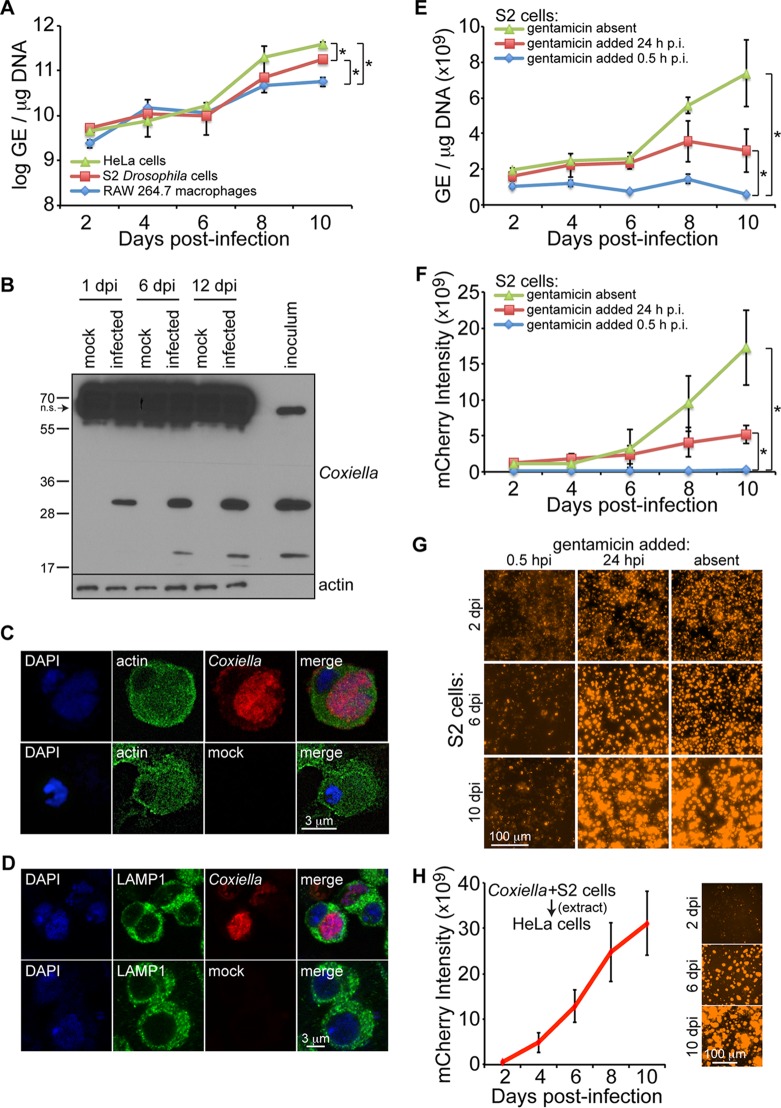
To address the question of whether Drosophila would be a suitable model to study Coxiella pathogenesis, we first investigated the ability of C. burnetii to infect and grow in Drosophila hemocyte-derived S2 cells in comparison to human HeLa cells and RAW264.7 mouse macrophages. Over a period of 2 to 8 days postinfection, no significant difference in bacterial genome equivalents (GE) was observed among Drosophila S2 cells, mouse RAW 267.4 macrophages, and HeLa cells. However, by 10 days postinfection, HeLa cells contained significantly higher GE levels than S2 cells, while macrophages had significantly lower GE levels than both S2 and HeLa cells (Fig. 1A). Coxiella antigens were detected in infected S2 cells at days 1, 6, and 12 postinfection, as demonstrated by immunoblotting using a rabbit anti-Coxiella polyclonal antibody and by observing specific bands at ∼20 and ∼30 kDa (Fig. 1B). Prominent nonspecific banding was also observed at ∼60 kDa, demonstrating that while the antibody is useful for Western blotting with Drosophila, it may not be useful for immunohistochemistry or immunoprecipitation experiments with Drosophila samples. Successful colonization of mammalian cells by Coxiella requires the formation of a specialized CCV (12, 16, 17). Confocal microscopy performed at 10 days postinfection in S2 cells infected with mCherry-expressing C. burnetii revealed the presence of a single large CCV (Fig. 1C). In addition, we monitored CCV formation using lysosomal-associated membrane protein 1 (LAMP1) as a marker for the late lysosome. LAMP1 surrounded the CCV at 4 days postinfection (Fig. 1D); however, to ascertain if LAMP1 is recruited to the vacuole, experiments to visualize the vacuolar membrane and determine if LAMP1 signal is enriched need to be performed. Nevertheless, these results indicate that C. burnetii is able to infect and replicate in a single large vacuole inside Drosophila hemocyte-derived S2 cells.
FIG 1.
C. burnetii replicates in Drosophila hemocyte-derived S2 cells. (A) Cells were infected (MOI = 100 GE/cell), and comparative growth kinetics of C. burnetii in insect and mammalian cells were determined by qPCR. The results are presented as log GE per microgram of DNA. (B) Immunoblotting detection of C. burnetii antigens in Drosophila S2 cells at 1, 6, and 12 days postinfection (dpi) using a rabbit polyclonal antibody against Coxiella. Nonspecific (n.s.) banding in S2 cell lysates is denoted by the arrow. (C and D) Drosophila hemocyte-derived S2 cells were infected with C. burnetii expressing mCherry and prepared for confocal microscopy at 4 dpi. Nuclei were stained with DAPI and actin (C) or LAMP1 (D). (E to G) A gentamicin protection assay was performed to evaluate the growth of mCherry-expressing C. burnetii in Drosophila S2 cells. (E and F) At the indicated times postinfection, total DNA was collected to determine GE levels (E) or mCherry intensity was measured at five different locations of three independent wells at the indicated time points postinfection (F). (G) Representative images for each condition at 2, 6, and 10 days postinfection. (H) mCherry-expressing Coxiella was isolated from infected S2 cells and used to infect HeLa cells at an MOI of 100 GE/cell. The intensity of mCherry was measured over the course of 10 days, and representative images are shown. The asterisks denote statistical significance (*, P < 0.05). The error bars indicate standard deviations.
C. burnetii is considered an obligate intracellular bacterium, but its ability to grow in vitro in the absence of host cells has been recently demonstrated (42). Thus, we next performed a gentamicin protection assay to investigate the ability of C. burnetii to grow within Drosophila S2 cells. The gentamicin assay was performed using mCherry-expressing C. burnetii, and bacterial growth was monitored by measuring GE and mCherry intensity. Drosophila S2 cells were infected with mCherry-expressing C. burnetii (multiplicity of infection [MOI] = 100 GE/cell) in the absence of gentamicin, and then the antibiotic was added at 0.5 h or 24 h postinfection. At 10 days postinfection, significant bacterial growth was observed in cultures lacking gentamicin or to which gentamicin was added 24 h postinfection compared to cultures to which the antibiotic was added 0.5 h postinfection (Fig. 1E and F), indicating intracellular growth of C. burnetii in S2 cells. Representative mCherry images are shown in Fig. 1G. The lack of bacterial growth when gentamicin was added 0.5 h postinfection suggests that complete binding and invasion of Coxiella in S2 cells occurs within the first 30 min of infection. Finally, to determine if bacteria grown in Drosophila S2 cells remain infectious to mammalian cells, at 10 days postinfection, infected S2 cells were pelleted by centrifugation, followed by Dounce homogenization to lyse the S2 cells. The bacterial GE were quantified, and 100 GE/cell was used to infect HeLa cells. The level of mCherry signal was then measured over the course of 10 days (Fig. 1H). Taken together, these results show that C. burnetii replicates inside S2 cells in the presence of gentamicin-containing medium and is able to reinfect mammalian cells.
