ABSTRACT
In this study, we constructed a set of Ralstonia eutropha H16 strains with single, double, or triple deletions of the (p)ppGpp synthase/hydrolase (spoT1), (p)ppGpp synthase (spoT2), and/or polyhydroxybutyrate (PHB) depolymerase (phaZa1 or phaZa3) gene, and we determined the impact on the levels of (p)ppGpp and on accumulated PHB. Mutants with deletions of both the spoT1 and spoT2 genes were unable to synthesize detectable amounts of (p)ppGpp and accumulated only minor amounts of PHB, due to PhaZa1-mediated depolymerization of PHB. In contrast, unusually high levels of PHB were found in strains in which the (p)ppGpp concentration was increased by the overexpression of (p)ppGpp synthase (SpoT2) and the absence of (p)ppGpp hydrolase. Determination of (p)ppGpp levels in wild-type R. eutropha under different growth conditions and induction of the stringent response by amino acid analogs showed that the concentrations of (p)ppGpp during the growth phase determine the amount of PHB remaining in later growth phases by influencing the efficiency of the PHB mobilization system in stationary growth. The data reported for a previously constructed ΔspoT2 strain (C. J. Brigham, D. R. Speth, C. Rha, and A. J. Sinskey, Appl Environ Microbiol 78:8033–8044, 2012, https://doi.org/10.1128/AEM.01693-12) were identified as due to an experimental error in strain construction, and our results are in contrast to the previous indication that the spoT2 gene product is essential for PHB accumulation in R. eutropha.
IMPORTANCE Polyhydroxybutyrate (PHB) is an important intracellular carbon and energy storage compound in many prokaryotes and helps cells survive periods of starvation and other stress conditions. Research activities in several laboratories over the past 3 decades have shown that both PHB synthase and PHB depolymerase are constitutively expressed in most PHB-accumulating bacteria, such as Ralstonia eutropha. This implies that PHB synthase and depolymerase activities must be well regulated in order to avoid a futile cycle of simultaneous PHB synthesis and PHB degradation (mobilization). Previous reports suggested that the stringent response in Rhizobium etli and R. eutropha is involved in the regulation of PHB metabolism. However, the levels of (p)ppGpp and the influence of those levels on PHB accumulation and PHB mobilization have not yet been determined for any PHB-accumulating species. In this study, we optimized a (p)ppGpp extraction procedure and a high-performance liquid chromatography–mass spectrometry (HPLC-MS)-based detection method for the quantification of (p)ppGpp in R. eutropha. This enabled us to study the relationship between the concentrations of (p)ppGpp and the accumulated levels of PHB in the wild type and in several constructed mutant strains. We show that overproduction of the alarmone (p)ppGpp correlated with reduced growth and massive overproduction of PHB. In contrast, in the absence of (p)ppGpp, mobilization of PHB was dramatically enhanced.
KEYWORDS: PHB accumulation, PHB degradation, ppGpp, stringent response, Ralstonia eutropha, polyhydroxyalkanoate synthesis
INTRODUCTION
Polyhydroxybutyrate (PHB) and related polyhydroxyalkanoates (PHA) are storage materials in many prokaryotic species and help the bacteria to survive under adverse conditions (1–6). PHB can be used industrially for the production of biodegradable plastic (7) and medicinal components (8, 9). The model organism for PHB production is Ralstonia eutropha H16, a Gram-negative betaproteobacterium. Two of its properties make R. eutropha famous: on the one hand, R. eutropha is able to grow chemolithoautotrophically with hydrogen and carbon dioxide as the electron donor and acceptor, respectively (a Knallgas bacterium), and on the other hand, it accumulates large amounts (>80% [dry weight] of the cell) of PHB. PHA produced by R. eutropha or related species are commercially available biopolymers and can be used for different purposes (10, 11).
In vivo, PHB granules are supramolecular complexes termed carbonosomes (5, 12). Carbonosomes have a polymer core and a surface layer of as many as 14 proteins with different functions (13) but apparently lack a phospholipid membrane (14), in contrast to what had been assumed previously.
PHA is produced in R. eutropha in the presence of a surplus of a carbon source or under otherwise imbalanced culture conditions. The structural genes for the formation of PHA, as well as the genes responsible for the regulation and mobilization of PHA under C-limiting conditions, have been identified and biochemically characterized over the past few decades (for a selection of reviews, see references 3 and 15–20). Much evidence that the key enzymes of PHA synthesis (PHA synthase [PhaC]) and PHA mobilization (PHB depolymerases [PhaZs]) are constitutively expressed and are generally present on the carbonosome granules has been published (21–24). This raises the question of how the synthesis and mobilization of carbonosomes are regulated and how a futile cycle of simultaneous synthesis and degradation is avoided. One possibility would be allosteric regulation of the key players (PHB synthase, PHB depolymerase) by key metabolites. An alternative or additional possibility is that PHA synthesis and mobilization are regulated by signal molecules. A key signal molecule is the alarmone (p)ppGpp, which, when present at high concentrations, induces the stringent response in Escherichia coli and many other bacterial species (25). The number of publications showing a role for (p)ppGpp in the survival of bacteria under changing environmental conditions is continuously increasing. Alarmones can dramatically influence transcription during the stringent response in different bacteria (26, 27). Recent publications show that ppGpp is robustly, but transiently, induced in response to DNA damage so as to allow efficient nucleotide excision repair in E. coli (28). Alarmones have been shown to induce the stringent response upon nitrogen starvation (29). It has also been shown that (p)ppGpp production can be stimulated by the phosphotransferase (PTS) system in order to respond to metabolic cues and control cell cycle progression and cell growth in Caulobacter crescentus (30). Furthermore, elevated synthesis of the (p)ppGpp alarmone can lead to a full bypass of the d,d-transpeptidase activity of penicillin binding proteins (PBPs) and to a broad spectrum of β-lactam resistances (31, 32). Alarmones also play a pivotal role in the mechanisms of bacterial persistence during stress and antibiotic exposure.
Support for a connection between PHB metabolism and the stringent response in R. eutropha was recently published in two reports in which the authors identified an interaction of the PTS protein EIIANtr with the ppGpp synthase/hydrolase SpoT1 (33) and showed that an EIIANtr knockout mutant accumulated substantially more PHB than the wild type (34). Furthermore, the North American authors of this study claimed in a previous study that a mutant (Re2411) with a deletion in the H16_A1337 gene (ΔspoT2) had completely lost the ability to accumulate PHB, pointing to an essential function of SpoT2 for PHB metabolism (24). Another report on an essential function of the spoT gene for PHB accumulation has been published for Rhizobium etli (35). However, (p)ppGpp, the key molecule in the stringent response, was not measured in any of these studies. This prompted us to hypothesize a correlation between the formation and concentration of (p)ppGpp, on the one hand, and the accumulation of PHB, on the other. To investigate this hypothesis, we constructed strains in which one or both spoT genes of R. eutropha were deleted. In the course of these investigations, it turned out that the previously published ΔspoT2 mutant (Re2411) (24) was not correctly constructed and in fact was a PHB synthase (ΔphaC) mutant, which explains the PHB-negative phenotype. Nonetheless, the determination of ppGpp concentrations and PHB contents in the background of the wild type and true spoT mutants revealed a positive correlation between the level of ppGpp and the amount of accumulated PHB.
RESULTS
Development of a (p)ppGpp extraction and quantification method suited to R. eutropha.
To investigate a possible contribution of a (p)ppGpp-mediated stringent response to PHB metabolism, it was first necessary to develop a (p)ppGpp extraction and quantification method that was suitable to R. eutropha, a hard-to-disrupt soil bacterium. For the optimization and adaptation of the nucleotide extraction procedure, several combinations of protocols for (p)ppGpp extraction and high-performance liquid chromatography–mass spectrometry (HPLC-MS) quantification from Gram-positive (Staphylococcus aureus) and Gram-negative (E. coli) species were tested. For details of the different methods tested, see Document S1 in the supplemental material. The final method that worked sufficiently well for R. eutropha is summarized in Materials and Methods.
Induction of the stringent response by norvaline.
