Abstract
Background/Aims
Previous studies have observed disturbances in the 1H nuclear magnetic resonance (NMR) blood spectral profiles in malignancy. No study has metabotyped serum or plasma of hepatocellular carcinoma (HCC) patients from two diverse populations. We aimed to delineate the HCC patient metabotype from Nigeria (mostly hepatitis B virus infected) and Egypt (mostly hepatitis C virus infected) to explore lipid and energy metabolite alterations that may be independent of disease aetiology, diet and environment.
Methods
Patients with HCC (53) and cirrhosis (26) and healthy volunteers (19) were recruited from Nigeria and Egypt. Participants provided serum or plasma samples, which were analysed using 600 MHz 1H NMR spectroscopy with nuclear Overhauser enhancement spectroscopy pulse sequences. Median group spectra comparison and multivariate analysis were performed to identify regions of difference.
Results
Significant differences between HCC patients and healthy volunteers were detected in levels of low density lipoprotein (P = 0.002), very low density lipoprotein (P < 0.001) and lactate (P = 0.03). N-acetylglycoproteins levels in HCC patients were significantly different from both healthy controls and cirrhosis patients (P < 0.001 and 0.001).
Conclusion
Metabotype differences were present, pointing to disturbed lipid metabolism and a switch from glycolysis to alternative energy metabolites with malignancy, which supports the Warburg hypothesis of tumour metabolism.
Abbreviations: 1H NMR, proton nuclear magnetic resonance; HCC, Hepatocellular carcinoma; HBV, Hepatitis B virus; HCV, Hepatitis C virus; NOESY, Nuclear Overhauser enhancement spectroscopy; LDL, Low density lipoprotein; JUTH, Jos University Teaching Hospital; US, Ultrasonography; CT, Computed Tomography; MRI, Magnetic resonance imaging; WHO, World Health Organisation; EDTA, Ethylenediaminetetraacetic acid; ALT, Alanine transaminase; ALP, Alkaline phosphatase; AFP, α-fetoprotein; IQR, Interquartile ranges; 1-D, One-dimensional; RD, Relaxation delay; tm, Mixing time; FID, Free induction decays; PCA, Principal components analysis; PLS-DA, Partial least squared discriminant analysis; HBsAg, Hepatitis B surface antigen; ELISA, Enzyme-linked immunosorbent assay; VLDL, Very low density lipoprotein; ppm, Parts per million; PC, Principal component; PPARα, Peroxisome proliferator-activated receptor α; IDL, Intermediate density lipoprotein
Keywords: hepatocellular carcinoma, Nigeria, Egypt, proton nuclear magnetic resonance spectroscopy, serum metabotype
Hepatocellular carcinoma (HCC) is the second commonest cause of cancer-related death and bears a poor prognosis in developing countries due to late diagnosis.1, 2, 3 Curative treatment options, namely orthotopic liver transplantation and surgical resection, are limited to low-grade cancers that are identified early.4 The widely accepted HCC screening using serum alpha-fetoprotein (AFP), a foetal glycoprotein, has shown evidence of improvement in mortality and morbidity.5 Although most HCC tumours secrete AFP, the tumour marker has poor sensitivity and specificity of less than 70%.6, 7, 8 Furthermore, serum AFP testing is unavailable in many parts of Africa, where HCC is most prevalent.
“Metabonomics” is the study of global metabolic responses to physiological, drug and disease stimuli. The most commonly used method of metabolite characterisation is proton nuclear magnetic resonance (1H NMR) spectroscopy.9 There is a paucity of data concerning the value of blood profiling using 1H NMR in HCC, but previous studies have identified a number of altered metabolites, implicating changes in hepatic function, lipid metabolism and bile acid metabolism.10, 11, 12, 13 Heterogeneity in genotype, diet, environment, co-morbid status and liver disease aetiological factors in man, may influence the ability to translate these findings to human disease.14
One previous study, performed in a Chinese population utilised 1H NMR of serum to discriminate patients with HCC (n = 39) from patients with cirrhosis (n = 36).15 In this study, alterations were observed in levels of lipoproteins, amino acids, N-acetylglycoproteins, ketoacids and lipids. Unfortunately, no information was provided on age, gender or liver disease aetiology of the participants, which is particularly relevant when utilising this method to distinguish patients with cancer to those without. In 1986, Fossel and colleagues proposed using the line widths of methyl (CH3) and methylene ((CH2)n), measured by 400 MHz 1H NMR spectroscopy, as a sensitive test for cancer.16 Levels of these metabolites were found to be significantly elevated in patients with a variety of tumours (n = 81). A number of validation studies performed on similar cohorts of patients using similar or higher magnetic field strengths, refuted this finding, citing age, triglyceride content and number of freeze thaw cycles as confounding variables that were likely to have contributed to Fossel's original findings.17, 18, 19, 20
We have previously found discriminatory metabolites for HCC using urinary metabolic profiling with 1H NMR spectroscopy in Nigerian, Egyptian and Gambian populations.21, 22, 23 The aim of the study presented here was to investigate whether serum and plasma 1H NMR profiles, collected in parallel with the published urinary studies, are different in patients with HCC compared to patients with cirrhosis and healthy volunteers in well-characterised populations from Nigeria and Egypt. These study populations were subject to widely different environmental, dietary and aetiological factors.
Methods
Patient and Healthy Volunteer Selection
Subjects were recruited in two cohorts from Jos University Teaching Hospital (JUTH) in Nigeria and The National Liver Institute, Menoufiya University, Shebeen El Kom, Egypt. The Nigerian study protocol was approved by the research ethics committee of JUTH, Nigeria and the Egyptian protocol by Menoufiya University, Egypt. The metabolic profiling protocol was approved by the research ethics committee of Imperial College London, UK. All volunteers provided informed, signed consent.
Hepatocellular carcinoma was diagnosed by radiological measures: ultrasonography (US) and/or computed tomography (CT) in Nigeria, while in Egypt, CT or magnetic resonance imaging (MRI) was used. Cirrhosis was diagnosed on clinical findings, by the presence of portal hypertension (esophageal varices or ascites) and US or CT confirmation. Tumours were staged according to the Okuda system, which includes tumour size, the presence of ascites, bilirubin and albumin levels as its criteria.24 This scoring method was chosen out of necessity as other, more comprehensive scoring tools, such as the Barcelona Clinic Liver Cancer staging algorithm, require World Health Organisation (WHO) performance status, presence of portal vein invasion and encephalopathy as criteria, which were not recorded for most of the Nigerian patients in this study, owing to lack of axial imaging resources.