C. burnetii induces the expression of antimicrobial peptides in Drosophila S2 cells.
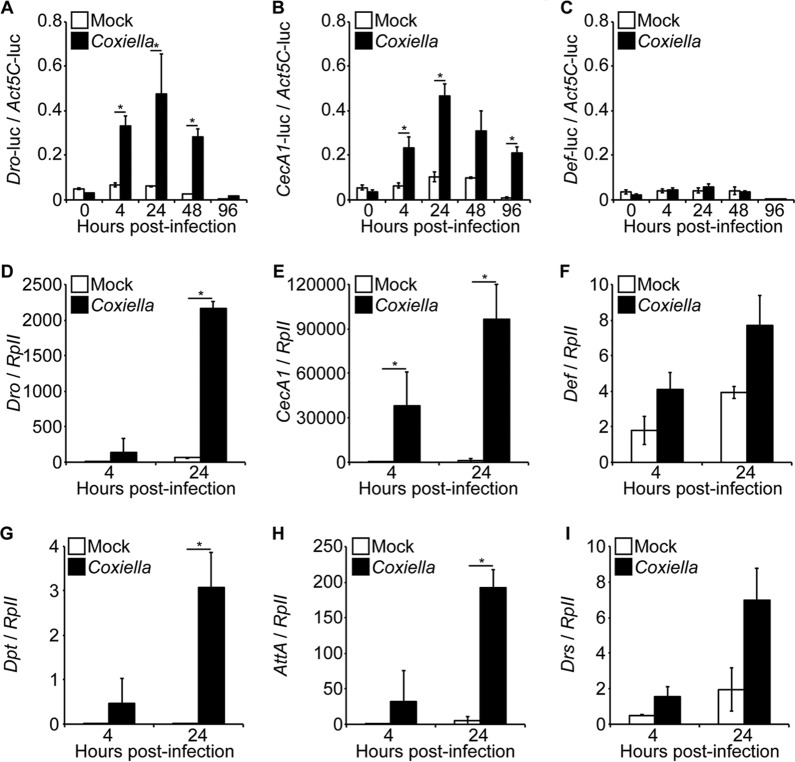
After demonstrating that C. burnetii infects and replicates in Drosophila S2 cells, we investigated its ability to induce an immune response in insect cells. Initially, we performed a luciferase reporter assay to investigate the activation of AMP promoters in infected insect cells. The results indicated significant activity of the CecropinA1 and Drosocin promoters (Fig. 2A and B). No significant activation of the Defensin promoter was observed (Fig. 2C). Next, we investigated AMP expression in S2 cells infected with C. burnetii. Similar to the promoter assay, Drosocin was significantly induced following Coxiella infection (Fig. 2D). Additionally, CecropinA1, Diptericin, and AttacinA (Fig. 2E, G, and H), AMP genes also regulated by the IMD pathway, were significantly upregulated in infected cells compared to uninfected controls. No significant upregulation of Defensin and Drosomycin (Fig. 2F and I), Toll pathway-specific AMPs, was observed by comparing infected and uninfected cells. These results indicate that an IMD-specific innate immune response in Drosophila S2 cells was activated upon infection with C. burnetii.
FIG 2.
C. burnetii induces the expression of AMPs in S2 cells. (A to C) A luciferase reporter assay was performed to investigate the activation of the Drosocin (Dro) (A), CecropinA1 (CecA1) (B), and Defensin (Def) (C) AMP promoters in S2 cells following infection. At 24 h posttransfection, the cells were infected with C. burnetii (MOI = 100 GE/cell), and luciferase (luc) activity was assessed at different times postinfection. The firefly luciferase activity of each sample was normalized to Actin5C-driven Renilla luciferase activity to correct for transfection efficiency. (D to I) Drosophila S2 cells were infected with C. burnetii (MOI = 100 GE/cell), and total RNA was collected at 4 h and 24 h postinfection to examine AMP expression. Gene expression levels for Drosocin (D), CecropinA1 (E), Defensin (F), Diptericin (G), AttacinA (H), and Drosomycin (I) were determined by qRT-PCR. The relative expression of AMP was normalized to Drosophila RpII. The asterisks denote statistical significance (*, P < 0.05). The error bars indicate standard deviations.
Adult Drosophila flies are susceptible to C. burnetii.
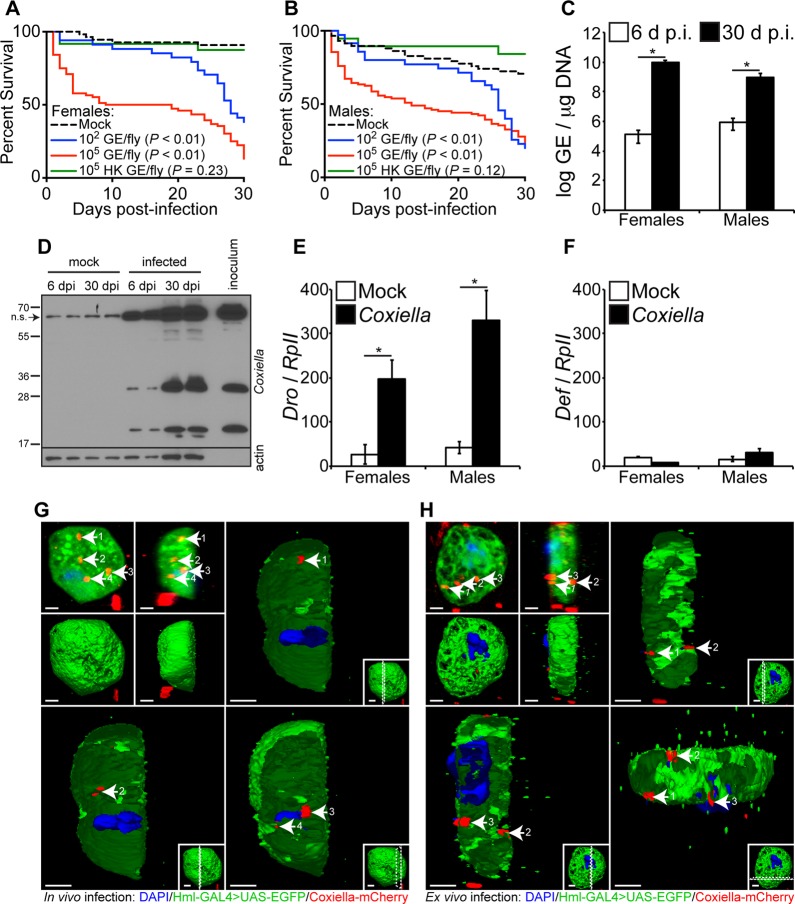
In vitro results using Drosophila S2 cells allowed us to frame a rationale for in vivo experiments using adult flies. Therefore, our next goal was to investigate the susceptibility of adult Drosophila flies to C. burnetii. Four-day-old Oregon-R flies were infected with live (102 or 105 GE/fly) or heat-killed (HK) (105 HK GE/fly) bacteria, and mortality was evaluated for a period of 30 days. The results demonstrated that both females and males are susceptible to infection (Fig. 3A and B). Mortality was dose dependent in females (P < 0.01), but not in males (P = 0.95). The data also showed that both male and female flies were resistant to HK C. burnetii, suggesting that mortality is associated with the presence of live bacteria (Fig. 3A and B, green curves).
FIG 3.
Adult Drosophila flies are susceptible to C. burnetii and elicit a host immune response. (A and B) Four-day-old Oregon-R female (A) and male (B) Drosophila flies were infected with live (102 or 105 GE/fly) or HK (105 GE/fly) C. burnetii, and survival was evaluated for 30 days. (C) Four-day-old adult Oregon-R flies were infected with C. burnetii (100 GE/fly), and bacterial levels were determined at 6 and 30 days postinfection by quantitative real-time PCR. (D) C. burnetii antigens were detected in infected flies at 6 and 30 days postinfection, as shown by immunoblotting using a rabbit polyclonal antibody against Coxiella. Biological duplicates are shown. Nonspecific (n.s.) banding from fly homogenates is denoted by the arrow. (E and F) Antimicrobial peptide levels of Drosocin (E) and Defensin (F) were determined in Oregon-R adults infected with C. burnetii (100 GE/fly) at 12 days postinfection. (G and H) Confocal microscopy showing mCherry-Coxiella invasion of hemocytes (white arrows) derived from 3rd-instar larvae infected in vivo (G) or ex vivo (H). The hemocytes expressed GFP, and the nuclei were stained with DAPI. Bars = 2 μm. Numbers by arrows designate the same Coxiella-mCherry signal among images in the same panel. Dotted lines in insets represent where the cross-section is made. The asterisks denote statistical significance (*, P < 0.05). The error bars indicate standard deviations.
After showing the susceptibility of Drosophila to C. burnetii, we investigated if the bacterial strain was able to replicate in adult flies. Four-day-old male and female Drosophila flies were infected with 100 GE/fly, and bacterial growth was investigated by quantitative real-time PCR (qPCR) and immunoblotting. A significant increase in Coxiella GE was observed from day 6 to day 30 postinfection in both female and male flies (Fig. 3C). In addition, Coxiella antigens were detected in infected flies at days 6 and 30 postinfection, as demonstrated by immunoblotting (Fig. 3D). Similar to the immunoblot from S2 cells, a nonspecific band was observed using adult flies. Collectively, these results indicate that C. burnetii is able to infect and replicate in adult Drosophila flies.