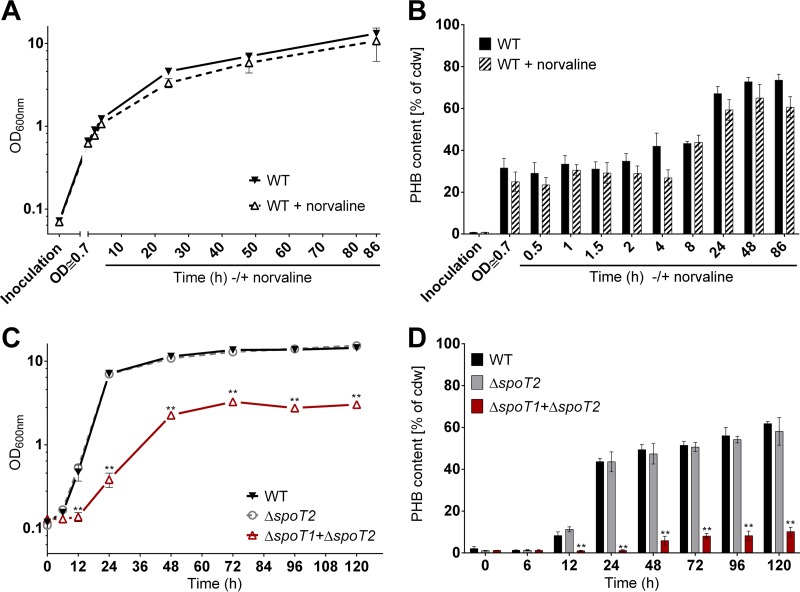
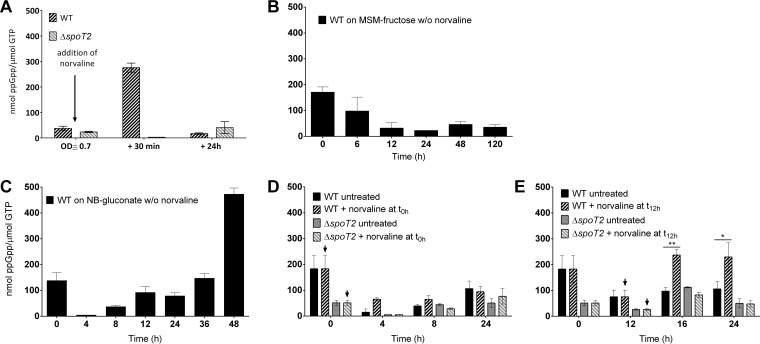
Previous analysis had suggested that the addition of norvaline—a substrate analog of the amino acid valine and a compound commonly used to induce the stringent response—to a growing culture of R. eutropha on fructose-mineral salts medium (MSM-fructose) could slightly increase the formation of PHB (24). However, the amount of (p)ppGpp was not determined in that study. We cultivated wild-type R. eutropha on MSM-fructose with or without the addition of norvaline under exactly the same conditions as those described previously (24). In this experiment, we followed the growth of R. eutropha and the formation of PHB, and we determined the (p)ppGpp concentration. The presence of norvaline slightly reduced the growth of R. eutropha (Fig. 1A). We could not detect a norvaline-mediated increase in PHB contents (Fig. 1B) such as that described previously (24). In our hands, the PHB contents of the norvaline-treated culture generally were rather lower than those of nontreated cultures. This finding could be a consequence of the reduced growth rate in the presence of norvaline; however, the observed differences were within the range of error. The concentration of (p)ppGpp at the time of norvaline addition (optical density at 600 nm [OD600], 0.7) was rather low [≈30 nmol (p)ppGpp/μmol GTP (Fig. 2A)] but increased approximately 9-fold, to ≈270 nmol (p)ppGpp/μmol GTP, 30 min after the addition of norvaline. At 24 h, the ppGpp level was decreased again, to values below 20 nmol ppGpp/μmol GTP. Such an increase in the ppGpp concentration during growth on MSM-fructose was never found in the absence of norvaline in a separate 120-h growth experiment on MSM-fructose (Fig. 2B). The values in the figures are given as the means of three biological triplicates and are expressed as the ratio of ppGpp to GTP. This avoids misinterpretation in case not all cells of a sample were broken by the cell disruption procedure. A high ppGpp-to-GTP ratio always correlated with high absolute amounts of ppGpp (data not shown). Our results confirmed that norvaline could induce the formation of ppGpp intermediately but had no positive effect on the accumulation of PHB.
FIG 1.
Growth and PHB accumulation of R. eutropha strains. (A and B) Growth (A) and PHB contents (B) of wild-type (WT) R. eutropha on MSM-fructose medium with or without the addition of 0.1% (wt/vol) norvaline. Inoculation was carried out with a 24-h NB-grown seed culture at an OD600 of ≈0.1. After 20 h, an OD600 of 0.7 was reached, and norvaline was added to one culture. PHB was quantified for samples taken at the indicated time points after norvaline addition. (C and D) Growth (C) and PHB contents (D) of wild-type R. eutropha and spoT mutants on MSM-fructose medium. Statistical significance was determined using the Holm-Sidak method (**, P < 0.01). All experiments were performed as biological triplicates. Data points or bars represent mean values; error bars, standard deviations.
FIG 2.
Effect of norvaline on the production of ppGpp. Wild-type (WT) R. eutropha alone (B and C) or both the WT and the ΔspoT2 mutant (A, D, and E) were grown on MSM-fructose medium (A and B) or on NB-gluconate medium (C to E) with (arrows) or without (w/o) norvaline. Norvaline (0.1%, wt/vol) was added when the OD600 had reached 0.7 (≈20 h after inoculation) (A) or at 0 h (D) or 12 h (E). ppGpp concentrations were determined in samples taken at the times indicated. Inoculations were carried out with NB-grown (stationary) seed cultures. All experiments were performed as biological triplicates. Bars represent mean values; error bars, standard deviations. Statistical significance was determined using the Holm-Sidak method (*, P < 0.05; **, P < 0.01).
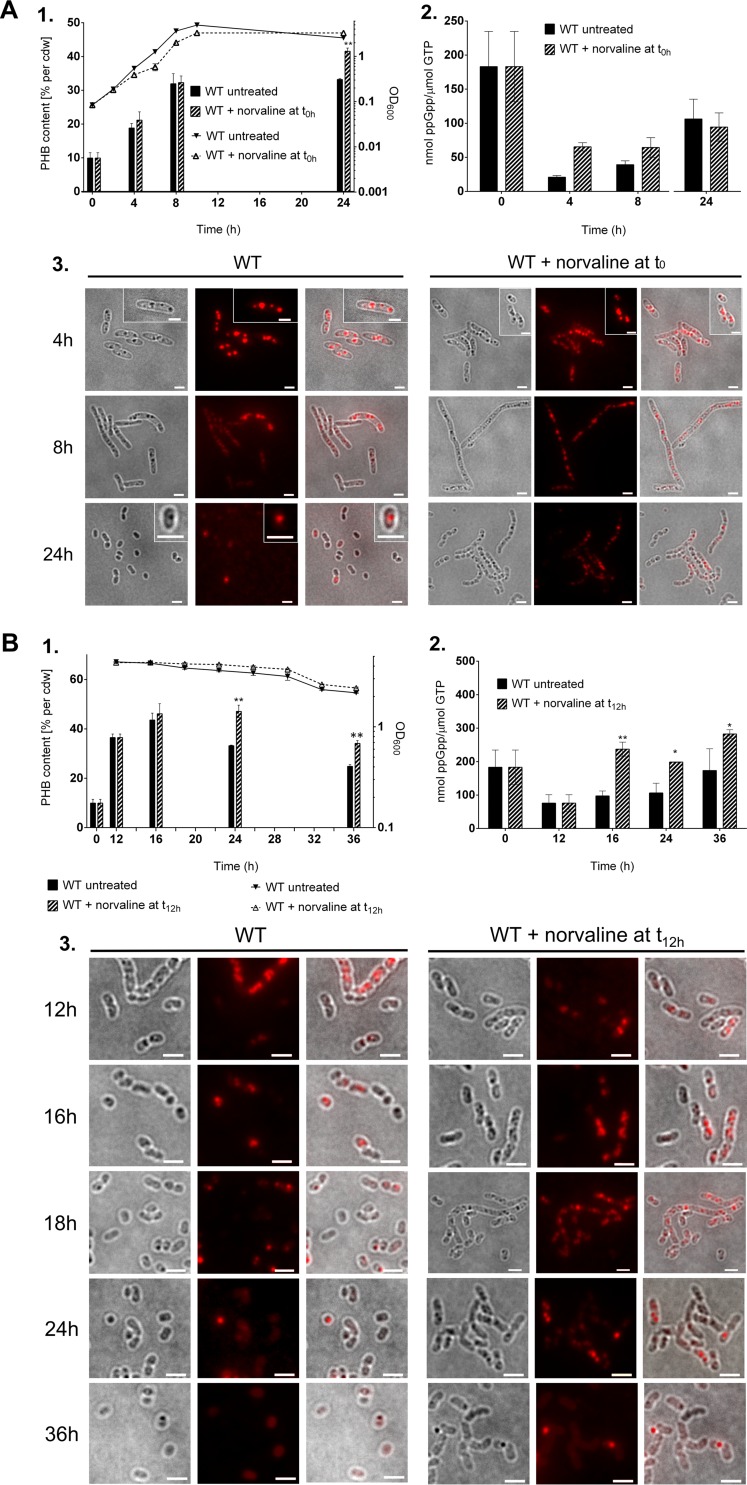
Next, we performed similar experiments in a complex medium. To this end, we cultivated R. eutropha in nutrient broth (NB) medium that had been supplemented with 0.2% gluconate to promote PHB accumulation by increasing the C/N ratio of the medium. Right at the beginning, 0.1% norvaline was added. Growth and PHB contents were followed by OD600 determination and fluorescence microscopic analysis after staining of the cells with Nile red (Fig. 3). At selected time points, samples were taken for the determination of growth, ppGpp concentrations, and PHB contents. As shown in Fig. 3A1, the presence of norvaline caused a slight slowdown of growth from that of a control without norvaline. However, after 24 h, the cultures with norvaline had caught up, and almost-identical OD values were found. Microscopic examination of the cultures at different times of growth revealed that the presence of norvaline had an impact on cell morphology in the late-exponential and stationary phases (Fig. 3A3). Treated cells were substantially longer (some were longer than 10 μm), presumably because cell division was inhibited or delayed in the presence of the amino acid analog. Examination of Nile red-stained cells also revealed an effect on PHB accumulation: norvaline-treated cells harvested at 8 h or 24 h had slightly more PHB granules than untreated cells. When we determined the PHB content qualitatively and quantitatively by gas chromatography, we confirmed a slight increase in the PHB contents in the stationary phase in the presence of norvaline (Fig. 3A1). However, we cannot tell whether the increased amount of PHB is due to the effect of the amino acid analog on PHB mobilization or is simply a consequence of the slower growth of the cultures. Slower growth gives rise to nutrient deprivation at a later time point and thus causes a later onset of PHB mobilization. A PHB homopolyester was accumulated, and no evidence for the presence of substantial amounts of 3-hydroxyvalerate (3HV) was obtained.