Sample Collection
5 mL fasted blood samples were venesected into either plain serum or ethylenediaminetetraacetic acid (EDTA)-containing sterile tubes and placed immediately on ice or into a refrigerator at 4 °C. Samples were centrifuged within 1–2 h at 4 °C, 1000 rpm for 10 min. The supernatant was then transferred as 2 mL aliquots into 2 mL microvial tubes and stored at −80 °C undergoing no freeze thaw-cycles until analysis. Forty-eight of 56 Nigerian samples were collected into tubes containing EDTA as an anticoagulant. The remainder of samples were collected into plain serum tubes. All of the Egyptian samples were collected into plain serum tubes, with no additives. Previous studies have reported similar 1H NMR metabolic profiles from serum and plasma, allowing the two to be compared with relative assurance.25, 26, 27 These studies highlight the fact that clinical differences between groups were profoundly more influential than spectral differences between EDTA plasma and plain serum samples.
Blood Laboratory Tests
For the Nigerian samples, serum urea, creatinine, alanine transaminase (ALT), alkaline phosphatase (ALP), total bilirubin and albumin levels were measured using automated techniques (Abbott™ Architect Ci16200 Analyser, UK) at St Mary's Hospital, London. Serum AFP was measured using an automated Siemens™ Immulite 2500 Analyser, (Deerfield, USA). For the Egyptian samples, serum AFP, creatinine, ALT, aspartate aminotransferase (AST), bilirubin and albumin were measured at the time of collection in Egypt using a Cobas Integer 400-Autoanalyzer, (Roche, Germany). Median and interquartile ranges (IQR) were calculated for each assay and median levels were compared using unpaired Mann–Whitney tests of significance.
Sample Preparation
Samples were prepared according to standard validated protocols.28 Samples were thawed at room temperature and 200 μL were transferred into 1.5 mL Eppendorf (Cambridge, UK) tubes to which 400 μL NaCl/D2O (90%/10%) were added. External reference standards, such as 3-trimethylsilyl-(2,2,3,3-2H4)-1-propionate (TSP), were not added, as in blood they may bind to protein, resulting in a final NMR signal that is reduced and has a very broad linewidth. The mixture underwent centrifugation for 5 min at 13,000 rpm and 550 μL of supernatant were transferred to Norell 5 mm 507-HP-7 NMR tubes (Norell, Landisville, New Jersey, USA) ready for 1H NMR analysis. Samples were analysed on the same day as preparation.
1H NMR Spectroscopy
All samples were run in a random, non-grouped order. All samples were run at the Department of Biomolecular Medicine, Imperial College London on two Bruker Ultrashield Plus™ 600 NMR systems operating at 600.29–600.44 MHz 1H frequency (Bruker Biospin, Rheinstetten, Germany).29 The systems were tuned, matched and frequency-locked on to 1H as the nucleus of interest. A representative sample was utilised to set shim gradients to ensure a homogenous magnetic field across the sample, a 90° pulse length and the water suppression offset parameters. These settings were saved and utilised for the whole sample set. Spectra were acquired using nuclear Overhauser enhancement spectroscopy (NOESY) 1-D pulse sequence with water presaturation, during the relaxation delay (RD) and mixing time (tm) using the following pulse programme: -RD-90°_t-90°-tm-90°-acquire; where RD = 2.0 s and tm = 0.1 s. For each sample, 128 free induction decays (FIDs) were collected into 32,000 data points with a spectral width of 20 parts per million (ppm). A line broadening function of 0.3–1.0 Hz was applied prior to Fourier transformation. Spectra were manually phased, baseline corrected and referenced to the α-glucose doublet at 5.23 ppm in TOPSPIN v2.0 (Bruker Biospin, Rheinstetten, Germany). Spectral peaks were assigned with reference to the literature.30, 31, 32
Data Pre-processing
Spectra were exported to MATLAB R2010 (MathWorks, Natick, Massachusetts, USA) and the water region from 4.5 to 6 ppm was excluded. As the concentration of EDTA varied between the serum and plasma samples, regions were excluded where it resonated, to avoid modelling differences between EDTA concentrations between samples. In a recent analysis of the effect of EDTA on metabolic profiling information recovery, it was reported that the resonances EDTA obscures commonly resonate elsewhere in the spectrum with few exceptions. Furthermore, the effect of EDTA on other molecules, in terms of spectral resonance or peak shift was found to be negligible.33 Data were normalised to median fold-change and median spectra for all groups were generated to allow visual comparison of spectra and allow the selection of regions that were divergent for use in multivariate and univariate analyses.
Univariate Analysis
Data were exported to GraphPad Prism (La Jolla, California, USA) for univariate analysis in the form of Mann–Whitney U-tests comparison of medians between groups, assuming non-parametric distribution of data. P-values of <0.05 were considered significant.
Multivariate Analysis
Median spectra of each group (HCC, cirrhosis and healthy volunteer) were compared in a combined analysis of Nigerian and Egyptian data. Regions that were visually divergent were selected for multivariate analysis. These areas are recorded in Table 1. The integral areas of these regions were recorded in a data matrix and exported to SIMCA (Umetrics, Umea, Sweden). Data were mean-centred and principle components analysis (PCA) was performed first to model overall variation and identify outliers. Only mean-centred data were used for further analysis. After outliers were identified and excluded, partial least squared discriminant analysis (PLS-DA) was performed to identify the discriminant strength of the metabolite based model and to generate a loadings plot from which metabolites could be identified which most greatly contributed to differences between the groups. In SIMCA-P v12, PLS-DA models were generated through seven-fold cross validation. In this method, every 7th sample was excluded (1st, 7th, 14th, 21st and so on), a model generated from the remaining samples and the excluded “training set” predicted back into the model. This was repeated for all the samples (grouping the 2nd, 9th, 16th and 3rd, 10th, 17th and so on) until all the samples were excluded once. The results were averaged to produce a model which was externally cross-validated. Spectral peaks which contributed most to PLS-DA models, and those visually different on median spectra comparison, were selected for peak integration. All data were mean centred prior to multivariate analysis. Country-specific and male-only analyses were performed to ensure that findings were due to metabolite characteristics secondary to HCC and not due to population or gender disparities between groups.
Table 1.