Next, we investigated the innate immune response elicited by C. burnetii infection in adult Drosophila flies. Expression of Drosocin and Defensin is a marker for the activation of the IMD and Toll pathways, respectively. Therefore, we determined the pattern of expression of Drosocin and Defensin in 4-day-old female and male flies infected with C. burnetii (100 GE/fly). The results demonstrated that Drosocin was significantly upregulated 12 days postinfection in females and males compared to controls (Fig. 3E). No significant upregulation of Defensin was observed in females and males (Fig. 3F). These results suggest that the IMD pathway mediates the innate immune response of adult flies to C. burnetii. Collectively, the results demonstrate that adult flies are susceptible to C. burnetii and that the bacterial strain is able to replicate in flies, despite the activation of the IMD pathway.
Since previous results indicated that Drosophila hemocyte-derived S2 cells, as well as Drosophila animals, were capable of being infected with Coxiella and exhibited host responses, we next asked whether hemocytes isolated from the animals were capable of being infected with Coxiella. To this end, we utilized flies carrying the reporter Hml-GAL4;UAS-EGFP (upstream activation sequence-enhanced green fluorescent protein), which causes hemocytes to express green fluorescent protein (GFP). Third-instar larvae were infected with mCherry-Coxiella, and 24 h postinfection, hemocytes were extracted from the animals and processed for confocal microscopy (Fig. 2G). Imaging showed that Coxiella infects hemocytes in vivo. Additionally, hemocytes were extracted from third-instar larvae and subsequently infected with mCherry-Coxiella (Fig. 2H), showing that hemocytes can be infected ex vivo.
Drosophila mutants for PGRP-LC and Relish are more susceptible to C. burnetii.
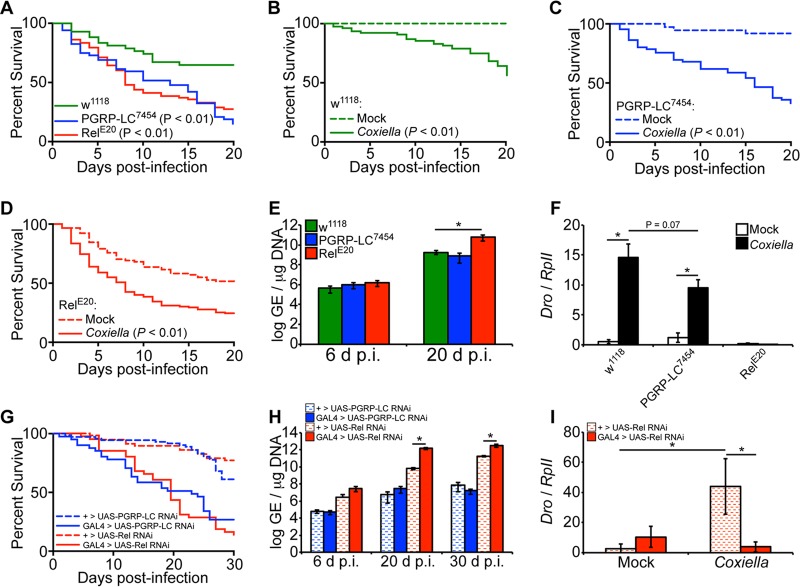
Considering that the IMD pathway is activated during infection, we next investigated the susceptibility of Drosophila containing loss-of-function peptidoglycan recognition protein (PGRP) LC (PGRP-LC) or the transcription factor Relish. While PGRP-LC is the upstream receptor that initiates the IMD pathway during bacterial infection, Relish is the downstream IMD transcription factor that ultimately leads to induction of AMPs. Flies containing point mutations in PGRP-LC, PGRP-LC7454 (43), and in Relish, RelE20 (44), were used in these experiments. The results showed that PGRP-LC7454 and RelE20 flies exhibited increased susceptibility to C. burnetii compared to control w1118 flies (Fig. 4A). Control w1118, PGRP-LC7454, and RelE20 flies were also significantly more susceptible (P < 0.01) to infection with C. burnetii (100 GE/fly) than their respective mock-infected controls (Fig. 4B to D). Interestingly, by 20 days postinfection, we observed a significant increase in bacterial load only in Relish mutant flies, but not PGRP-LC mutant flies, compared to w1118 flies (Fig. 4E). We then investigated the levels of expression of Drosocin in infected PGRP-LC7454 and RelE20 flies to evaluate the role of the AMP in susceptibility. Control w1118 and PGRP-LC7454 flies expressed significantly higher levels of Drosocin at 12 days postinfection than in mock infection, while RelE20 flies did not exhibit induction of Drosocin compared to mock infection (Fig. 4F). To further corroborate these results, we utilized transgenic flies carrying RNA interference (RNAi) cassettes for PGRP-LC or Relish, ubiquitously driven by Actin5C-GAL4. Compared to control flies lacking the Actin5C-GAL4 driver, both PGRP-LC and Relish knockdown flies exhibited increased mortality, and the bacterial load was significantly increased only in Relish RNAi flies (Fig. 4G and H). Similar to the results in Relish mutant flies, Relish RNAi flies exhibited reduced levels of Drosocin expression compared to controls (Fig. 4I). Finally, we performed partial-rescue experiments for the PGRP-LC and Relish mutations by crossing each of the mutant lines with the w1118 control line. Compared to flies carrying homozygous mutations in PGRP-LC or Relish, flies that were heterozygous for the mutations exhibited decreased mortality during C. burnetii infection (see Fig. S1 in the supplemental material). Altogether, the data indicate that flies with a loss-of-function mutation in PGRP-LC or Relish are more susceptible to C. burnetii than control flies, suggesting that the presence of a functional IMD pathway protects the animals from mortality during infection. Additionally, decreased expression of Drosocin in infected loss-of-function or knockdown Relish flies was correlated with an increased bacterial load.
FIG 4.
Drosophila PGRP-LC7454 and RelE20 mutants are more susceptible to C. burnetii NMII clone 4. (A to F) Adult w1118, PGRP-LC7454, and RelE20 male flies, 4 days of age, were mock infected or infected with C. burnetii (100 GE/fly). Percent survival was evaluated for a period of 20 days, comparing infected flies to one another (A) or mock- and Coxiella-infected flies for each genotype (B to D). (E) Bacterial loads were determined at 6 and 20 days postinfection by qPCR. (F) Expression of Drosocin in w1118, PGRP-LC7454, and RelE20 flies was determined at 12 days postinfection by reverse transcriptase quantitative real-time PCR, and the results were normalized to the Drosophila RpII transcripts. (G to I) Four-day-old sibling adult flies carrying a UAS-induced dsRNA cassette targeting Relish (TRiP.HMS00070) or PGRP-LC (TRiP.HMS00259) with an Actin5C-driven GAL4 element (GAL4 > UAS) or lacking the GAL4 element (+ > UAS) were infected with C. burnetii (100 GE/fly). (G) Percent survival was evaluated for a period of 30 days. (H) The bacterial loads were determined at 6, 20, and 30 dpi by qPCR. (I) Expression of Drosocin was determined at 12 dpi. The asterisks denote statistical significance (*, P < 0.05). The error bars indicate standard deviations.
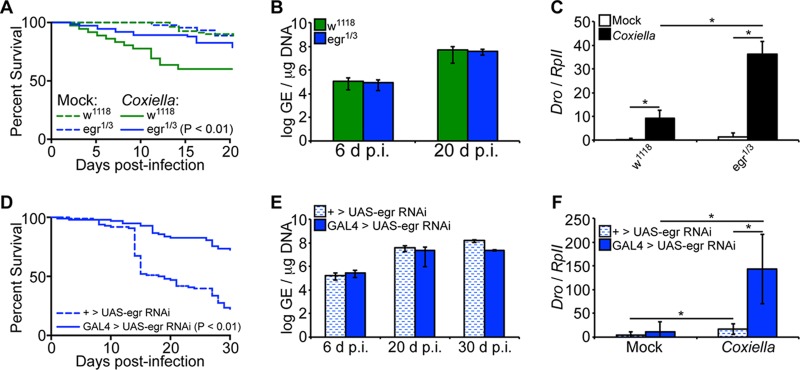
Eiger-deficient Drosophila flies are less susceptible to C. burnetii.