FIG 3.
Effects of norvaline on PHB content, cell morphology, and ppGpp concentration. Wild-type (WT) R. eutropha was grown on NB-gluconate medium with or without the addition of 0.1% (wt/vol) norvaline at 0 h (A) or at 12 h (B). Samples were taken at the indicated times. (A1 and B1) OD600 values (graphs) and PHB contents (bars). cdw, dry weight of cells. (A2 and B2) ppGpp concentrations. (A3 and B3) Cell morphologies and the formation of PHB granules were visualized by bright-field microscopy (left panels) and by fluorescence microscopy after staining of the cells with Nile red (center panels). Images are merged in the panels on the right. Bars, 2 μm. Statistical significance was determined using the Holm-Sidak method (*, P < 0.05; **, P < 0.01).
To determine whether the increase in the PHB content resulted from reduced growth caused by the presence of norvaline or from the induction of the stringent response, we repeated the experiment under modified conditions: this time, norvaline was added 12 h after inoculation. At this time point, the cells were already in the stationary-growth phase (Fig. 3B1). The effect of norvaline addition at 12 h on the PHB content was substantially stronger than that of norvaline addition at 0 h, and a PHB content of ≈50% of cells (dry weight) was found at 24 h, whereas the PHB content of the control culture without norvaline had dropped to ≈33% (Fig. 3B1). The number of elongated cells was also increased in the norvaline-treated culture (Fig. 3B3).
To find direct evidence that norvaline induces the stringent response by the formation of alarmones, we determined the ppGpp contents of cells for the control culture and for the cultures that were treated with norvaline at 0 h or 12 h. As shown in Fig. 2C, the wild type in the absence of norvaline contained only very small amounts of ppGpp in the exponential phase at 4 h (0 to 20 nmol ppGpp/μmol GTP). About 40 nmol ppGpp/μmol GTP was detected at 8 h; this concentration increased to ≈100 nmol ppGpp/μmol GTP at 12 and 24 h and further increased in the late-stationary phase to 400 to 500 nmol ppGpp/μmol GTP at 48 h. We assume that one or several amino acids of the complex medium were consumed, which, in turn, limited growth and induced the stringent response via SpoT2 (RelA) (for further evidence, see the experiment with the ΔspoT2 mutant described below). The high concentration of ppGpp after 48 h (>400 nmol ppGpp/μmol GTP) also explains the detection of substantial levels of ppGpp at the very beginning of the experiment (Fig. 2B to E and 3A2 and B2), because the seed cultures were also grown on NB medium until the stationary phase. When norvaline was present in the growth medium from the beginning, the levels of ppGpp found were substantially higher at 4 h and 8 h (60 to 70 nmol ppGpp/μmol GTP) than those for the control without norvaline (Fig. 3A2). At later time points (≥24 h), no differences in the ppGpp levels could be detected. When norvaline was added at the transition from exponential growth to stationary growth (at 12 h), a strong effect on the ppGpp concentration was found, and the ppGpp levels at 16 h to 36 h were ≈2 times as high (≥200 nmol ppGpp/μmol GTP) as those in the absence of norvaline and were even further increased at 36 h, to 280 nmol ppGpp/μmol GTP (Fig. 3B2). Similar results in terms of increased ppGpp levels and PHB contents in the stationary phase were found when another amino acid analog, serine hydroxamate, was added to an NB-gluconate culture (see Fig. S2 in the supplemental material). In summary, our data demonstrate a correlation between the presence of norvaline, the formation of ppGpp, and the accumulation of PHB during growth on a complex medium. Since the bacteria no longer multiply and hardly synthesize PHB de novo after 12 h (Fig. 3B1), the differences in PHB contents are likely a consequence of differences in the PHB mobilization system between treated and untreated cells. We assume that the addition of norvaline transiently increased the ppGpp concentration, which, in turn, directly or indirectly slowed down the mobilization of PHB. However, there must be other factors that also have an impact on the mobilization of PHB, because in the late-stationary phase on complex medium, the ppGpp levels are highest (Fig. 2C), and PHB has been completely consumed. We therefore decided to study the relationship between PHB metabolism and the stringent response by constructing mutants in which one or both of the genes responsible for ppGpp synthesis (spoT) were deleted. These deletions should affect the cells throughout the growth phase and not only at certain time points during growth.
Deletion of spoT2 has no detectable effect on growth, PHB synthesis, or PHB mobilization.
R. eutropha H16 has two enzymes for the synthesis of (p)ppGpp, which are called SpoT1 (H16_A0955; orthologous to SpoT in E. coli) and SpoT2 (H16_A1337; orthologous to RelA in E. coli). In E. coli, SpoT is an alarmone synthase and hydrolase that is able to phosphorylate GDP to ppGpp and GTP to pppGpp or to hydrolyze these alarmone nucleotides to GDP and GTP, respectively (36). RelA has only alarmone synthase activity and is unable to hydrolyze (p)ppGpp. To determine whether the two spoT genes of R. eutropha were expressed, we inserted a gene coding for the enhanced yellow fluorescent protein (eyfp) in frame to the 3′ end of the spoT1 or the spoT2 gene of the chromosome. Fluorescence microscopic examination of the strains expressing either spoT1-eyfp or spoT2-eyfp showed very weak yellow-green fluorescence that was homogeneously distributed in the cytoplasm (see Fig. S3 in the supplemental material). This result confirmed that both spoT genes were expressed, although at low levels, in wild-type R. eutropha.