Spectral Regions Selected for Multi- and Univariate Analyses.
| Region (ppm) | Molecule Moeity |
||
|---|---|---|---|
| 1 | 0.8–0.85 | Low density lipoprotein | CH3 |
| 2 | 0.85–0.88 | Very low density lipoprotein | CH3 |
| 3 | 1.21–1.24 | Low density lipoprotein | -(CH2)n- |
| 4 | 1.25–1.30 | Very low density lipoprotein | -(CH2)n- |
| 5 | 1.31–1.32 | Lactate | CH3 |
| 6 | 2.02–2.05 | N-Acetylglycoproteins | NHCOCH3 |
| 7 | 2.22–2.23 | Acetoacetate | CH3 |
| 8 | 4.098–4.108 | Lactate | CH |
| 9 | 8.445–8.45 | Formate | CH |
Results
Subject Selection and Demographics
A total of 98 volunteers were recruited for study, 56 from Nigeria and 42 from Egypt. Subjects were recruited in three cohorts, 53 patients with ultrasound or computed tomography proven HCC (29 Nigerian + 24 Egyptian, median age: 50, 70% male); 26 patients with clinically-confirmed cirrhosis with features of portal hypertension, but no HCC (12 Nigerian + 14 Egyptian, median age: 48.5, 69% male); and 19 healthy subjects with no history of liver disease (15 Nigerian + 4 Egyptian, median age: 40, 42% male). All patients, except one, in the Nigerian HCC and cirrhosis groups were hepatitis B surface antigen (HBsAg) positive. The single non-HBV patient with cirrhosis was also anti-hepatitis C virus (HCV) antibody negative and was therefore classified as having idiopathic liver disease. In the Egyptian cohort, all the patients with cirrhosis and 23/24 patients with HCC had chronic HCV. The single HCC patient without HCV had idiopathic liver disease. All healthy volunteers were HBsAg and anti-HCV antibody negative with no history of liver disease.
There was no significant difference between the ages of all three groups, although patients in the healthy volunteer group had a median age of 40 years, compared to that of 50 years for patients with HCC (p = 0.09). There were fewer males in the healthy volunteer group (42% versus 70% in the HCC group, p = 0.052). The biochemical analyses of the patients are outlined in Table 2. Median serum AFP levels were significantly higher (1198 IU mL−1) in patients with HCC, compared to those with cirrhosis and to healthy volunteers (5.61 and 1.44, p < 0.001). Of note, if an AFP cut-off value of 400 IU mL−1 was used for HCC diagnosis, 19 tumours would have not been diagnosed (64% sensitivity). Creatinine levels were comparable across groups, but serum ALT, bilirubin and albumin were deranged in the HCC and cirrhosis groups in comparison to healthy controls. HCC was staged according to the Okuda criteria, which showed 8 patients were Stage 1, 25 patients were Stage 2 and 16 patients were Stage 3. Four Nigerian patients were unable to be staged accurately, owing to a lack of clinical data.
Table 2.
Biochemical Analysis of All Patients.
| Test (range) | HCC | Cirrhosis | Healthy controls | P-values (Mann–Whitney) |
|---|---|---|---|---|
| Serum samples (n) | 53 | 26 | 19 | - |
| AFP (IU mL−1) | 1198 | 5.61+ | 1.44+ | aandb<0.001* |
| Creatinine (mmol L−1) | 63.0 | 82.5 | 70.0 | a0.39 and b0.04* |
| ALT (IU L−1) | 52.5+ | 32.5 | 22.0 | a<0.001* and b0.04* |
| Bilirubin (μmol L−1) | 29.0+ | 36.8 | 6.9 | a<0.001* and b0.43 |
| Albumin (g L−1) | 26.6+ | 23.8 | 45.7 | a<0.001* and b0.16 |
Key:+Some data missing. Mann–Whitney non-parametric comparisons of HCC versus healthy (a) and versus cirrhosis (b).
1H NMR Spectroscopy
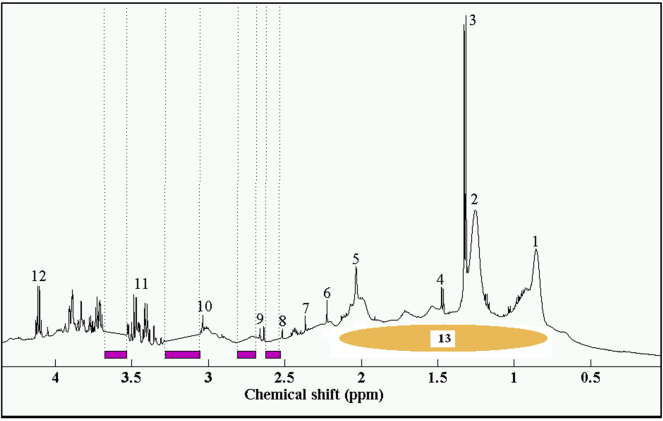
A representative NOESY plasma spectrum is displayed in Figure 1 with indication of which regions were excluded due to EDTA resonances. The area of exclusion is, therefore, relatively small in comparison to the whole spectrum. The resolution between the overlapping peaks of low density lipoprotein (LDL) at 0.8 ppm and 1.21 ppm and very low density lipoprotein (VLDL) at 0.85 ppm and 1.25 ppm was poor, although could discernibly be distinguished.
Figure 1.
Representative plasma spectrum with EDTA exclusion. Key: (1) and (2) LDL/VLDL; (3) lactate (CH3); (4) alanine; (5) N-acetylglycoproteins; (6) acetoacetate; (8) and (9) citrate; (10) creatinine; (11) glucose resonances; (12) lactate (CH); (13) albumin and albumin-bound fatty acids (Nicholson et al., 1995).31 Purple bars indicate areas of EDTA resonance exclusion.