It has been demonstrated in mammals that the pathogenesis of Coxiella is associated, in part, with overexpression of proinflammatory cytokines, such as TNF-α and interleukin 1β (IL-1β) (45). While no Drosophila homologs have been identified for IL-1β, Eiger has been identified as the only known TNF superfamily ligand homolog in the flies (36). Therefore, we next infected Eiger mutant Drosophila flies with C. burnetii to investigate the underlying mechanism of susceptibility. Eiger mutant males, egr1/3, a cross between the point mutation Eiger mutants egr1 and egr3 previously described (36), were infected with C. burnetii NMII clone 4 (100 GE/fly), and mortality was evaluated for a period of 20 days. No significant mortality was observed in the Coxiella-infected egr1/3 flies compared to mock-infected controls (P = 0.26), but infected Eiger mutant flies showed decreased mortality compared to infected control w1118 flies (Fig. 5A). Interestingly, no significant difference in the bacterial load was observed between w1118 and Eiger mutant flies (Fig. 5B), suggesting a dissociation between mortality and bacterial load. Nevertheless, levels of Drosocin expression were significantly upregulated (P < 0.05) in Eiger mutant flies at 12 days postinfection compared to control w1118 flies (Fig. 5C), indicating activation of the IMD pathway in Eiger mutant flies. To support these results, we utilized transgenic flies carrying an RNAi cassette for Eiger, ubiquitously driven by Actin5C-GAL4. Compared to control flies lacking the Actin5C-GAL4 driver, Eiger knockdown flies exhibited reduced mortality yet similar levels of bacterial load (Fig. 5D and E). Similar to the results in egr1/3 flies, Eiger knockdown flies exhibited increased levels of Drosocin induction compared to control flies (Fig. 5F). Finally, levels of Eiger induction were not significantly altered during C. burnetii infection (see Fig. S2 in the supplemental material), suggesting that the effects of Eiger on the host during infection are posttranscriptional, similar to those observed during S. Typhimurium infection (37). Collectively, the data indicate that Eiger mutant flies are more resistant to C. burnetii infection. Considering the absence of mortality in infected Eiger mutant flies and the fact that no difference in bacterial load was observed between Eiger mutant and wild-type flies, the data suggest that Eiger mutant flies were able to limit the impact of infection and display tolerance for C. burnetii infection.
FIG 5.
Eiger mutant Drosophila flies display tolerance for C. burnetii. (A to C) Adult w1118 and Eiger mutant (egr1/3) male flies, 4 days of age, were mock infected or infected with C. burnetii (100 GE/fly). (A) Mortality was significantly increased (P < 0.01) in w1118 flies compared to Eiger mutant flies. (B) Coxiella GE was quantified at 6 and 20 days postinfection by qPCR. (C) Levels of Drosocin were measured in Eiger mutant flies and control w1118 flies at 12 days postinfection. (D to F) Four-day-old sibling adult flies carrying a UAS-induced dsRNA cassette targeting Eiger (TRiP.HMC03963) with an Actin5C-driven GAL4 element (GAL4 > UAS-egr RNAi) or lacking the GAL4 element (+ > UAS-egr RNAi) were infected with C. burnetii (100 GE/fly). (D) Percent survival was evaluated for a period of 30 days. (E) Bacterial loads were determined at 6, 20, and 30 days postinfection by qPCR. (F) Expression of Drosocin was determined at 12 days postinfection. The asterisks denote statistical significance (*, P < 0.05). The error bars indicate standard deviations.
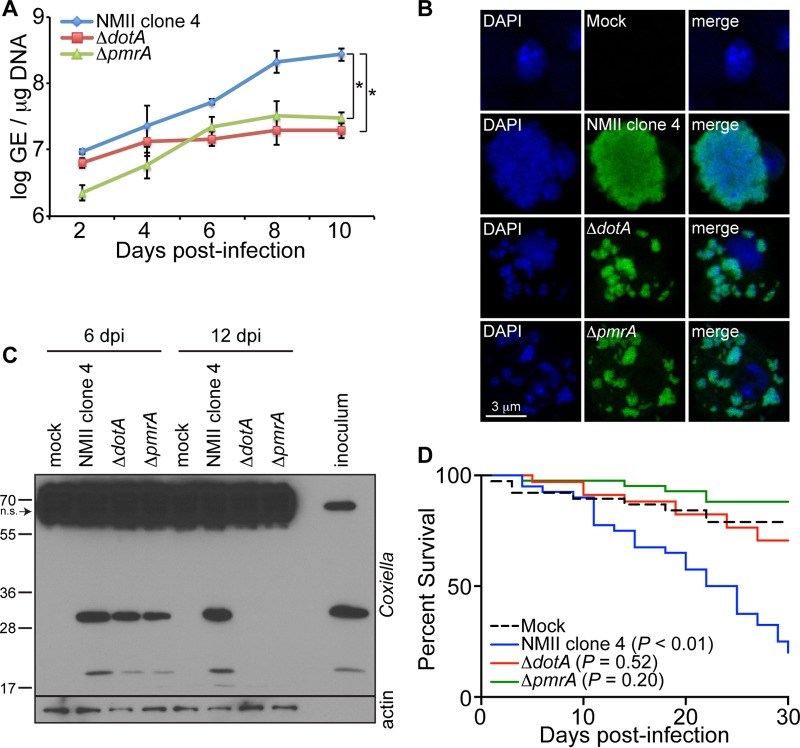
The T4SS of Coxiella is implicated in the establishment of infection in Drosophila.
Successful colonization of mammalian cells by C. burnetii requires the formation of a single CCV that is actively controlled by the bacterial T4SS and its secreted factors (46–48). Consequently, the T4SS and its secreted factors have been described as a novel virulence factor of Coxiella in mammals (16, 17, 49). We used Drosophila S2 cells and adult flies to investigate the role of the T4SS during C. burnetii infection. We infected S2 cells with the control background strain of C. burnetii (NMII clone 4) or the ΔdotA or ΔpmrA mutant and evaluated bacterial growth. While dotA encodes structural components of the T4SS, pmrA acts as a regulatory element for the proper expression of Dot/Icm genes (16, 17). At 10 days postinfection, both ΔdotA and ΔpmrA mutants exhibited reduced levels in S2 cells compared to the control background strain (Fig. 6A). However, ΔpmrA growth from baseline was comparable to that of the control strain. The differences in GE observed between the ΔdotA and ΔpmrA mutants may be due to the fact that DotA is a structural component of the T4SS while PmrA is a regulatory factor. Additionally, fluorescence microscopy of S2 cells infected with GFP-expressing C. burnetii or mutant bacteria revealed that the ΔdotA and ΔpmrA mutants were localized in small, dispersed vacuoles, which is in contrast to infection with the control background strain, which localizes in a single large vacuole (Fig. 6B). In addition, Coxiella antigens were not detected in S2 cells infected with ΔdotA and ΔpmrA mutants by 12 days postinfection, as demonstrated by immunoblotting (Fig. 6C). Together these results show that a pathogen-specific process, namely, the T4SS, is required for the formation of a large CCV and sustained bacterial replication. To investigate the role of the T4SS in vivo, we infected adult Oregon-R flies with the C. burnetii background strain or the ΔdotA or ΔpmrA mutant (100 GE/fly), and mortality was evaluated. The flies succumbed to infection with the background strain; however, no significant mortality was observed in flies infected with the ΔdotA or ΔpmrA mutant, indicating that the T4SS is critical for the establishment of infection in vivo in arthropods (Fig. 6D). While Fig. 6A shows that the ΔpmrA mutant is less infectious at early time points in S2 cells and grows as rapidly as the control strain, it does not grow in a single large vacuole, intracellular antigens are ultimately cleared, and it does not cause mortality in wild-type flies, similar to ΔdotA. Altogether, considering that ΔdotA and ΔpmrA mutants do not have a functional T4SS, our results indicate that this Coxiella secretion system is essential for the formation of the CCV and establishment of infection in arthropods.
FIG 6.
The C. burnetii type 4 secretion system is essential for establishment of infection in Drosophila. (A) S2 cells were infected with NMII clone 4 or the ΔdotA or ΔpmrA mutant (MOI = 100 GE/cell), and bacterial growth was assessed by qPCR. (B) S2 cells were infected with NMII clone 4 or the ΔdotA and ΔpmrA mutant expressing GFP. CCV formation was observed by confocal microscopy at 6 days postinfection. (C) Coxiella antigens were examined in S2 cells by immunoblotting using an anti-Coxiella polyclonal antibody at 6 and 12 days following infection with NMII clone 4 or the ΔdotA and ΔpmrA mutants. Nonspecific (n.s.) banding in S2 cell lysates is denoted by the arrow. (D) Four-day-old adult Oregon-R flies were infected with 100 GE/fly of NMII clone 4 or the ΔdotA or ΔpmrA mutant, and mortality was monitored for 30 days. The asterisks denote statistical significance (*, P < 0.05). The error bars indicate standard deviations.