A spoT2 (H16_A1337) deletion strain of R. eutropha (named Re2411) had been constructed in a previous investigation by the North American authors of this study (24). Remarkably, Re2411 was unable to form any detectable PHB on MSM-fructose, indicating that the stringent response could be essential for PHB metabolism. A new R. eutropha ΔspoT2 strain was constructed in the lab of the University of Stuttgart authors because the previously published Re2411 strain was not yet available at the beginning of this project. However, when the growth and PHB accumulation of the newly constructed ΔspoT2 strain were determined, no significant defect in growth or PHB accumulation could be detected in comparison to the wild type (Fig. 2; see also Fig. 4 and explanation below). This result showed that SpoT2 did not detectably influence the accumulation of PHB in MSM-fructose or NB-gluconate medium. Nevertheless, reexamination of strain Re2411 (ΔspoT2), which had meanwhile become available, reproducibly confirmed the PHB-negative phenotype that had been described previously (24). To resolve this contradiction, the spoT2 and phaC1 (PHB synthase) genes of the ΔspoT2 mutant of this study, the Re2411 (ΔspoT2) mutant of the previous study, and wild-type R. eutropha were PCR amplified. As shown in Fig. S4 in the supplemental material, full-length PCR products of the spoT2 gene were obtained from chromosomal DNA of the wild type and the Re2411 (ΔspoT2) mutant. Only a short PCR product was found for the newly constructed ΔspoT2 mutant, and DNA sequencing confirmed the deletion of the spoT2 gene in the ΔspoT2 mutant and the presence of the full-length spoT2 gene in the PCR product of the Re2411 mutant. This result clearly showed that the assumed ΔspoT2 mutant (Re2411) in fact has a wild-type spoT2 locus and that thus, the absence of PHB in that mutant cannot be caused by the absence of spoT2. When we amplified the phaC1 genes of all strains by PCR, we obtained DNA fragments corresponding to full-length phaC1 for the wild type and the new ΔspoT2 mutant and confirmed the expected presence of the PHB synthase gene. Notably, the phaC1 locus-specific PCR product was considerably smaller for the Re2411 mutant than for the wild type (Fig. S4), and DNA sequencing showed that this locus corresponded to a precise deletion of the phaC1 gene. This result explains the PHB-negative phenotype of the Re2411 mutant, which in fact has a phaC1 deletion and not a spoT2 deletion. The absence of PHB in the Re2411 mutant is not a consequence of a deleted spoT2 gene. These results appear to contradict the statement made in the previous publication on the supposedly essential function of SpoT2 for PHB accumulation in R. eutropha (24).
FIG 4.
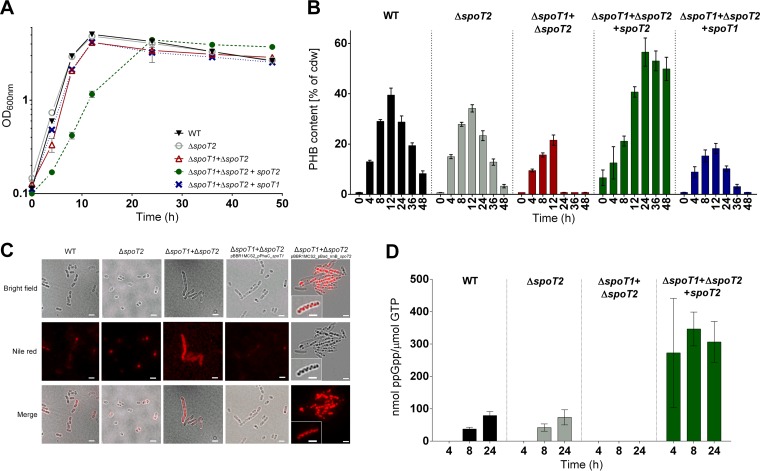
Growth and PHB accumulation in R. eutropha spoT mutants. Shown are growth curves on NB-gluconate medium (with growth expressed as the OD600) (A), PHB contents (B), cell morphology and the formation of PHB granules (C), and ppGpp contents (D) for R. eutropha strains with the indicated genotypes. All experiments were performed as biological triplicates. The data points or bars represent means; error bars, standard deviations. Note that in panel D, no ppGpp was detected for the wild type (WT) at 4 h or for the ΔspoT1 ΔspoT2 double mutant throughout growth. Two complemented mutants were included in the experiment: the ΔspoT1 ΔspoT2 double mutant harboring a plasmid with a spoT2 gene under the control of the pBAD promoter (in the presence of arabinose) and the same mutant harboring a plasmid with a spoT1 gene under the control of the phaC promoter. Bar, 2 μm.
Deletion of spoT2 abolishes ppGpp formation under stringent-response conditions.
The results presented above suggested that (p)ppGpp is somehow important for PHB metabolism but excluded the possibility that SpoT2 has a prominent function in PHB metabolism. By analogy to E. coli, we suspected that the physiological function of SpoT2 is to act as a sensor for amino acid starvation. To find experimental evidence for this, we compared the responses of wild-type R. eutropha and the ΔspoT2 mutant to the addition of norvaline. To this end, we added norvaline to an MSM-fructose culture and to an NB-gluconate culture and determined the amounts of ppGpp before and after the addition of norvaline. As is evident from Fig. 2A, D, and E, norvaline effectuated a strong increase in the concentration of ppGpp in the wild type. This effect was detectable on both culture media and was independent of the time of norvaline addition. No increase in the concentration of ppGpp in the ΔspoT2 mutant was found within 24 h after the addition of norvaline. This result is in agreement with a RelA-related function of SpoT2 in the stringent response.
Deletion of spoT1 leads to severe growth defects in R. eutropha.
To investigate potential effects of the spoT1 gene on (p)ppGpp formation and PHB metabolism, we tried to construct a ΔspoT1 mutant by a procedure analogous to that used for the construction of the spoT2 mutant. Unfortunately, it was not possible to produce a ΔspoT1 strain despite multiple attempts. Since the spoT1 and spoT2 genes are annotated as a bifunctional (p)ppGpp synthase/hydrolase and a monofunctional (p)ppGpp synthase, respectively, we assume that the (p)ppGpp synthase activity of SpoT2 in a ΔspoT1 background would result in permanent accumulation of (p)ppGpp. Permanently high levels of ppGpp can lead to arrest of the cell cycle, as has been described for other species (36), and can explain the failure to isolate a spoT1 mutant. Therefore, investigation of the phenotype of a spoT1 deletion in a wild-type spoT2 background was not possible. To analyze a potential function of SpoT1 in PHB metabolism, we constructed a spoT1 deletion in a ΔspoT2 background of R. eutropha. The deletion of spoT1 in a spoT2 background did not impair growth on NB or on MSM-fructose medium. The absence of both spoT genes abolishes (p)ppGpp-synthesizing activity, as revealed by the inability to detect any ppGpp under conditions under which the wild type readily produced ppGpp (Fig. 4D). Apparently, the absence of (p)ppGpp is less problematic for the cells than the permanent presence of high levels of (p)ppGpp.
A ΔspoT1 ΔspoT2 double-deletion mutant shows strong impairment in the accumulation of PHB.
The construction of a R. eutropha ΔspoT1 ΔspoT2 double-deletion mutant was successful (a confirmation of the genotype of the ΔspoT1 ΔspoT2 double mutant is shown in Fig. S5 in the supplemental material). A much lower growth rate was determined for the ΔspoT1 ΔspoT2 double mutant on MSM-fructose, while no detectable growth defect was found for the ΔspoT2 single mutant (Fig. 1C), relative to the wild type. Most remarkably, the ability of the ΔspoT1 ΔspoT2 double-deletion mutant to accumulate PHB under permissive conditions (MSM-fructose) was strongly reduced (Fig. 1D). At 24 h of growth, the wild type and the ΔspoT2 mutant had already accumulated >40% PHB (dry weight of cells), while the PHB content of the ΔspoT1 ΔspoT2 double-deletion mutant at the same time point was near the detection limit (<5%). A PHB content of only 5 to 10% was determined for the ΔspoT1 ΔspoT2 double mutant in the stationary phase between 48 and 120 h. This is considerably lower than the PHB contents of the wild type and the ΔspoT2 single mutant at the same time points (50 to 60% PHB) (Fig. 1D). Our result shows that PHB synthesis is still functioning in the ΔspoT1 ΔspoT2 double mutant. However, as indicated below, the PHB mobilization system apparently is much more active in the double mutant than in the wild type.
PHB mobilization is regulated by the presence of ppGpp.
The PHB mobilization activity of R. eutropha was much higher in cultures grown on a complex medium (NB plus gluconate) than in cultures grown on MSM-fructose (see above; see also Fig. S6 in the supplemental material). To study the effect of spoT gene deletion on PHB mobilization, we performed growth experiments on nutrient broth to which 0.2% (wt/vol) sodium gluconate had been added to increase the C/N ratio for the purpose of increasing the amount of accumulated PHB. Under this condition, wild-type R. eutropha accumulates as much as ≈40% of PHB in the exponential phase and degrades it in the stationary phase (37).
To simulate the knockout of spoT1 alone, we reintroduced a functional copy of a plasmid-borne spoT2 gene under the control of an arabinose-inducible promoter into the ΔspoT1 ΔspoT2 double mutant. This enabled us to study the effect of a single spoT1 deletion by cultivating the transconjugant in the presence of arabinose. Figure 4 shows the growth and PHB accumulation results for the wild type and the spoT deletion mutants on NB-gluconate medium: the growth of the ΔspoT2 single mutant, the ΔspoT1 ΔspoT2 double mutant, and the ΔspoT1 ΔspoT2 double mutant harboring an intact spoT1 copy on a plasmid was undistinguishable from that of the wild type (Fig. 4A). Only the simulated single ΔspoT1 deletion strain (the ΔspoT1 ΔspoT2 mutant with spoT2 and arabinose added) showed a lag phase of several hours and severe growth defects. This indicated that the presence of SpoT2 alone is not lethal but leads to severe growth defects. The reason why we were unable to isolate a growing single ΔspoT1 deletion strain remains unknown.