Univariate Statistical Analysis
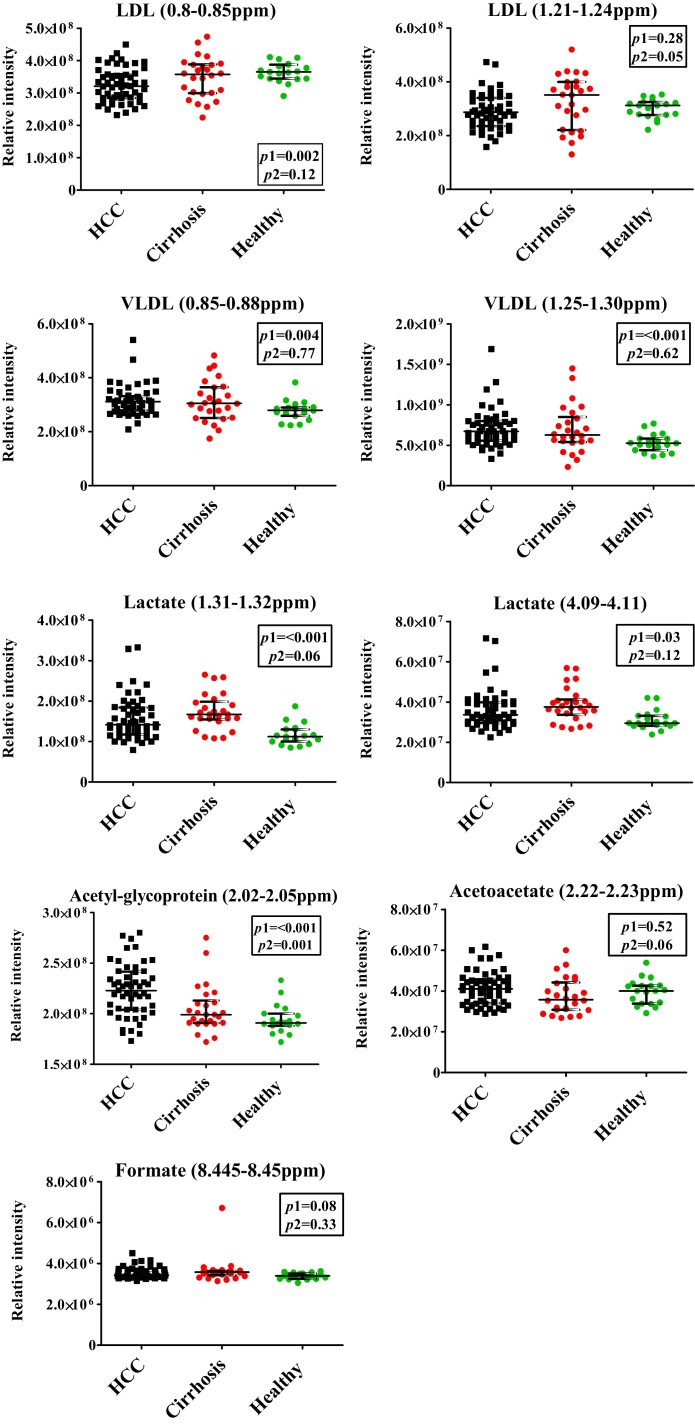
Univariate analyses, using the spectral integral values of one peak, which corresponds to one metabolite, were performed (Figure 5 and Table 3). The most prominent spectral peaks, arising from LDL and VLDL molecules, showed significant difference between the groups. LDL levels were reduced in patients with HCC, compared to both healthy volunteers (p = 0.28 and 0.002) and cirrhosis (p = 0.12 and 0.05). VLDL levels were raised in patients with HCC, compared to healthy volunteers (p = 0.004 and <0.001), but not when compared to patients with cirrhosis (p = 0.77 and 0.62). Lactate levels, both at 1.31 ppm (doublet) and 4.11 ppm (quadruplet), were significantly raised in patients with HCC, compared to healthy controls (p = <0.001 and 0.03), but not when compared to patients with cirrhosis (p = 0.06 and 0.12). N-Acetylglycoproteins levels were significantly raised in patients with HCC compared to both healthy volunteers and patients with cirrhosis (p < 0.001 and 0.001), while acetoacetate was non-significantly raised (p = 0.52 and 0.06). Finally, formate levels, although visually appearing altered between group median spectra, displayed no significant differences between the groups.
Figure 5.
Univariate analysis of discriminatory metabolites. Key:P1 = P-value of HCC versus healthy control analyses; P2 = P-value of HCC versus cirrhosis analyses. Mann–Whitney tests of significance used for generation of P-values.
Table 3.
Metabolite Differences Between Groups.
| Metabolite | Moiety | Chemical shift (ppm) | HCC vs. Healthy | HCC vs. Cirrhosis | Pathway |
|---|---|---|---|---|---|
| LDL | CH3 | 0.8–0.85 | ↓* | ↓ | Lipid production/use |
| LDL | -(CH2)n- | 1.21–1.24 | ↓ | ↓ | |
| VLDL | CH3 | 0.85–0.88 | ↑* | ↑ | |
| VLDL | -(CH2)n- | 1.25–1.30 | ↑* | ↑ | |
| Lactate | CH3 | 1.31–1.32 | ↑* | ↓ | Inflammation |
| Lactate | CH | 4.098–4.108 | ↑* | ↓ | |
| N-Acetyl-glycoproteins | NHCOCH3 | 2.02–2.05 | ↑* | ↑* | |
| Acetoacetate | CH3 | 2.22–2.23 | ↑ | ↑* | Lipid metabolism |
| Formate | CH | 8.445–8.45 | ↑ | ↑ | 1-carbon pathway |
Key: ↑↓Indicates increased or decreased in patients with HCC. *P-value <0.05.