DISCUSSION
In the present study, we describe the use of D. melanogaster as a model to investigate host and bacterial factors implicated in C. burnetii infection. By using this model, we demonstrate that adult flies are susceptible to the BSL2 Nine Mile phase II clone 4 strain of C. burnetti. We also show that this Coxiella strain replicates in flies, despite the activation of the IMD pathway, a canonical immune pathway of Drosophila implicated in the control of infection with Gram-negative bacteria. Our data indicate that Eiger, a Drosophila TNF superfamily homolog, contributes to mortality of adult flies infected with C. burnetii and that Eiger mutant flies are less susceptible to infection. We also demonstrate that the Coxiella T4SS is essential for CCV formation and the establishment of infection in the Drosophila model.
A variety of animal models have been used to investigate the pathogenesis of Coxiella, including mice, guinea pigs, and nonhuman primates (50–55). Considering that immunocompetent hosts are resistant to the Coxiella phase II strains, most animal studies require the use of phase I virulent strains, which requires BSL3 facilities (4). Other animal and avirulent bacterial models, particularly those suitable for BSL2, represent safer alternatives to investigate how host and bacterial factors interface and affect the pathogenesis of C. burnetii. Here, we present Drosophila as a genetically tractable host model to study Coxiella infection that complements previous work performed in mammalian and other invertebrate models (23–25). The malleability of the C. elegans and Drosophila models makes them applicable to studies in mammalian systems, and Drosophila can be used to identify novel arthropod genetic variants implicated in susceptibility to C. burnetii infection that have homologous mammalian counterparts. An additional advantage of this model shown in our study is that wild-type immunocompetent Drosophila flies succumb to C. burnetii NMII clone 4, the only strain of Coxiella exempt from BSL3 regulations. Therefore, Drosophila, in conjunction with C. burnetii, emerges as an in vitro and in vivo system to study both host and bacterial factors implicated in infection.
The Drosophila IMD signaling pathway is activated in flies to respond to infection with Gram-negative bacteria (27). Bacterial peptidoglycans are sensed by PGRPs, such as PGRP-LC and PGRP-LE. This signal activates the IMD pathway nuclear factor Relish, which translocates to the nucleus, leading to the expression of AMPs, particularly Drosocin, Cecropin, Attacin, and Diptericin (43, 56). Here, we show that C. burnetii activates the IMD pathway, which led to significant induction of Drosocin in infected cells and adult flies. It has been demonstrated that continuous activation of AMPs under the control of the IMD signaling pathway leads to relative resistance to F. tularensis, a facultative intracellular Gram-negative bacterium that is closely related to C. burnetii (57). The study demonstrated that flies defective in the IMD pathway succumb rapidly to Francisella infection (34). Here, we show similar results, as the PGRP-LC7454 and RelE20 mutant flies showed significantly more susceptibility to infection than control w1118 flies. However, the Coxiella load was affected only in Relish mutants, which was correlated with a significant decrease in Drosocin expression in Relish mutant and RNAi flies. Previously, it was shown that Francisella is sensitive to Drosophila AMPs and grows to higher titers in Relish mutant flies (34). Similarly, we found that the Coxiella load was increased in Relish mutant flies, which exhibited a loss of AMP expression. However, PGRP-LC mutant flies did not exhibit increased bacterial loads or as significant a decrease in AMP induction. This suggests that once Coxiella is replicating intracellularly, the PGRP-LC pathway is less active and Coxiella activates Relish for subsequent AMP induction through an alternative mechanism.
It has been shown that phase I and phase II strains of C. burnetii show similar growth rates in mammalian cells (12–17). We demonstrate that C. burnetii shows growth kinetics in Drosophila S2 cells similar to those in HeLa cells and mouse macrophages. Drosophila hemocytes have been shown to be an appropriate, genetically amenable model for analyzing phagosome maturation (58). Localization of LAMP1 in Leishmania-containing vacuoles has been shown in Drosophila S2 cells, confirming that the fly cells maintain the Leishmania parasite within compartments that share characteristics of phagolysosomes, as previously shown in mammalian cells (59, 60). In Coxiella infection of mammalian cells, following internalization, the nascent CCV proceeds through the default endocytic pathway and ultimately fuses with the lysosomal compartment (61). The mature CCV is then decorated with late vacuolar markers, such as Rab7, LAMP1, LAMP2, and LAMP3, and autophagosome markers, such as LC3 and Rab24 (61–65). We show that, similar to mammalian cells, LAMP1 surrounds the CCV at 4 days postinfection, suggesting that the default endocytic pathway of infected Drosophila S2 cells was not disturbed by infection. Further experimentation is needed to determine definitively if LAMP1 is recruited to the CCV membrane. Our results also validate Drosophila S2 cells as a hemocyte system to investigate the intracellular trafficking of Coxiella in arthropod cells.
The Drosophila host factor Eiger, the only known TNF homolog in Drosophila, contributes to pathology induced during infection with S. Typhimurium (38). Eiger activates the JNK pathway and induces the expression of apoptosis genes implicated in the susceptibility of flies to infection (36–38). We found that the Drosophila Eiger mutants did not succumb to infection with C. burnetii, suggesting that the TNF homolog may contribute to pathogenesis and consequently mortality in the fly model. It was shown that knocking down Eiger expression in the fat body leads to an increase in survival after S. Typhimurium infection, but it had no effect on the bacterial load, indicating an increase in host tolerance (39). Interestingly, no difference in Coxiella growth was observed in Eiger mutant flies compared to control w1118 flies, indicating dissociation between mortality and bacterial growth in these animals. Two recent studies have shown that in mouse bone marrow-derived macrophages there is production of TNF (66, 67). Additionally, cells lacking TLR2 or its downstream signaling components exhibited reduced TNF production and increased levels of Coxiella (66). While we did not observe induction of Eiger during infection, we observed an increase in Drosocin induction in Eiger mutant flies compared to the control flies, perhaps as a compensatory mechanism, similar to that observed during S. Typhimurium infection (38). Together, our data suggest that Eiger mutant flies were able to limit pathogenesis by becoming tolerant of C. burnetii, which was associated with increased AMP induction.
In mammalian cells, the T4SS system and its secreted factors are required for intracellular replication of C. burnetii (13, 16, 17, 47). It was also recently shown that the Coxiella T4SS is required for bacterial replication in hemocytes of the greater wax moth, G. mellonella (24). Here, we expanded that knowledge by demonstrating that ΔdotA and ΔpmrA mutants, both of which lack a functional T4SS, do not establish a productive infection in vitro and in vivo in the Drosophila model. Our results indicate that following infection, the T4SS mutants locate in small, dispersed vacuoles inside Drosophila hemocyte-derived S2 cells. In contrast, wild-type bacteria form a single large intracellular CCV. Taken together, these data indicate that the Coxiella T4SS is essential for efficient formation of the CCV in arthropod cells, as previously described for mammalian cells (13, 16, 17, 47).
In conclusion, this work demonstrates the usefulness of D. melanogaster as a novel model to investigate host and bacterial components implicated in Coxiella infection. Our results using Drosophila corroborated relevant aspects of Coxiella infection previously shown in G. mellonella, C. elegans, and tick cells (24–26), such as CCV formation and the role of the T4SS in replication in an arthropod model. Using adult flies, we were able to demonstrate that the Drosophila TNF homolog, Eiger, is implicated in susceptibility to infection. We also demonstrated that Eiger mutant flies were able to tolerate high levels of C. burnetii, similar to the levels in control flies, while exhibiting increased survival and AMP induction similar to that observed during S. Typhimurium infection (38). Thus, Drosophila serves as a valuable genetically tractable model for investigating host and bacterial mechanisms associated with pathogenesis and the control of infection. This model is applicable and complementary to studies in mammalian systems to decipher the host response and life cycle of Coxiella in the arthropod host.
MATERIALS AND METHODS
C. burnetii, insect cells, and mammalian cells.
Wild-type and GFP- or mCherry-expressing C. burnetii NMII clone 4 RSA439 bacteria, generous gifts from Robert A. Heinzen (Rocky Mountain Laboratories, NIH, Hamilton, MT), were propagated in acidified citrate cysteine medium 2 as previously described (42). C. burnetii mutants for dotA (ΔdotA) and pmrA (ΔpmrA), encoding two components of the bacterial T4SS, expressing GFP were also provided by R. A. Heinzen. The mutant strains were propagated as previously described (16, 17). All Coxiella infections utilized the avirulent NMII clone 4 strain, which is exempt from the U.S. CDC select agent regulations and suitable for work at BSL2. C. burnetii stocks were quantified by measuring bacterial GE using qPCR as previously described (12, 68). The rabbit polyclonal antibody against Coxiella phase II antigens that was used for immunoblots in this study was provided by R. A. Heinzen. Drosophila hemocyte-derived S2 cells were maintained at 28°C in tissue culture flasks containing Schneider's Drosophila medium (Gibco, Waltham, MA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (HyClone), 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 0.25 μg/ml of amphotericin B (Fungizone) antimycotic (Life Technologies, Waltham, MA). RAW267.4 mouse macrophages and HeLa cells (ATCC) were maintained at 37°C and 5% CO2 in tissue culture flasks containing Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% heat-inactivated FBS, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 0.25 μg/ml of amphotericin B antimycotic.