The most remarkable results were obtained when we determined the amount of accumulated PHB during growth on NB-gluconate (Fig. 4B): as with the MSM-fructose culture, no significant differences in PHB content were detected between the wild type and the single ΔspoT2 mutant, either during the PHB accumulation phase (0 to 12 h) or in the PHB mobilization phase (12 to 48 h). In contrast, the ΔspoT1 ΔspoT2 double mutant produced only half as much PHB (≈20%) as the wild type (≈40%), and the degradation of PHB was much faster. After 24 h, the cells were devoid of PHB. This result corresponded to the extremely small amounts of accumulated PHB in the ΔspoT1 ΔspoT2 double mutant on MSM-fructose (Fig. 1D). The phenotype of strongly reduced PHB accumulation (or strongly enhanced PHB mobilization) could be rescued to almost-wild-type levels by complementing the ΔspoT1 ΔspoT2 knockout mutant with an intact spoT1 copy on a plasmid (Fig. 4B).
The cells of the R. eutropha ΔspoT1 ΔspoT2 double mutant carrying an arabinose-inducible spoT2 copy on a plasmid not only grew more slowly than the wild type (Fig. 4A) but also produced much more PHB (as much as ≈ 60%, a value that had never been observed for the wild type on NB–0.2% gluconate medium). Furthermore, the cells showed reduced PHB mobilization in the stationary phase. These data were verified by gas chromatography (Fig. 4B) and by fluorescence microscopy after staining of the cells with Nile red (Fig. 4C). Microscopic examination revealed altered morphology for the cells of the ΔspoT1 ΔspoT2 double mutant: the mutant cells were much longer and became almost filamentous (Fig. 4C).
Next, we determined the ppGpp and GTP contents in cell extracts of all strains by HPLC-MS in the exponential-growth phase (4 h), at the beginning of the transition from the exponential- to the stationary-growth phase (8 h), and in the stationary phase (24 h) (Fig. 4D). Wild-type R. eutropha and its ΔspoT2 mutant had no detectable ppGpp in the exponential phase (4 h), but the ppGpp/GTP ratio became detectable during later stages of growth and increased up to ≈80 nmol ppGpp/μmol GTP in the stationary phase (24 h). As expected, the ΔspoT1 ΔspoT2 double mutant was unable to produce ppGpp at any time, confirming the assumption that SpoT1 and SpoT2 are the only (p)ppGpp-synthesizing enzymes in R. eutropha (Fig. 4D). The cells of the R. eutropha ΔspoT1 ΔspoT2 double mutant carrying the arabinose-inducible copy of spoT2 had a high ppGpp/GTP ratio at all time points of growth. Even in the exponential phase at 4 and 8 h, a ppGpp/GTP ratio of 250 to 350 ppGpp nmol/μmol GTP was determined. This result is in agreement with the absence of ppGpp hydrolase activity in SpoT2. We conclude that permanently high levels of (p)ppGpp correlate with high PHB accumulation.
PhaZa1 mobilizes PHB in the absence of ppGpp.
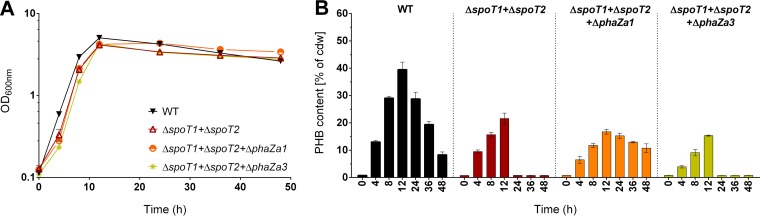
R. eutropha has seven PHB depolymerase genes (37). However, the individual physiological functions of these PHB depolymerase isoenzymes are not precisely known. The most important depolymerase responsible for PHB degradation is probably PhaZa1 (21, 37–40). (For the designations of R. eutropha PHB depolymerases, see Fig. S7 in the supplemental material.) PHB degradation was very fast and efficient in the absence of ppGpp (Fig. 4B). To find out whether the PHB depolymerase PhaZa1 was responsible for the low PHB content of the ΔspoT1 ΔspoT2 double mutant, we deleted the phaZa1 gene in the background of the ΔspoT1 ΔspoT2 double mutant. For purposes of comparison, a deletion of a gene encoding a PHB depolymerase predicted to be less important than PhaZa1, PhaZa3, was introduced in a ΔspoT1 ΔspoT2 background. Figure 5 shows that for all three strains, growth on NB-gluconate medium was not substantially changed. The ΔspoT1 ΔspoT2 ΔphaZa3 triple-knockout mutant behaved exactly like the ΔspoT1 ΔspoT2 double-knockout mutant: in both strains, only about half (20%) of the PHB content of the wild type (40%) was accumulated in the first 12 h, and this small amount of PHB was rapidly degraded in the stationary phase, much more rapidly than in the wild type. Interestingly, the ΔspoT1 ΔspoT2 ΔphaZa1 triple-knockout mutant also produced less PHB than the wild type, but the accumulated PHB was only very slowly and incompletely degraded in the stationary-growth phase. Even after 48 h, the cells still contained at least 10% PHB (dry weight), whereas the PHB contents of the ΔspoT1 ΔspoT2 double mutant and the ΔspoT1 ΔspoT2 ΔphaZa3 triple mutant were already at or below the detection limit after 24 h. This result indicated that PhaZa1 is the principal depolymerase responsible for the degradation of PHB in the absence of ppGpp (Fig. 5B). Since R. eutropha is able to accumulate PHB even in the absence of (p)ppGpp (ΔspoT1 ΔspoT2 double mutant), we suggest that (p)ppGpp either indirectly or directly inhibits PHB mobilization.
FIG 5.
The PHB depolymerase PhaZa1 is responsible for the mobilization of PHB in the ΔspoT1 ΔspoT2 double mutant. Shown are the growth curves (with growth expressed as the OD600) (A) and PHB contents (B) of wild-type (WT) R. eutropha H16 and the indicated mutants. All experiments were performed as biological triplicates. The data points or bars represent means; error bars, standard deviations.
DISCUSSION
It is known that R. eutropha intermediately synthesizes PHB in a complex medium such as nutrient broth (NB) in the exponential-growth phase and subsequently degrades (mobilizes) PHB in the stationary phase. In contrast, when grown on MSM-fructose, R. eutropha does not degrade PHB as long as a suitable carbon source, such as fructose or gluconate, is present (see Fig. S6 in the supplemental material). Only when the medium is replaced by fresh medium without a carbon source but with an excess of a nitrogen source (ammonium) is PHB degraded (37, 41). However, even under such “mobilization conditions,” only a portion of the previously accumulated PHB is reutilized. This indicates that PHB synthesis and PHB mobilization are differentially regulated in NB-grown and MSM-fructose-grown cells.
NB and MSM differ in amino acid content: all 20 amino acids are present in NB medium and are completely absent in MSM. Since E. coli and presumably many other bacterial species respond to a (sudden) lack of amino acids with the (p)ppGpp-mediated stringent response, we assumed that the availability of all 20 amino acids from NB medium results in a shutdown of the expression of all amino acid biosynthetic pathways. Accordingly, (p)ppGpp levels and the stringent response should be low during the exponential growth of R. eutropha on NB medium but should increase as soon as the first amino acids of the nutrient broth are exhausted. Determination of (p)ppGpp levels during growth on NB-gluconate medium confirmed this expectation. Hardly any (p)ppGpp was detected during exponential growth (4 h) on NB-gluconate medium, but (p)ppGpp levels increased in the transition phase from exponential growth to the stationary phase and strongly increased in the late-stationary-growth phase. Thus, (p)ppGpp levels reflect the successful consumption of the amino acids of the growth medium. On the other hand, growth on MSM requires that the biosynthetic pathways for the formation of all amino acids be turned on. As a consequence, the intracellular levels of amino acids should be sufficiently high to allow for protein biosynthesis but are probably lower than those in an exponentially growing culture in nutrient broth medium. Indeed, the concentration of (p)ppGpp was low during exponential growth but substantially higher than in NB-gluconate medium and only slightly increased (doubled) in the stationary-growth phase under nitrogen-limited conditions.