Multivariate Statistical Analysis
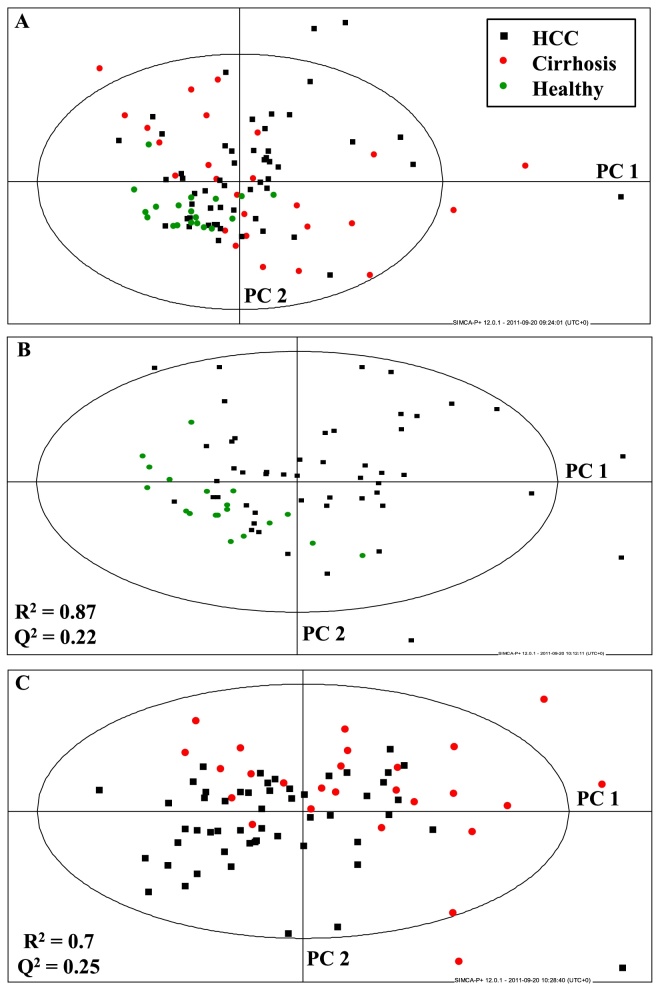
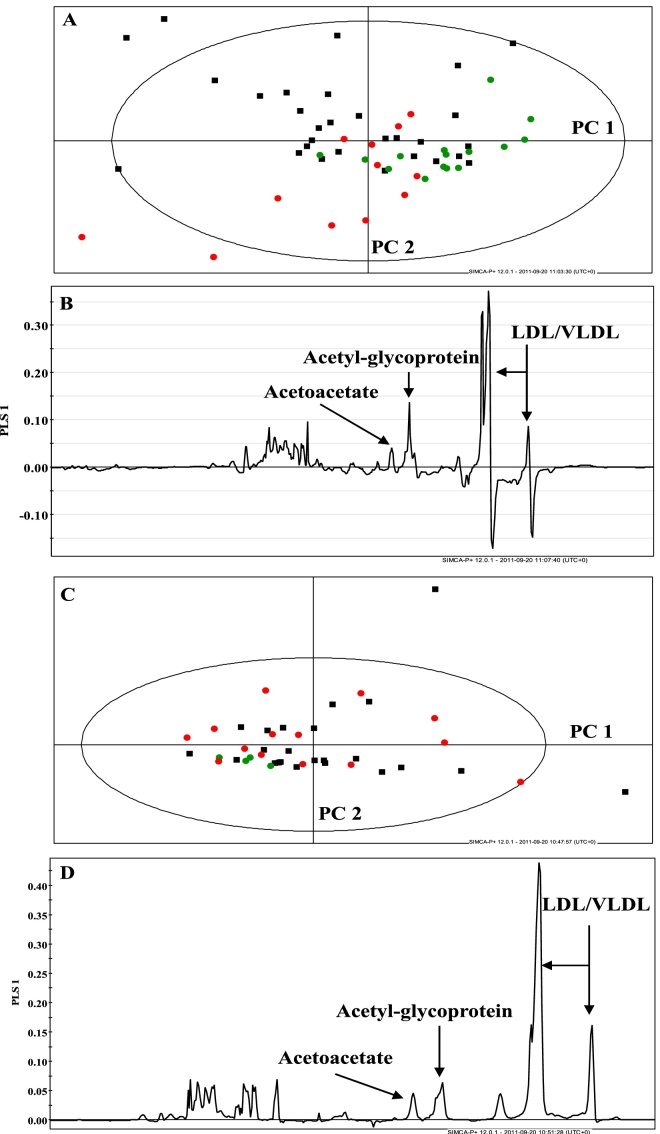
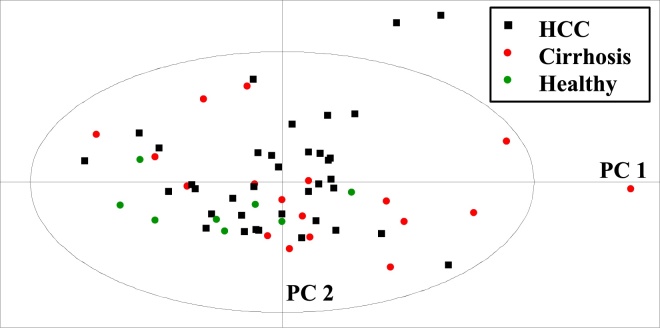
Principal components analysis and PLS-DA were performed on the data matrix, consisting of those spectral regions that appeared most divergent between patient and control groups. Nine regions were identified, which are tabulated (Table 1). Principal components analysis of all groups is shown in Figure 2A. Supervised PLS-DA was undertaken and is displayed for HCC and healthy volunteer and HCC and cirrhosis groups in Figure 2B and C. The fit of the models was good (R2 = 0.87 and 0.7). However, the goodness of prediction or Q2 levels was low: 0.22 and 0.25. Figure 3A-D displays the separate multivariate analyses for the Nigerian and Egyptian cohorts. These analyses confirm that the combined analyses reflect the country-specific results, with metabolites such as LDL, VLDL, N-acetylglycoproteins and acetoacetate as contributing most to discrimination between patients and healthy volunteer groups. Finally, male-only analyses were performed using both Nigerian and Egyptian data. This is represented in a PCA plot in Figure 4. The data displayed similar clustering to combined plots and the metabolites contributing most to discrimination between group remained very similar, confirming that gender disparities between disease and healthy volunteer groups were not confounding multivariate results (Figure 5).
Figure 2.
Multivariate analyses of combined Nigerian and Egyptian samples. (A) PCA scatter plot of all groups; (B) PLS-DA scatter plot of HCC and healthy volunteer samples; (C) PLS-DA scatter plot of HCC and cirrhosis samples.
Figure 3.
Multivariate analysis plots of Nigerian and Egyptian data. (A) and (B) PCA and PLS-DA loadings plot of Nigerian data; (C) and (D) PCA and PLS-DA loadings plot of Egyptian data.
Figure 4.
Principal components analysis of male volunteer samples.