Bacterial infections in vitro.
S2 cells were plated at 2 × 105 cells per well in 24-well plates containing Schneider's Drosophila medium with 10% FBS without antibiotic, and RAW267.4 mouse macrophages or HeLa cells were plated at 105 cells per well in 24-well plates containing DMEM-10% FBS without antibiotic. The following day, the cells were infected with C. burnetii (MOI = 100 GE/cell) in DMEM containing 2% FBS without antibiotics. The insect and mammalian cells were collected at different time points postinfection to investigate bacterial growth by qPCR, as previously described (12, 68). Total RNA from the insect cells was also collected at different time points postinfection for quantitative reverse transcriptase PCR (qRT-PCR) to assess the expression of AMPs. For the gentamicin protection assay, S2 cells were plated at 2 × 105 cells in 96-well plates containing Schneider's Drosophila medium with 10% FBS without antibiotic. The following day, the cells were infected with C. burnetii expressing mCherry (MOI = 100 GE/cell) in Schneider's medium containing 2% FBS without antibiotics. Gentamicin (10 μg/ml; Gibco) was then added at 0.5 h or 24 h postinfection. The gentamicin-containing medium was replaced every 2 days, and bacterial growth was assessed by examining mCherry intensity using a Cytation 3 imaging reader (Biotek, Winooski, VT).
Luciferase reporter assay.
Activation of CecropinA1, Drosocin, and Defensin promoters in S2 cells following C. burnetii infection was performed by luciferase reporter assay. The cells were transiently transfected with the CecropinA1 (69), Drosocin (70), or Defensin (71) promoter cloned into pGL4.10 (Promega), along with the promoter for Actin5C cloned into pRL (Promega, Madison, WI) as an internal transfection control, using Cellfectin II (Life Technologies). Six hours posttransfection, the medium was replaced with fresh growth medium, and 16 h following the medium change, the cells were infected with C. burnetii (MOI = 100 GE/cell) diluted in medium containing 2% FBS, and the firefly luciferase activity was assessed at different time points postinfection.
Confocal microscopy.
The infected cells were fixed for 1 h in 2% formaldehyde, followed by permeabilization for 10 min in 0.1% Triton X-100. The cells were blocked in phosphate-buffered saline (PBS) containing 10% FBS and incubated with antibodies against actin (Sigma A2066) or LAMP-1 (Abcam 30687) for 1 h at room temperature. The cells were washed and incubated with Alexa Fluor-488 (Thermo Fisher A-11008)-conjugated secondary antibodies for 1 h at room temperature. The cells were washed, incubated with DAPI (4′,6-diamidino-2-phenylindole) (Sigma D9542) for 15 min, and mounted onto microscope slides. Images were obtained using a Leica SP8-X White Light Laser point scanning confocal microscope and analyzed using Leica Application Suite X.
D. melanogaster and infections.
The Drosophila w1118, Oregon-R, Hml-EGFP driver (w1118;P{wHml-GAL4.Δ}2,P{UAS-2xEGFP}AH2), Act5C-GAL4 driver (y1w*;P{Act5C-GAL4}25FO1/CyO), tub-GAL4 driver (y1w*;P{tubP-GAL4}LL7/TM3,Sb1,Ser1), Double balancer (w*;KrIf-1/CyO;D1/TM3,Ser1), Relish RNAi (y1v1;;P{yTRiP.HMS00070}attP2), Eiger RNAi (y1sc*v1;P{TRiP.HMC03963}attP40), PGRP-LC RNAi (y1sc*v1;;P{TRiP.HMS00259}attP2), PGRP-LC7454 (43), RelE20 (44), Eiger1, and Eiger3 (36) strains were used in this study. RNAi knockdown was performed using sibling progeny from crosses between the parental Act5C-GAL4 driver line and the corresponding RNAi lines. Progeny flies carrying the CyO balancer were used as control flies. All the fly strains were grown in standard meal agar fly food and maintained at 23°C and 68% humidity. Fly stocks were cleared of Wolbachia infection by feeding two generations with standard fly food containing 0.05 mg/ml tetracycline (Sigma).
Adult flies were injected with live bacteria (102 or 105 GE/fly) or HK (98°C for 1 h) bacteria (105 GE/fly). For injections, flies were anesthetized with CO2 and injected with 23 nl of bacteria or PBS using a pulled 0.53-mm glass needle and an automatic nanoliter injector (Drummond Scientific, Broomall, PA). Individual flies were injected at the ventrolateral surface of the fly thorax and placed into new vials. Unless otherwise noted, adult male flies were used for all experiments. Third-instar larvae from Hml-GAL4>UAS-EGFP flies were infected with mCherry-expressing bacteria using a 0.001-mm tungsten needle while the larvae were in a pool of 109 GE/ml of bacteria for 1 h. Hemocytes were isolated by mechanical dissection as previously described (72). After the injections, the adult flies were monitored daily for mortality and collected at different times postinfection to assess the bacterial load and expression of AMPs. Survival curves were performed using a minimum of 80 flies per condition, including at least two experimental replicates. The bacterial load was determined by qPCR in 3 biological replicates of flies homogenized in PBS as described previously (12, 68).
Expression of antimicrobial peptides.
The relative expression of AMPs in Drosophila S2 cells and in adult flies was determined by qRT-PCR. For S2 cells, total RNA was extracted using the GeneJet RNA purification kit (Thermo Scientific). Samples were treated with DNase I (Invitrogen), and cDNA was synthesized using the iScript Reverse Transcriptase kit (Bio-Rad, Hercules, CA). For adult Drosophila flies, total RNA from infected and uninfected flies was isolated at different time points postinfection from at least 2 biological replicates containing 3 flies in each sample. The flies were homogenized in solution D (4 M guanidinium thiocyanate, 25 mM sodium citrate, 0.5% sarcosyl, 0.1 M 2-mercaptoethanol). RNA extraction, DNase treatment, and cDNA synthesis were performed as described above. Reverse transcriptase quantitative real-time PCR was performed using SsoFast SYBR green PCR master mix (Bio-Rad) in an ABI 7500 Fast thermocycler using 60°C as the annealing temperature. Expression of the following genes was evaluated using the specific primers given in parentheses: Drosomycin (5-CGTGAGAACCTTTTCCAATATGATG-3 and 5-TCCCAGGACCACCAGCAT-3), Diptericin (5-GCTGCGCAATCGCTTCTACT-3 and 5-TGGTGGAGTGGGCTTCATG-3), AttacinA (5-CACAATGTGGTGGGTCAGG-3 and 5-GGCACCATGACCAGCATT-3), Drosocin (5-GCACAATGAAGTTCACCATCGT-3 and 5-CCACACCCATGGCAAAAAC-3), Defensin (5-GCCAGAACGCAGCCACAT-3 and 5-CGGTGTGGTTCCAGTTCCA-3), CecropinA1 (5-GGACAATCGGAAGCTGGTT-3 and 5-TGTGCTGACCAACACGTTC-3), and Eiger (5-GATGGTCTGGATTCCATTGC-3 and 5-TAGTCTGCGCCAACATCATC-3) (37). The Drosophila RNA polymerase II gene (RpII) (5-TTGACGTAAGCATCACCTG-3 and 5-GAAGCGTTTCTCCAAACGAG-3) was utilized as an internal control for gene induction.
Immunoblotting.
Flies were homogenized in radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 1 mM Na3VO4, 1 mM NaF, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μM aprotinin, 5 μg/ml leupeptin, 1 μg/ml pepstatin A). Total protein was determined using the bicinchoninic acid (BCA) assay (Pierce, Waltham, MA). Equal amounts of protein were subjected to SDS-PAGE. The proteins were transferred to a polyvinylidene difluoride (PVDF) membrane and blocked in 5% bovine serum albumin (BSA) in 0.1% Tween 20–Tris-buffered saline. The membrane was incubated with rabbit anti-Coxiella phase II antibody (1:10,000) overnight at 4°C. Antibody-bound proteins were detected using anti-rabbit secondary antibodies conjugated to horseradish peroxidase. The blots were developed by chemiluminescence using luminol enhancer solution (ThermoFisher).
Statistics.
A two-tailed Student t test assuming unequal variance was utilized to compare means of quantitative data. Mortality curves were analyzed by the log-rank (Mantel-Cox) test using GraphPad Prism (GraphPad Software, Inc.).