In this study, we showed that the time course of PHB accumulation and PHB mobilization was strongly influenced by the concentration of the alarmone ppGpp. We could show that SpoT2 resembles RelA and is responsible for the stringent response in R. eutropha in the early-stationary-growth phase of NB-grown cells. The simulated ΔspoT1 mutant (the ΔspoT1 ΔspoT2 double mutant complemented with spoT2 under arabinose control), in which (p)ppGpp synthesis was permanently turned on in the presence of arabinose, accumulated extremely large amounts of PHB during growth. The high PHB content could have one of two explanations. (i) It could be an indirect effect of the reduced growth caused by high (p)ppGpp levels, which also affect many other cellular processes (36). In the presence of an unlimited carbon source and a simultaneous reduction in carbon-consuming reactions, the surplus of “unused” carbon metabolites could be channeled to PHB synthesis, leading to the high levels of PHB observed. (ii) High levels of PHB could also be a direct consequence of a (p)ppGpp-mediated downregulation of PHB-mobilizing activities. The first assumption, of increased PHB accumulation caused by high concentrations of carbon metabolites that cannot be efficiently used for anabolic processes at high (p)ppGpp levels, appears unlikely, because the PHB content did not decrease in the late-stationary-growth phase of the mutant cells on NB-gluconate medium (Fig. 4A and B). Despite the slower growth of the ΔspoT1 ΔspoT2 double mutant, the cells had caught up to the level of wild-type cells at 24 h. At this time, the nutrients must have been consumed by both strains to comparable extents. For the wild type, the intermediately accumulated PHB is consumed in the stationary phase, and after 48 h, only residual amounts of PHB (<10% of cells [dry weight]) can be detected. For the ppGpp-overproducing mutant, only a marginal decrease in the PHB content, from ≈55% to 50% of the cells (dry weight), was determined. This finding supports the assumption that the PHB-mobilizing activity is switched off almost completely in the presence of unusually high ppGpp concentrations. Unfortunately, PHB depolymerase activity is difficult to determine accurately in cell extracts, since many isoenzymes of PHB depolymerases can be present, PHB depolymerases can detach from PHB granules during cell extract preparation, and PHB depolymerase activity might depend on the PHB granule surface and PHB granule composition (42, 43).
If (permanently) high levels of ppGpp inhibit PHB mobilization activity, the opposite effect could be expected in the absence of ppGpp. This was indeed observed by the determination of low PHB contents in the ppGpp-free ΔspoT1 ΔspoT2 double mutant under PHB permissive culture conditions on NB-gluconate medium (Fig. 4A and B). Moreover, these (low) PHB contents of the ΔspoT1 ΔspoT2 double mutant were rapidly degraded to levels near or even below the detection limit in the stationary phase, confirming a highly active PHB-mobilizing system. The PHB depolymerase PhaZa1 was identified as the most important PHB depolymerase for mobilization in the stationary phase of NB-gluconate cultures (Fig. 5). Interestingly, similar yet slightly different results were obtained when the PHB content of the ΔspoT1 ΔspoT2 double mutant was followed on MSM-fructose. In this case, hardly any PHB (≤5% of cells [dry weight]) was accumulated during growth, though levels increased slightly in the stationary phase, to about 10% (Fig. 1C and D). Although the difference in PHB content from the wild type (50 to 70%) is enormous, the PHB content in the double mutant increased in the stationary phase on MSM-fructose, while it decreased in NB-gluconate medium. Apparently, the balance between PHB-synthesizing and PHB-mobilizing activity in the absence of ppGpp in MSM-grown cells is different from that in NB-grown cells. In future studies, it will be necessary to investigate how the specific activities of the PHB-synthesizing and PHB-mobilizing enzymes are regulated. Since PHB synthase and PHB depolymerase(s) are constitutively expressed in R. eutropha, we assume that a combination of allosteric regulation and covalent modification of the involved enzymes are present. The assumption of a covalent modification such as phosphorylation would be in line with previous data. Karstens and coworkers previously showed that in R. eutropha, SpoT1 but not SpoT2 interacted with the nonphosphorylated form of the EIIANtr component of the PTS system (33). This indicates a possible phosphorylation cascade from the carbon source uptake system to SpoT1, which could, in turn, influence the activity of the PHB mobilization system. A connection between the nitrogen-related PTSNtr system and (p)ppGpp alarmone synthesis has also been described for C. crescentus (30). Since PHB accumulation in most PHB-accumulating bacteria is highest under conditions of nitrogen limitation or otherwise imbalanced growth conditions, the connection between the PTSNtr system and PHB accumulation might be broadly distributed among PHA-accumulating species.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. R. eutropha H16 strains were grown aerobically in nutrient broth (NB; 0.8% [wt/vol]) with or without sodium gluconate (0.2% [wt/vol]) at 30°C. Alternatively, R. eutropha was grown in mineral salts medium (MSM) (44) with fructose (2% [wt/vol]) as a carbon source at 30°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| JM109 (SN528) | For cloning and plasmid storage | DSMZ 3423 |
| TOP10 (SN5931) | For cloning and plasmid storage | 48 |
| S17-1 (SN490) | For conjugation | 49 |
| Ralstonia eutropha | ||
| H16 (SN1342) | R. eutropha wild-type strain | DSMZ 428 |
| H16 ΔspoT2 (SN5351) | R. eutropha H16 knockout of spoT2 | This study |
| Re2411 (SN5784) | See Results | 24 |
| H16 ΔspoT1 ΔspoT2 (SN5991) | R. eutropha H16 knockout of spoT2 and spoT1 | This study |
| H16 ΔspoT1 ΔspoT2 ΔphaZa1 (SN6171) | R. eutropha H16 knockout of spoT1, spoT2, and phaZa1 | This study |
| H16 ΔspoT1 ΔspoT2 ΔphaZa3 (SN6172) | R. eutropha H16 knockout of spoT1, spoT2, and phaZa3 | This study |
| H16 SpoT1-eYFP (SN6476) | Chromosomal integration of eyfp downstream of spoT1 | This study |
| H16 SpoT2-eYFP (SN6477) | Chromosomal integration of eyfp downstream of spoT2 | This study |
| Plasmids | ||
| pLO3 | Deletion vector; Tcr sacB | 50 |
| pLO3_phaZa1 (SN6135) | Plasmid storage strain for phaZa1 knockout; Tcr | This study |
| pLO3_phaZ5 (SN6136) | Plasmid storage strain for phaZ5 knockout; Tcr | This study |
| pLO3_spoT1 (SN5309) | Conjugation strain for spoT1 knockout; Tcr | This study |
| pLO3_spoT2 (SN5310) | Conjugation strain for spoT2 knockout; Tcr | This study |
| pLO3_spoT1-eyfp (SN6474) | Chromosomal integration of eyfp fused to spoT1; Tcr | This study |
| pLO3_spoT2-eyfp (SN6475) | Chromosomal integration of eyfp fused to spoT2; Tcr | This study |
| pBBR1MCS2 | Broad-host-range vector; Kmr | 49 |
| pBBR1MCS2-PphaC-eyfp-c1 | Universal vector for construction of fusions C-terminal to eYFP under the control of the PphaC1 promoter | 51 |
| pBBR1MCS2-PphaC-eyfp-n1 | Universal vector for construction of fusions N-terminal to eYFP under the control of the PphaC1 promoter | 51 |
| pBBR1MCS2-PphaC-spoT1 (SN6166) | Complementation of SpoT1; Kmr | This study |
| pBBR1MCS2-PBAD-rrnB-spoT2 (SN6204) | Inducible complementation of SpoT2; Kmr | This study |
| pBBR1MCS2 -PphaC-spoT1-eyfp-N (SN6129) | N-terminal fusion of SpoT1 to eYFP | This study |
| pBBR1MCS2-PphaC-spoT2-eyfp-N (SN6130) | N-terminal fusion of SpoT2 to eYFP | This study |
| pBBR1MCS2-PphaC-eyfp-C-spoT1 (SN6131) | C-terminal fusion of SpoT1 to eYFP | This study |
| pBBR1MCS2- PphaC-eyfp-C-spoT2 (SN6132) | C-terminal fusion of SpoT2 to eYFP | This study |
Tcr, tetracycline resistance; Kmr, kanamycin resistance.
Extraction and quantitative measurement of intracellular nucleotide pools.