Discussion
This is the first study to characterise the metabolic changes in serum and plasma due to HCC in two completely diverse populations with different genetics, diet and underlying disease aetiology. Multivariate analysis displayed reasonable separation of disease and healthy groups, while comparison of median group spectra, combined with univariate analyses identified several metabolites elevated or reduced in the blood of patients with HCC. Furthermore, combined analyses, of subjects from Nigerian and Egypt, revealed similar results to country-specific analyses. Given that the majority of patients from Nigeria were HBV-infected and those from Egypt were HCV-infected, this would suggest that blood metabolite profiles in the presence of HCC are dependent on the tumour effects, rather than aetiology of liver disease.30
There have been several previous studies that utilised serum 1H NMR for HCC identification.12, 13, 14, 15, 34 Assi and colleagues utilised a large 1H NMR study to associate lifestyle exposure with metabolomic signals of HCC in a European cohort of the European Prospective Investigation into Cancer and Nutrition (EPIC) study.14 The study highlighted the presence of a complex interaction of dietary and lifestyle factors leading to metabolic changes that may contribute to HCC. A study by Liu and colleagues identified potential biomarkers by comparing 43 HCC patients with 42 cirrhosis patients and 18 healthy volunteers. There were significant elevations in beta-hydroxybutyrate, glycerol and oxaloacetate in the HCC group, and fatty acid elevation in the cirrhosis group, including isobutyrate, linoelaidic acid and linoleic acid, compared with the healthy volunteers.34 Nahon and colleagues compared the serum data of patients with compensated biopsy-proven alcoholic cirrhosis, of whom 93 had cirrhosis without HCC, 28 had small HCC and 33 had large HCC determined by the Milan criteria.12 The study showed significant increase in glutamate, acetate and N-acetyl glycoproteins in large HCC compared to the cirrhotic group without HCC. The significance of the results is debatable due to the various metabolic effect of chronic alcoholism. Wei and colleagues compared patients with HCC with those with HCV, and identified significant alteration in choline, valine and creatinine in the HCC groups.13 Overexpression of metabolites, such as choline, has been found to be raised with a series of different tumours, and these are likely to represent non-specific serum markers.35 Furthermore, these studies did not offer metabolic comparison with healthy controls. Gao and colleagues utilised 1H NMR of serum from patients with HCC in comparison to patients with liver cirrhosis and healthy volunteers,15 the results showing some similarities to those reported here. The Gao study did not clarify the patient age, gender or aetiology of liver disease. However, the metabolite signatures that we report here and those of the Gao study infer a significant influence of HCC upon lipid metabolism. Blood VLDL levels were elevated in patients with HCC, both in comparison to cirrhosis and healthy states in both studies.15
Low density lipoprotein levels, conversely, were reduced in our study. Acetoacetate, a by-product of fatty acid oxidation, was elevated in patients with HCC. It is increasingly recognised that the liver, as a central hub of lipid metabolism, may alter its production of VLDL as a result of disease.36 This is of particular importance in the presence of HCV particles which utilise altered VLDL particles as a transport and translocation facilitator thereby affecting blood levels.37 It is less well documented how HCC may affect this pathway. Intuitively, it would be expected that as a tumour grows in an already diseased cirrhotic liver, functionality decreases and lipid production does so as well. The results in this study are therefore counter-intuitive, with raised VLDL and reduced LDL levels. Gao and colleagues offer little explanation of why this would occur in their study, stating that HCC and cirrhosis merely enhance lipid metabolism. The genetic changes that occur in HCC are diverse and can affect many pathways.38 It is possible that one of these affected pathways may affect lipid metabolism and promote the production of VLDL. A candidate may be Peroxisome proliferator-activated receptor α (PPARα), a nuclear transcription factor, which, if activated, is known to decrease hepatic VLDL secretion and enhance clearance.39 It is also plausible that peripheral VLDL breakdown, via the lipolytic pathway, is reduced. If this were the case then less LDL would be formed, as seen here. This may be affected through a down-regulation of lipolytic enzymes, such as hepatic or lipoprotein lipase, the interplay of which is highly complex in lipid metabolism.36
A more robust argument for the observed rise in metabolites that we observed may be explained by the Warburg phenomenon, which highlights the preferential metabolism of glucose by anaerobic glycolysis in tumour cells.40 Glycolysis produces energy at a higher rate than oxidative phosphorylation in cancer cells, albeit at the compromise of metabolic efficiency.41 The heightened rate of anaerobic metabolism may be a favourable trait for a rapidly proliferating tumour.42 The shift in metabolism causes a rise in by-products of anaerobic respiration, such as lactate, which was significantly raised in the HCC group.
The increase in VLDL that we noted may be a consequential effect of alternative energy metabolism in HCC. Hepatic VLDL is produced by fatty acid esterification with glycerophosphate, a by-product of anaerobic glycolysis.43 Hepatic VLDL secretion may be the inappropriate response from the tumour's anaerobic respiration, leading to global lipid mobilisation for the lipolytic pathway. The result of the pathway is supported by the observed increase in acetoacetate in the HCC group. Acetoacetate is a ketone body, which together with acetone and beta-hydroxybutyrate, is formed as a by-product of beta-oxidation.44 The rise in ketone bodies was also observed in the Liu study and may indicate a globally heightened lipolytic pathway in the HCC group as a consequence of abnormal anaerobic respiration.34
In our study, formate levels were elevated in patients with HCC. This metabolite is produced from the folate cycle in hepatic embryonic cells. In conjunction with the abnormal rise of AFP, an embryonic glycoprotein detectable in HCC, the increase in formate is an unsurprising result of liver tumorigenesis.45
N-Acetylglycoproteins were increased in patients with HCC in our study. These represent “acute phase protein” fragments of glycoproteins, such as α1-acid glycoprotein, haptoglobin, transferrin and fibrinogen. Hepatocytes are known to secrete these molecules under a number of different stressful stimuli including cancer.46, 47 A NMR study by Bell and colleagues, comparing patients with different malignancies to matched healthy controls, observed this resonance to display large variations in amplitude in the blood of cancer patients, compared to healthy volunteers.17 In HCC on the background of a cirrhotic liver, it may be that hepatic function is preserved to an extent, so as to secrete this molecule as a stress response.
Our study characterises a certain metabolic trait in patients with HCC and cirrhosis, which can be distinguished from the healthy population. This finding is extremely relevant to the current investigative changes for HCC in clinical practice. If the metabolites that characterise HCC are incorporated into a testable profile, they may be given a simple scoring system to identify both the presence and severity of HCC by a blood test. While identifying AFP is no longer the recommended guideline for HCC diagnosis, a minimally invasive serum marker for cancer is extremely useful, particularly in a developing world scenario where management of advanced tumours is limited. Early identification through a simple serum investigation may be an important step in addressing the global HCC burden.
In conclusion, this study has produced results which may provide insight into the altered lipid pathways induced by Warburg's phenomenon of anaerobic respiration in HCC. This is the first blood profiling NMR study to look at two ethnically diverse patient populations and find results that independent of genetics, diet and underlying disease aetiology, all factors that have limited the meaningfulness and translatability of previous studies. Our previous urinary metabolic profiling studies in the same populations have thrown up a different series of metabolites present in the urine, which delineate HCC distinct from cirrhosis and non-cirrhotic liver disease.21, 22, 23 This highlights that parallel investigations on different body fluids are valuable in the search for new biomarkers of liver cancer, and that ultimately, a combined biomarker panel from blood, urine and possibly stool may be a useful avenue for future research.