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael E. Konkel and Laura R. H. Ahlers for critical review of the manuscript. We thank Anders Omsland for propagating Coxiella strains.
This investigation was supported by funds from Washington State University and National Institutes of Health Public Health Service grant R00AI106963 (to A.G.G.).
The contents are solely our responsibility and do not necessarily represent the official views of the NIH.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00218-17.
REFERENCES
- 1.Maurin M, Raoult D. 1999. Q fever. Clin Microbiol Rev 12:518–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schimmer B, Morroy G, Dijkstra F, Schneeberger PM, Weers-Pothoff G, Timen A, Wijkmans C, van der Hoek W. 2008. Large ongoing Q fever outbreak in the south of The Netherlands, 2008. Euro Surveill 13:18939. [PubMed] [Google Scholar]
- 3.Sondgeroth KS, Davis MA, Schlee SL, Allen AJ, Evermann JF, McElwain TF, Baszler TV. 2013. Seroprevalence of Coxiella burnetii in Washington State domestic goat herds. Vector Borne Zoonotic Dis 13:779–783. doi: 10.1089/vbz.2013.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madariaga MG, Rezai K, Trenholme GM, Weinstein RA. 2003. Q fever: a biological weapon in your backyard. Lancet Infect Dis 3:709–721. doi: 10.1016/S1473-3099(03)00804-1. [DOI] [PubMed] [Google Scholar]
- 5.Hackstadt T, Peacock MG, Hitchcock PJ, Cole RL. 1985. Lipopolysaccharide variation in Coxiella burnetti: intrastrain heterogeneity in structure and antigenicity. Infect Immun 48:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoover TA, Culp DW, Vodkin MH, Williams JC, Thompson HA. 2002. Chromosomal DNA deletions explain phenotypic characteristics of two antigenic variants, phase II and RSA 514 (crazy), of the Coxiella burnetii nine mile strain. Infect Immun 70:6726–6733. doi: 10.1128/IAI.70.12.6726-2733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang G, Russell-Lodrigue KE, Andoh M, Zhang Y, Hendrix LR, Samuel JE. 2007. Mechanisms of vaccine-induced protective immunity against Coxiella burnetii infection in BALB/c mice. J Immunol 179:8372–8380. doi: 10.4049/jimmunol.179.12.8372. [DOI] [PubMed] [Google Scholar]
- 8.Andoh M, Russell-Lodrigue KE, Zhang G, Samuel JE. 2005. Comparative virulence of phase I and II Coxiella burnetii in immunodeficient mice. Ann N Y Acad Sci 1063:167–170. doi: 10.1196/annals.1355.026. [DOI] [PubMed] [Google Scholar]
- 9.Andoh M, Naganawa T, Hotta A, Yamaguchi T, Fukushi H, Masegi T, Hirai K. 2003. SCID mouse model for lethal Q fever. Infect Immun 71:4717–4723. doi: 10.1128/IAI.71.8.4717-4723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Islam A, Lockhart M, Stenos J, Graves S. 2013. The attenuated nine mile phase II clone 4/RSA439 strain of Coxiella burnetii is highly virulent for severe combined immunodeficient (SCID) mice. Am J Trop Med Hyg 89:800–803. doi: 10.4269/ajtmh.12-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochoa-Reparaz J, Sentissi J, Trunkle T, Riccardi C, Pascual DW. 2007. Attenuated Coxiella burnetii phase II causes a febrile response in gamma interferon knockout and Toll-like receptor 2 knockout mice and protects against reinfection. Infect Immun 75:5845–5858. doi: 10.1128/IAI.00901-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howe D, Shannon JG, Winfree S, Dorward DW, Heinzen RA. 2010. Coxiella burnetii phase I and II variants replicate with similar kinetics in degradative phagolysosome-like compartments of human macrophages. Infect Immun 78:3465–3474. doi: 10.1128/IAI.00406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, Roy CR. 2014. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog 10:e1004286. doi: 10.1371/journal.ppat.1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez E, Cantet F, Fava L, Norville I, Bonazzi M. 2014. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PLoS Pathog 10:e1004013. doi: 10.1371/journal.ppat.1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larson CL, Beare PA, Voth DE, Howe D, Cockrell DC, Bastidas RJ, Valdivia RH, Heinzen RA. 2015. Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication. Infect Immun 83:661–670. doi: 10.1128/IAI.02763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beare PA, Sandoz KM, Larson CL, Howe D, Kronmiller B, Heinzen RA. 2014. Essential role for the response regulator PmrA in Coxiella burnetii type 4B secretion and colonization of mammalian host cells. J Bacteriol 196:1925–1940. doi: 10.1128/JB.01532-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. 2011. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2:e00175-11. doi: 10.1128/mBio.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carey KL, Newton HJ, Luhrmann A, Roy CR. 2011. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog 7:e1002056. doi: 10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mediannikov O, Fenollar F, Socolovschi C, Diatta G, Bassene H, Molez JF, Sokhna C, Trape JF, Raoult D. 2010. Coxiella burnetii in humans and ticks in rural Senegal. PLoS Negl Trop Dis 4:e654. doi: 10.1371/journal.pntd.0000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halajian A, Palomar AM, Portillo A, Heyne H, Luus-Powell WJ, Oteo JA. 2016. Investigation of Rickettsia, Coxiella burnetii and Bartonella in ticks from animals in South Africa. Ticks Tick Borne Dis 7:361–366. doi: 10.1016/j.ttbdis.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Noda AA, Rodriguez I, Miranda J, Contreras V, Mattar S. 2016. First molecular evidence of Coxiella burnetii infecting ticks in Cuba. Ticks Tick Borne Dis 7:68–70. doi: 10.1016/j.ttbdis.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Duron O, Sidi-Boumedine K, Rousset E, Moutailler S, Jourdain E. 2015. The importance of ticks in Q fever transmission: what has (and has not) been demonstrated? Trends Parasitol 31:536–552. doi: 10.1016/j.pt.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Duron O, Noel V, McCoy KD, Bonazzi M, Sidi-Boumedine K, Morel O, Vavre F, Zenner L, Jourdain E, Durand P, Arnathau C, Renaud F, Trape JF, Biguezoton AS, Cremaschi J, Dietrich M, Leger E, Appelgren A, Dupraz M, Gomez-Diaz E, Diatta G, Dayo GK, Adakal H, Zoungrana S, Vial L, Chevillon C. 2015. The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii. PLoS Pathog 11:e1004892. doi: 10.1371/journal.ppat.1004892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norville IH, Hartley MG, Martinez E, Cantet F, Bonazzi M, Atkins TP. 2014. Galleria mellonella as an alternative model of Coxiella burnetii infection. Microbiology 160:1175–1181. doi: 10.1099/mic.0.077230-0. [DOI] [PubMed] [Google Scholar]
- 25.Battisti JM, Watson LA, Naung MT, Drobish AM, Voronina E, Minnick MF. 2017. Analysis of the Caenorhabditis elegans innate immune response to Coxiella burnetii. Innate Immun 23:111–127. doi: 10.1177/1753425916679255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann JA, Hetru C, Reichhart JM. 1993. The humoral antibacterial response of Drosophila. FEBS Lett 325:63–66. doi: 10.1016/0014-5793(93)81414-U. [DOI] [PubMed] [Google Scholar]
- 27.Lemaitre B, Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu Rev Immunol 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 28.Dionne MS, Schneider DS. 2008. Models of infectious diseases in the fruit fly Drosophila melanogaster. Dis Model Mech 1:43–49. doi: 10.1242/dmm.000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panayidou S, Ioannidou E, Apidianakis Y. 2014. Human pathogenic bacteria, fungi, and viruses in Drosophila: disease modeling, lessons, and shortcomings. Virulence 5:253–269. doi: 10.4161/viru.27524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey UB, Nichols CD. 2011. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol Rev 63:411–436. doi: 10.1124/pr.110.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann JA. 2003. The immune response of Drosophila. Nature 426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 32.Dionne MS, Ghori N, Schneider DS. 2003. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect Immun 71:3540–3550. doi: 10.1128/IAI.71.6.3540-3550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanji T, Hu X, Weber AN, Ip YT. 2007. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol Cell Biol 27:4578–4588. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vonkavaara M, Telepnev MV, Ryden P, Sjostedt A, Stoven S. 2008. Drosophila melanogaster as a model for elucidating the pathogenicity of Francisella tularensis. Cell Microbiol 10:1327–1338. doi: 10.1111/j.1462-5822.2008.01129.x. [DOI] [PubMed] [Google Scholar]