The procedure for the extraction of nucleotides was adapted from the work of Ihara and coworkers (45). Cells were grown either in 100 ml of NB (0.8% [wt/vol]) with or without the addition of 0.2% (wt/vol) sodium gluconate or in 100 ml of mineral salts medium (with 2% fructose and 0.1% [wt/vol] NH4Cl) at 30°C. One hundred milliliters of culture was harvested by centrifugation in Falcon tubes half-filled with ice (7 min, 5,000 rpm, 4°C). The pellets were flash-frozen in liquid nitrogen and were stored at −80°C until extraction. The pellets were resuspended in 3 ml of 2 M formic acid during thawing on ice. Samples were incubated for 30 min on ice before the addition of 3 ml of 50 mM ammonium acetate (pH 4.5). Oasis WAX 1 cc Vac cartridges (60 mg sorbent per cartridge; particle size, 30 μm) were equilibrated first with 3 ml methanol and then with 3 ml of 50 mM ammonium acetate by centrifugation in a 15-ml Falcon tube (5 min, 5,000 rpm, 4°C). The samples were loaded onto the equilibrated cartridges and were centrifuged (5 min, 5,000 rpm, 4°C). The cartridges were washed first with 3 ml of 50 mM ammonium acetate and then with 3 ml methanol. The samples were eluted in 1 ml of a methanol–H2O–NH4OH (20:70:10) solution, transferred to a fresh Falcon tube, and lyophilized overnight.
Lyophilized samples were dissolved in 100 μl of water and were centrifuged (5 min, 14,000 rpm, 4°C), and 5 μl of each sample was used for liquid chromatography-mass spectrometry (LC-MS) analysis, using an UltiMate 3000 HPLC system (Dionex), coupled to an electrospray ionization–time of flight (ESI-TOF) mass spectrometer (MicrOTOF II; Bruker), operated in negative-ion mode. Samples were separated by use of a SeQuant ZIC-pHILIC column (PEEK; length, 150 mm; inside diameter, 2.1 mm; particle size, 5 μm; Merck) at 30°C. The gradient and flow rate were those described previously (46). The Data Analysis program (Bruker) was used to present the nucleotide masses as extracted ion chromatograms, and the peak areas were calculated and quantified with Prism, version 6 (GraphPad), with a baseline set to 100. Dilution series of commercial GTP (m/z 521.98) and ppGpp (m/z 601.95) (TriLink BioTechnologies, San Diego, CA, USA) nucleotides were used for calibration to quantify the nucleotides. The concentrations of pppGpp were generally too low to be reliably quantified in R. eutropha samples.
Construction of gene deletions and gene fusions.
Precise genomic deletions of R. eutropha genes were constructed using the sacB-sucrose selection method (10% sucrose used for selection) and pLO3 as a deletion vector as described previously (37). The same procedure was used to construct chromosomal integrations of eyfp into the genome. The genotypes of all constructs and mutants generated were verified by PCR of the respective DNA regions and determination of their DNA sequences.
Microscopic methods.
PHB granules were visualized by fluorescence microscopy of Nile red-stained cells using standard filters (excitation wavelengths, 562 and 40 nm; emission wavelength, 594 nm [long-pass filter]) as described previously (13, 14). Insoluble inclusions could also be visualized by bright-field microscopy and appeared as dark spheres.
Other methods.
Quantitative analysis of PHA contents was performed using GC after acid methanolysis of lyophilized cells according to reference 47. DNA manipulation and construction of plasmids were done by standard molecular biological methods and according to the supplier's instructions. DNA was amplified by PCR using commercial oligonucleotides of at least 16 bp that were identical in sequence to the corresponding region of the R. eutropha genome. DNA sequencing was performed using the following primers: H16_Seq_PhaC1_fwd (CCATTCGGATAGCATCTCCCCATGCAAAGTGCCG), p_YFP_n1_seq (CAGCTCCTCGCCCTTGCTCA), pBBR_pBad_Seq_F (GCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATAC), pBBR_pBad_eY_Seq_F (GCTCCTCGCCCTTGCTCACCAT), pLO3_fwd (GCAAGCAGCAGATTACGCG), pLO3_rev (CCGAGGATGACGATGAGCG), pBBR1MCS2_fwd (CAGCTATGACCATGATTACGCC), pBBR1MCS2_rev (ACGTTGTAAAACGACGGCC), and pBBR1MCS2_f2-new (TATTATACGCAAGGCGACAAGG).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (Je 152/16-1 and GRK1708).
We thank Anna Sznajder, Simone Reinhardt, and Anna Schweter for support in some experiments. The input of Martin Siemann-Herzberg, Benjamin Kästle, and Alexander Klotz in the development of a method for ppGpp extraction is gratefully acknowledged.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00755-17.
REFERENCES
- 1.Anderson AJ, Dawes EA. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev 54:450–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madison LL, Huisman GW. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev 63:21–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinbüchel A, Aerts K, Babel W, Follner C, Liebergesell M, Madkour MH, Mayer F, Pieper-Furst U, Pries A, Valentin HE. 1995. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can J Microbiol 41(Suppl 1):S94–S105. [DOI] [PubMed] [Google Scholar]
- 4.Prieto A, Escapa IF, Martinez V, Dinjaski N, Herencias C, de la Peña F, Tarazona N, Revelles O. 2016. A holistic view of polyhydroxyalkanoate metabolism in Pseudomonas putida. Environ Microbiol 18:341–357. doi: 10.1111/1462-2920.12760. [DOI] [PubMed] [Google Scholar]
- 5.Jendrossek D, Pfeiffer D. 2014. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ Microbiol 16:2357–2373. doi: 10.1111/1462-2920.12356. [DOI] [PubMed] [Google Scholar]
- 6.Shively JM, Cannon GC, Heinhorst S, Bryant DA, DasSarma S, Bazylinski D, Preiss J, Steinbüchel A, Docampo R, Dahl C. 15 December 2011 Bacterial and archaeal inclusions. In eLS. John Wiley & Sons, Ltd, Chichester, United Kingdom. doi: 10.1002/9780470015902.a0000302.pub3. [DOI] [Google Scholar]
- 7.Riedel SL, Jahns S, Koenig S, Bock MCE, Brigham CJ, Bader J, Stahl U. 2015. Polyhydroxyalkanoates production with Ralstonia eutropha from low quality waste animal fats. J Biotechnol 214:119–127. doi: 10.1016/j.jbiotec.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Zonari A, Martins TMM, Paula ACC, Boeloni JN, Novikoff S, Marques AP, Correlo VM, Reis RL, Goes AM. 2015. Polyhydroxybutyrate-co-hydroxyvalerate structures loaded with adipose stem cells promote skin healing with reduced scarring. Acta Biomater 17:170–181. doi: 10.1016/j.actbio.2015.01.043. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q, Wang Y, Chen G-Q. 2009. Medical application of microbial biopolyesters polyhydroxyalkanoates. Artif Cells Blood Substit Immobil Biotechnol 37:1–12. doi: 10.1080/10731190802664429. [DOI] [PubMed] [Google Scholar]
- 10.Chen G-Q. 2009. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem Soc Rev 38:2434–2446. doi: 10.1039/b812677c. [DOI] [PubMed] [Google Scholar]
- 11.Dinjaski N, Prieto MA. 2015. Smart polyhydroxyalkanoate nanobeads by protein based functionalization. Nanomedicine (Lond) 11:885–899. doi: 10.1016/j.nano.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jendrossek D. 2009. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191:3195–3202. doi: 10.1128/JB.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sznajder A, Pfeiffer D, Jendrossek D. 2015. Comparative proteome analysis reveals four novel polyhydroxybutyrate (PHB) granule-associated proteins in Ralstonia eutropha H16. Appl Environ Microbiol 81:1847–1858. doi: 10.1128/AEM.03791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bresan S, Sznajder A, Hauf W, Forchhammer K, Pfeiffer D, Jendrossek D. 2016. Polyhydroxyalkanoate (PHA) granules have no phospholipids. Sci Rep 6:26612. doi: 10.1038/srep26612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rehm BHA. 2003. Polyester synthases: natural catalysts for plastics. Biochem J 376:15–33. doi: 10.1042/bj20031254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stubbe J, Tian J, He A, Sinskey AJ, Lawrence AG, Liu P. 2005. Nontemplate-dependent polymerization processes: polyhydroxyalkanoate synthases as a paradigm. Annu Rev Biochem 74:433–480. doi: 10.1146/annurev.biochem.74.082803.133013. [DOI] [PubMed] [Google Scholar]
- 17.Rehm BHA. 2010. Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol 8:578–592. doi: 10.1038/nrmicro2354. [DOI] [PubMed] [Google Scholar]