Conflicts of Interest
The authors have none to declare.
Financial Support
The study was supported by project grants from the Associations of Physicians of Great Britain and Ireland. MIFS and NGL were supported by personal grants from the Royal College of Physicians of London, the University of London and the Trustees of the London Clinic, London, UK. MMEC is supported by a Fellowship from the Sir Halley Stewart Trust (Cambridge, United Kingdom). MMEC and SDT-R hold grants from the United Kingdom Medical Research Council. AIG was supported by a doctorate grant from the Egyptian Ministry of Higher Education.
Author's Contributions
The study was conceived and overseen by SDT-R, EO, IW, IJC, RW and EH. MIFS, NGL, AIG and MMEC conducted the study, while NGL, AIG, EB and MMEC were responsible for sample collection in-country, transport and processing. MIFS undertook the analyses, which were verified by NGL, EH and SDT-R. The paper was written primarily by MIFS, JUK and SDT-R, but all authors contributed to the writing of the manuscript and approved the final version
Acknowledgements
All authors acknowledge the support of the National Institute for Health Research Biomedical Research Centre at Imperial College London for infrastructure support.
References
- 1.El-Serag H.B., Kanwal F. Epidemiology of hepatocellular carcinoma in the United States: Where are we? Where do we go? Hepatology. 2014;60:1767–1775. doi: 10.1002/hep.27222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan S.A., Taylor-Robinson S.D., Toledano M.B., Beck A., Elliott P., Thomas H.C. Changing international trends in mortality rates for liver, biliary and pancreatic tumours. J Hepatol. 2002;37:806–813. doi: 10.1016/s0168-8278(02)00297-0. [DOI] [PubMed] [Google Scholar]
- 3.Taylor-Robinson S.D., Foster G.R., Arora S., Hargreaves S., Thomas H.C. Increase in primary liver cancer in the UK, 1979-94. Lancet. 1997;350:1142–1143. doi: 10.1016/S0140-6736(05)63789-0. [DOI] [PubMed] [Google Scholar]
- 4.Llovet J.M., Bru C., Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 5.Yuen M.F., Cheng C.C., Lauder I.J., Lam S.K., Ooi C.G., Lai C.L. Early detection of hepatocellular carcinoma increases the chance of treatment: Hong Kong experience. Hepatology. 2000;31:330–335. doi: 10.1002/hep.510310211. [DOI] [PubMed] [Google Scholar]
- 6.Furui J., Furukawa M., Kanematsu T. The low positive rate of serum alpha-fetoprotein levels in hepatitis C virus antibody-positive patients with hepatocellular carcinoma. Hepatogastroenterology. 1995;42:445–449. [PubMed] [Google Scholar]
- 7.Nguyen M.H., Keeffe E.B. Screening for hepatocellular carcinoma. J Clin Gastroenterol. 2002;35:S86–S91. doi: 10.1097/00004836-200211002-00004. [DOI] [PubMed] [Google Scholar]
- 8.Peng Y.C., Chan C.S., Chen G.H. The effectiveness of serum alpha-fetoprotein level in anti-HCV positive patients for screening hepatocellular carcinoma. Hepatogastroenterology. 1999;46:3208–3211. [PubMed] [Google Scholar]
- 9.Nicholson J.K., Lindon J.C. Systems biology: Metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 10.Gao H., Dong B., Liu X., Xuan H., Huang Y., Lin D. Metabonomic profiling of renal cell carcinoma: high-resolution proton nuclear magnetic resonance spectroscopy of human serum with multivariate data analysis. Anal Chim Acta. 2008;624:269–277. doi: 10.1016/j.aca.2008.06.051. [DOI] [PubMed] [Google Scholar]
- 11.Yin P., Wan D., Zhao C. A metabonomic study of hepatitis B-induced liver cirrhosis and hepatocellular carcinoma by using RP-LC and HILIC coupled with mass spectrometry. Mol Biosyst. 2009;5:868–876. doi: 10.1039/b820224a. [DOI] [PubMed] [Google Scholar]
- 12.Nahon P., Amathieu R., Triba M.N. Identification of serum proton NMR metabolomic fingerprints associated with hepatocellular carcinoma in patients with alcoholic cirrhosis. Clin Cancer Res. 2012;18:6714–6722. doi: 10.1158/1078-0432.CCR-12-1099. [DOI] [PubMed] [Google Scholar]
- 13.Wei S., Suryani Y., Gowda G.A., Skill N., Maluccio M., Raftery D. Differentiating hepatocellular carcinoma from hepatitis C using metabolite profiling. Metabolites. 2012;2:701–716. doi: 10.3390/metabo2040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Assi N., Fages A., Vineis P. A statistical framework to model the meeting-in-the-middle principle using metabolomic data: application to hepatocellular carcinoma in the EPIC study. Mutagenesis. 2015 doi: 10.1093/mutage/gev045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao H., Lu Q., Liu X. Application of 1H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 2009;100:782–785. doi: 10.1111/j.1349-7006.2009.01086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fossel E.T., Carr J.M., McDonagh J. Detection of malignant tumors. Water-suppressed proton nuclear magnetic resonance spectroscopy of plasma. N Engl J Med. 1986;315:1369–1376. doi: 10.1056/NEJM198611273152201. [DOI] [PubMed] [Google Scholar]
- 17.Bell J.D., Brown J.C., Norman R.E., Sadler P.J., Newell D.R. Factors affecting 1H NMR spectra of blood plasma: cancer, diet and freezing. NMR Biomed. 1988;1:90–94. doi: 10.1002/nbm.1940010206. [DOI] [PubMed] [Google Scholar]
- 18.Holmes K.T., Mackinnon W.B., May G.L. Hyperlipidemia as a biochemical basis of magnetic resonance plasma test for cancer. NMR Biomed. 1988;1:44–49. doi: 10.1002/nbm.1940010108. [DOI] [PubMed] [Google Scholar]
- 19.Okunieff P., Zietman A., Kahn J. Lack of efficacy of water-suppressed proton nuclear magnetic resonance spectroscopy of plasma for the detection of malignant tumors. N Engl J Med. 1990;322:953–958. doi: 10.1056/NEJM199004053221403. [DOI] [PubMed] [Google Scholar]
- 20.Wilding P., Senior M.B., Inubushi T., Ludwick M.L. Assessment of proton nuclear magnetic resonance spectroscopy for detection of malignancy. Clin Chem. 1988;34:505–511. [PubMed] [Google Scholar]