- 35.Vonkavaara M, Pavel ST, Holzl K, Nordfelth R, Sjostedt A, Stoven S. 2013. Francisella is sensitive to insect antimicrobial peptides. J Innate Immun 5:50–59. doi: 10.1159/000342468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, Aigaki T, Miura M. 2002. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J 21:3009–3018. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandt SM, Dionne MS, Khush RS, Pham LN, Vigdal TJ, Schneider DS. 2004. Secreted bacterial effectors and host-produced Eiger/TNF drive death in a Salmonella-infected fruit fly. PLoS Biol 2:e418. doi: 10.1371/journal.pbio.0020418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider DS, Ayres JS, Brandt SM, Costa A, Dionne MS, Gordon MD, Mabery EM, Moule MG, Pham LN, Shirasu-Hiza MM. 2007. Drosophila Eiger mutants are sensitive to extracellular pathogens. PLoS Pathog 3:e41. doi: 10.1371/journal.ppat.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mabery EM, Schneider DS. 2010. The Drosophila TNF ortholog Eiger is required in the fat body for a robust immune response. J Innate Immun 2:371–378. doi: 10.1159/000315050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J, Hopkins K, Sabin L, Yasunaga A, Subramanian H, Lamborn I, Gordesky-Gold B, Cherry S. 2013. ERK signaling couples nutrient status to antiviral defense in the insect gut. Proc Natl Acad Sci U S A 110:15025–15030. doi: 10.1073/pnas.1303193110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bou Sleiman MS, Osman D, Massouras A, Hoffmann AA, Lemaitre B, Deplancke B. 2015. Genetic, molecular and physiological basis of variation in Drosophila gut immunocompetence. Nat Commun 6:7829. doi: 10.1038/ncomms8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, Porcella SF, Heinzen RA. 2009. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci U S A 106:4430–4434. doi: 10.1073/pnas.0812074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. 2002. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 44.Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, Hultmark D. 1999. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell 4:827–837. doi: 10.1016/S1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 45.Capo C, Zugun F, Stein A, Tardei G, Lepidi H, Raoult D, Mege JL. 1996. Upregulation of tumor necrosis factor alpha and interleukin-1 beta in Q fever endocarditis. Infect Immun 64:1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. 2004. Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol 186:7344–7352. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, Dealing CM, Roman VA, Banga S, Tan Y, Luo ZQ, Samuel JE. 2013. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol 195:3914–3924. doi: 10.1128/JB.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moffatt JH, Newton P, Newton HJ. 2015. Coxiella burnetii: turning hostility into a home. Cell Microbiol 17:621–631. doi: 10.1111/cmi.12432. [DOI] [PubMed] [Google Scholar]
- 49.Beare PA, Sandoz KM, Omsland A, Rockey DD, Heinzen RA. 2011. Advances in genetic manipulation of obligate intracellular bacterial pathogens. Front Microbiol 2:97. doi: 10.3389/fmicb.2011.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonder JC, Kishimoto RA, Kastello MD, Pedersen CE Jr, Larson EW. 1979. Cynomolgus monkey model for experimental Q fever infection. J Infect Dis 139:191–196. doi: 10.1093/infdis/139.2.191. [DOI] [PubMed] [Google Scholar]
- 51.Kishimoto RA, Johnson JW, Kenyon RH, Ascher MS, Larson EW, Pedersen CE Jr. 1978. Cell-mediated immune responses of guinea pigs to an inactivated phase I Coxiella burnetii vaccine. Infect Immun 19:194–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kishimoto RA, Gonder JC, Johnson JW, Reynolds JA, Larson EW. 1981. Evaluation of a killed phase I Coxiella burnetii vaccine in cynomolgus monkeys (Macaca fascicularis). Lab Anim Sci 31:48–51. [PubMed] [Google Scholar]
- 53.Baumgartner W, Bachmann S. 1992. Histological and immunocytochemical characterization of Coxiella burnetii-associated lesions in the murine uterus and placenta. Infect Immun 60:5232–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stein A, Louveau C, Lepidi H, Ricci F, Baylac P, Davoust B, Raoult D. 2005. Q fever pneumonia: virulence of Coxiella burnetii pathovars in a murine model of aerosol infection. Infect Immun 73:2469–2477. doi: 10.1128/IAI.73.4.2469-2477.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russell-Lodrigue KE, Zhang GQ, McMurray DN, Samuel JE. 2006. Clinical and pathologic changes in a guinea pig aerosol challenge model of acute Q fever. Infect Immun 74:6085–6091. doi: 10.1128/IAI.00763-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kurata S. 2014. Peptidoglycan recognition proteins in Drosophila immunity. Dev Comp Immunol 42:36–41. doi: 10.1016/j.dci.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Champion MD. 2011. Host-pathogen o-methyltransferase similarity and its specific presence in highly virulent strains of Francisella tularensis suggests molecular mimicry. PLoS One 6:e20295. doi: 10.1371/journal.pone.0020295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shandala T, Lim C, Sorvina A, Brooks DA. 2013. A Drosophila model to image phagosome maturation. Cells 2:188–201. doi: 10.3390/cells2020188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peltan A, Briggs L, Matthews G, Sweeney ST, Smith DF. 2012. Identification of Drosophila gene products required for phagocytosis of Leishmania donovani. PLoS One 7:e51831. doi: 10.1371/journal.pone.0051831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lodge R, Descoteaux A. 2006. Phagocytosis of Leishmania donovani amastigotes is Rac1 dependent and occurs in the absence of NADPH oxidase activation. Eur J Immunol 36:2735–2744. doi: 10.1002/eji.200636089. [DOI] [PubMed] [Google Scholar]
- 61.Heinzen RA, Scidmore MA, Rockey DD, Hackstadt T. 1996. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun 64:796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ghigo E, Honstettre A, Capo C, Gorvel JP, Raoult D, Mege JL. 2004. Link between impaired maturation of phagosomes and defective Coxiella burnetii killing in patients with chronic Q fever. J Infect Dis 190:1767–1772. doi: 10.1086/425041. [DOI] [PubMed] [Google Scholar]
- 63.Sauer JD, Shannon JG, Howe D, Hayes SF, Swanson MS, Heinzen RA. 2005. Specificity of Legionella pneumophila and Coxiella burnetii vacuoles and versatility of Legionella pneumophila revealed by coinfection. Infect Immun 73:4494–4504. doi: 10.1128/IAI.73.8.4494-4504.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Romano PS, Gutierrez MG, Beron W, Rabinovitch M, Colombo MI. 2007. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell Microbiol 9:891–909. doi: 10.1111/j.1462-5822.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 65.Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, Colombo MI. 2005. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol 7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 66.Bradley WP, Boyer MA, Nguyen HT, Birdwell LD, Yu J, Ribeiro JM, Weiss SR, Zamboni DS, Roy CR, Shin S. 2016. Primary role for Toll-like receptor-driven tumor necrosis factor rather than cytosolic immune detection in restricting Coxiella burnetii phase II replication within mouse macrophages. Infect Immun 84:998–1015. doi: 10.1128/IAI.01536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cockrell DC, Long CM, Robertson SJ, Shannon JG, Miller HE, Myers L, Larson CL, Starr T, Beare PA, Heinzen RA. 2017. Robust growth of avirulent phase II Coxiella burnetii in bone marrow-derived murine macrophages. PLoS One 12:e0173528. doi: 10.1371/journal.pone.0173528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shannon JG, Howe D, Heinzen RA. 2005. Virulent Coxiella burnetii does not activate human dendritic cells: role of lipopolysaccharide as a shielding molecule. Proc Natl Acad Sci U S A 102:8722–8727. doi: 10.1073/pnas.0501863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kylsten P, Samakovlis C, Hultmark D. 1990. The cecropin locus in Drosophila; a compact gene cluster involved in the response to infection. EMBO J 9:217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Charlet M, Lagueux M, Reichhart JM, Hoffmann D, Braun A, Meister M. 1996. Cloning of the gene encoding the antibacterial peptide drosocin involved in Drosophila immunity. Expression studies during the immune response. Eur J Biochem 241:699–706. [DOI] [PubMed] [Google Scholar]
- 71.Dimarcq JL, Hoffmann D, Meister M, Bulet P, Lanot R, Reichhart JM, Hoffmann JA. 1994. Characterization and transcriptional profiles of a Drosophila gene encoding an insect defensin. A study in insect immunity. Eur J Biochem 221:201–209. [DOI] [PubMed] [Google Scholar]
- 72.Petraki S, Alexander B, Bruckner K. 2015. Assaying blood cell populations of the Drosophila melanogaster larva. J Vis Exp doi: 10.3791/52733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.