- 18.Jendrossek D, Handrick R. 2002. Microbial degradation of polyhydroxyalkanoates. Annu Rev Microbiol 56:403–432. doi: 10.1146/annurev.micro.56.012302.160838. [DOI] [PubMed] [Google Scholar]
- 19.Jendrossek D, Schirmer A, Schlegel HG. 1996. Biodegradation of polyhydroxyalkanoic acids. Appl Microbiol Biotechnol 46:451–463. doi: 10.1007/s002530050844. [DOI] [PubMed] [Google Scholar]
- 20.Mezzina MP, Pettinari MJ. 2016. Phasins, multifaceted polyhydroxyalkanoate granule-associated proteins. Appl Environ Microbiol 82:5060–5067. doi: 10.1128/AEM.01161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence AG, Schoenheit J, He A, Tian J, Liu P, Stubbe J, Sinskey AJ. 2005. Transcriptional analysis of Ralstonia eutropha genes related to poly-(R)-3-hydroxybutyrate homeostasis during batch fermentation. Appl Microbiol Biotechnol 68:663–672. doi: 10.1007/s00253-005-1969-3. [DOI] [PubMed] [Google Scholar]
- 22.Pötter M, Steinbüchel A. 2006. Biogenesis and structure of polyhydroxyalkanoate granules, p 109–136. In Shively JM. (ed), Microbiology monographs, vol 1 Inclusions in prokaryotes. Springer, Berlin, Germany. [Google Scholar]
- 23.Peplinski K, Ehrenreich A, Doering C, Boemeke M, Reinecke F, Hutmacher C, Steinbüchel A. 2010. Genome-wide transcriptome analyses of the “Knallgas” bacterium Ralstonia eutropha H16 with regard to polyhydroxyalkanoate metabolism. Microbiology (Reading, Engl) 156:2136–2152. doi: 10.1099/mic.0.038380-0. [DOI] [PubMed] [Google Scholar]
- 24.Brigham CJ, Speth DR, Rha C, Sinskey AJ. 2012. Whole-genome microarray and gene deletion studies reveal regulation of the polyhydroxyalkanoate production cycle by the stringent response in Ralstonia eutropha H16. Appl Environ Microbiol 78:8033–8044. doi: 10.1128/AEM.01693-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao NN, Gómez-García MR, Kornberg A. 2009. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem 78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 26.Ross W, Sanchez-Vazquez P, Chen AY, Lee J-H, Burgos HL, Gourse RL. 2016. ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol Cell 62:811–823. doi: 10.1016/j.molcel.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eymann C, Homuth G, Scharf C, Hecker M. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J Bacteriol 184:2500–2520. doi: 10.1128/JB.184.9.2500-2520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamarthapu V, Epshtein V, Benjamin B, Proshkin S, Mironov A, Cashel M, Nudler E. 2016. ppGpp couples transcription to DNA repair in E. coli. Science 352:993–996. doi: 10.1126/science.aad6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown DR, Barton G, Pan Z, Buck M, Wigneshweraraj S. 2014. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat Commun 5:4115. doi: 10.1038/ncomms5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronneau S, Petit K, De Bolle X, Hallez R. 2016. Phosphotransferase-dependent accumulation of (p)ppGpp in response to glutamine deprivation in Caulobacter crescentus. Nat Commun 7:11423. doi: 10.1038/ncomms11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hugonnet J-E, Mengin-Lecreulx D, Monton A, den Blaauwen T, Carbonnelle E, Veckerlé C, Brun YV, van Nieuwenhze M, Bouchier C, Tu K, Rice LB, Arthur M. 2016. Factors essential for l,d-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli. eLife 5:e19469. doi: 10.7554/eLife.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harms A, Maisonneuve E, Gerdes K. 2016. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 33.Karstens K, Zschiedrich CP, Bowien B, Stülke J, Görke B. 2014. Phosphotransferase protein EIIANtr interacts with SpoT, a key enzyme of the stringent response, in Ralstonia eutropha H16. Microbiology (Reading, Engl) 160:711–722. doi: 10.1099/mic.0.075226-0. [DOI] [PubMed] [Google Scholar]
- 34.Kaddor C, Steinbüchel A. 2011. Effects of homologous phosphoenolpyruvate-carbohydrate phosphotransferase system proteins on carbohydrate uptake and poly(3-hydroxybutyrate) accumulation in Ralstonia eutropha H16. Appl Environ Microbiol 77:3582–3590. doi: 10.1128/AEM.00218-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calderón-Flores A, Du Pont G, Huerta-Saquero A, Merchant-Larios H, Servín-González L, Durán S. 2005. The stringent response is required for amino acid and nitrate utilization, Nod factor regulation, nodulation, and nitrogen fixation in Rhizobium etli. J Bacteriol 187:5075–5083. doi: 10.1128/JB.187.15.5075-5083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 37.Sznajder A, Jendrossek D. 2014. To be or not to be a poly(3-hydroxybutyrate) (PHB) depolymerase: PhaZd1 (PhaZ6) and PhaZd2 (PhaZ7) of Ralstonia eutropha, highly active PHB depolymerases with no detectable role in mobilization of accumulated PHB. Appl Environ Microbiol 80:4936–4946. doi: 10.1128/AEM.01056-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.York GM, Lupberger J, Tian JM, Lawrence AG, Stubbe J, Sinskey AJ. 2003. Ralstonia eutropha H16 encodes two and possibly three intracellular poly[d-(−)-3-hydroxybutyrate] depolymerase genes. J Bacteriol 185:3788–3794. doi: 10.1128/JB.185.13.3788-3794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchino K, Saito T, Jendrossek D. 2008. Poly(3-hydroxybutyrate) (PHB) depolymerase PhaZa1 is involved in mobilization of accumulated PHB in Ralstonia eutropha H16. Appl Environ Microbiol 74:1058–1063. doi: 10.1128/AEM.02342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eggers J, Steinbüchel A. 2013. Poly(3-hydroxybutyrate) degradation in Ralstonia eutropha H16 is mediated stereoselectively to (S)-3-hydroxybutyryl coenzyme A (CoA) via crotonyl-CoA. J Bacteriol 195:3213–3223. doi: 10.1128/JB.00358-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Handrick R, Reinhardt S, Jendrossek D. 2000. Mobilization of poly(3-hydroxybutyrate) in Ralstonia eutropha. J Bacteriol 182:5916–5918. doi: 10.1128/JB.182.20.5916-5918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gebauer B, Jendrossek D. 2006. Assay of poly(3-hydroxybutyrate) depolymerase activity and product determination. Appl Environ Microbiol 72:6094–6100. doi: 10.1128/AEM.01184-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jendrossek D. 2007. Peculiarities of PHA granules preparation and PHA depolymerase activity determination. Appl Microbiol Biotechnol 74:1186–1196. doi: 10.1007/s00253-007-0860-9. [DOI] [PubMed] [Google Scholar]
- 44.Schlegel HG, Gottschalk G, Von Bartha R. 1961. Formation and utilization of poly-beta-hydroxybutyric acid by Knallgas bacteria (Hydrogenomonas). Nature 191:463–465. doi: 10.1038/191463a0. [DOI] [PubMed] [Google Scholar]
- 45.Ihara Y, Ohta H, Masuda S. 2015. A highly sensitive quantification method for the accumulation of alarmone ppGpp in Arabidopsis thaliana using UPLC-ESI-qMS/MS. J Plant Res 128:511–518. doi: 10.1007/s10265-015-0711-1. [DOI] [PubMed] [Google Scholar]
- 46.Kästle B, Geiger T, Gratani FL, Reisinger R, Goerke C, Borisova M, Mayer C, Wolz C. 2015. rRNA regulation during growth and under stringent conditions in Staphylococcus aureus. Environ Microbiol 17:4394–4405. doi: 10.1111/1462-2920.12867. [DOI] [PubMed] [Google Scholar]
- 47.Brandl H, Gross RA, Lenz RW, Fuller RC. 1988. Pseudomonas oleovorans as a source of poly(beta-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl Environ Microbiol 54:1977–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beyer HM, Gonschorek P, Samodelov SL, Meier M, Weber W, Zurbriggen MD. 2015. AQUA cloning: a versatile and simple enzyme-free cloning approach. PLoS One 10(9):e0137652. doi: 10.1371/journal.pone.0137652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 50.Lenz O, Friedrich B. 1998. A novel multicomponent regulatory system mediates H-2 sensing in Alcaligenes eutrophus. Proc Natl Acad Sci U S A 95:12474–12479. doi: 10.1073/pnas.95.21.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfeiffer D, Wahl A, Jendrossek D. 2011. Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly(3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol Microbiol 82:936–951. doi: 10.1111/j.1365-2958.2011.07869.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.