- 21.Shariff M.I., Ladep N.G., Cox I.J. Characterization of urinary biomarkers of hepatocellular carcinoma using magnetic resonance spectroscopy in a Nigerian population. J Proteome Res. 2010;9:1096–1103. doi: 10.1021/pr901058t. [DOI] [PubMed] [Google Scholar]
- 22.Shariff M.I., Gomaa A.I., Cox I.J. Urinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: a validation study. J Proteome Res. 2011;10:1828–1836. doi: 10.1021/pr101096f. [DOI] [PubMed] [Google Scholar]
- 23.Ladep N.G., Dona A.C., Lewis M.R. Discovery and validation of urinary metabotypes for the diagnosis of hepatocellular carcinoma in West Africans. Hepatology. 2014;60:1291–1301. doi: 10.1002/hep.27264. [DOI] [PubMed] [Google Scholar]
- 24.Okuda K., Ohtsuki T., Obata H. Natural history of hepatocellular carcinoma and prognosis in relation to treatment. Study of 850 patients. Cancer. 1985;56:918–928. doi: 10.1002/1097-0142(19850815)56:4<918::aid-cncr2820560437>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 25.Deprez S., Sweatman B.C., Connor S.C., Haselden J.N., Waterfield C.J. Optimisation of collection, storage and preparation of rat plasma for 1H NMR spectroscopic analysis in toxicology studies to determine inherent variation in biochemical profiles. J Pharm Biomed Anal. 2002;30:1297–1310. doi: 10.1016/s0731-7085(02)00455-7. [DOI] [PubMed] [Google Scholar]
- 26.Teahan O., Gamble S., Holmes E. Impact of analytical bias in metabonomic studies of human blood serum and plasma. Anal Chem. 2006;78:4307–4318. doi: 10.1021/ac051972y. [DOI] [PubMed] [Google Scholar]
- 27.Wedge D.C., Allwood J.W., Dunn W. Is serum or plasma more appropriate for intersubject comparisons in metabolomic studies? An assessment in patients with small-cell lung cancer. Anal Chem. 2011;83:6689–6697. doi: 10.1021/ac2012224. [DOI] [PubMed] [Google Scholar]
- 28.Beckonert O., Keun H.C., Ebbels T.M. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 29.Keun H.C., Ebbels T.M., Antti H. Analytical reproducibility in (1)H NMR-based metabonomic urinalysis. Chem Res Toxicol. 2002;15:1380–1386. doi: 10.1021/tx0255774. [DOI] [PubMed] [Google Scholar]
- 30.Holmes E., Loo R.L., Stamler J. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholson J.K., Foxall P.J., Spraul M., Farrant R.D., Lindon J.C. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal Chem. 1995;67:793–811. doi: 10.1021/ac00101a004. [DOI] [PubMed] [Google Scholar]
- 32.Wishart D.S., Tzur D., Knox C. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barton R.H., Waterman D., Bonner F.W. The influence of EDTA and citrate anticoagulant addition to human plasma on information recovery from NMR-based metabolic profiling studies. Mol Biosyst. 2010;6:215–224. doi: 10.1039/b907021d. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y., Hong Z., Tan G. NMR and LC/MS-based global metabolomics to identify serum biomarkers differentiating hepatocellular carcinoma from liver cirrhosis. Int J Cancer. 2014;135:658–668. doi: 10.1002/ijc.28706. [DOI] [PubMed] [Google Scholar]
- 35.Ramirez de Molina A., Rodriguez-Gonzalez A., Gutierrez R. Overexpression of choline kinase is a frequent feature in human tumor-derived cell lines and in lung, prostate, and colorectal human cancers. Biochem Biophys Res Commun. 2002;296:580–583. doi: 10.1016/s0006-291x(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 36.Bassendine M.F., Sheridan D.A., Felmlee D.J., Bridge S.H., Toms G.L., Neely R.D. HCV and the hepatic lipid pathway as a potential treatment target. J Hepatol. 2011;55:1428–1440. doi: 10.1016/j.jhep.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen S.U., Bassendine M.F., Burt A.D., Martin C., Pumeechockchai W., Toms G.L. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J Virol. 2006;80:2418–2428. doi: 10.1128/JVI.80.5.2418-2428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wurmbach E., Chen Y.B., Khitrov G. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 39.Shah A., Rader D.J., Millar J.S. The effect of PPAR-alpha agonism on apolipoprotein metabolism in humans. Atherosclerosis. 2010;210:35–40. doi: 10.1016/j.atherosclerosis.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 40.Warburg O., Posener K., Negelein E. Ueber den stoffwechsel der tumoren. Biochem Z. 1924;152:319–344. [Google Scholar]
- 41.Ganapathy V., Thangaraju M., Prasad P.D. Nutrient transporters in cancer: relevance to Warburg hypothesis and beyond. Pharmacol Ther. 2009;121:29–40. doi: 10.1016/j.pharmthera.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 43.Goldberg R.B. Lipid disorders in diabetes. Diabetes Care. 1981;4:561–572. doi: 10.2337/diacare.4.5.561. [DOI] [PubMed] [Google Scholar]
- 44.Laffel L. Ketone bodies: a review of physiology, pathophysiology and application of monitoring to diabetes. Diabetes Metab Res. 1999;15:412–426. doi: 10.1002/(sici)1520-7560(199911/12)15:6<412::aid-dmrr72>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 45.Brumm C., Schulze C., Charels K., Morohoshi T., Klöppel G. The significance of alpha-fetoprotein and other tumour markers in differential immunocytochemistry of primary liver tumours. Histopathology. 1989;14:503–513. doi: 10.1111/j.1365-2559.1989.tb02186.x. [DOI] [PubMed] [Google Scholar]
- 46.Baumann H., Jahreis G.P., Gaines K.C. Synthesis and regulation of acute phase plasma proteins in primary cultures of mouse hepatocytes. J Cell Biol. 1983;97:866–876. doi: 10.1083/jcb.97.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y., Holmes E., Tang H. Experimental metabonomic model of dietary variation and stress interactions. J Proteome Res. 2006;5:1535–1542. doi: 10.1021/pr0504182. [DOI] [PubMed] [Google Scholar]