ABSTRACT
Integrative and conjugative elements (ICEs) are widespread chromosomal mobile genetic elements which can transfer autonomously by conjugation in bacteria. Thirteen ICEs with a conjugation module closely related to that of ICESt3 of Streptococcus thermophilus were characterized in Streptococcus salivarius by whole-genome sequencing. Sequence comparison highlighted ICE evolution by shuffling of 3 different integration/excision modules (for integration in the 3′ end of the fda, rpsI, or rpmG gene) with the conjugation module of the ICESt3 subfamily. Sequence analyses also pointed out a recombination occurring at oriT (likely mediated by the relaxase) as a mechanism of ICE evolution. Despite a similar organization in two operons including three conserved genes, the regulation modules show a high diversity (about 50% amino acid sequence divergence for the encoded regulators and presence of unrelated additional genes) with a probable impact on the regulation of ICE activity. Concerning the accessory genes, ICEs of the ICESt3 subfamily appear particularly rich in restriction-modification systems and orphan methyltransferase genes. Other cargo genes that could confer a selective advantage to the cell hosting the ICE were identified, in particular, genes for bacteriocin synthesis and cadmium resistance. The functionality of 2 ICEs of S. salivarius was investigated. Autonomous conjugative transfer to other S. salivarius strains, to S. thermophilus, and to Enterococcus faecalis was observed. The analysis of the ICE-fda border sequence in these transconjugants allowed the localization of the DNA cutting site of the ICE integrase.
IMPORTANCE The ICESt3 subfamily of ICEs appears to be widespread in streptococci and targets diverse chromosomal integration sites. These ICEs carry diverse cargo genes that can confer a selective advantage to the host strain. The maintenance of these mobile genetic elements likely relies in part on self-encoded restriction-modification systems. In this study, intra- and interspecies transfer was demonstrated for 2 ICEs of S. salivarius. Closely related ICEs were also detected in silico in other Streptococcus species (S. pneumoniae and S. parasanguinis), thus indicating that diffusion of ICESt3-related elements probably plays a significant role in horizontal gene transfer (HGT) occurring in the oral cavity but also in the digestive tract, where S. salivarius is present.
KEYWORDS: integrative and conjugative elements, Streptococcus salivarius, accretion, bacteriocin, cadmium resistance, conjugation, gene transfer, restriction-modification systems
INTRODUCTION
Acquisition of genes by horizontal gene transfer (HGT) is a major driving force for evolution of bacterial genomes (1, 2). The main mechanism is conjugation, a process that enables transfer of large DNA fragments without requiring any similarity between sequences, thus being naturally broad host range (3). Besides conjugative transfer of extrachromosomal elements (plasmids), recent analyses have revealed that many integrated mobile elements, called integrative and conjugative elements (ICEs), also encode their own transfer by conjugation (4, 5). Like all other mobile genetic elements (MGEs) (6), ICEs have a modular structure. Three modules, a recombination module, a conjugation module, and a regulation module, together control and ensure the excision and transfer of the element (7). ICEs are able to excise from the bacterial chromosome generally by site-specific recombination, to transfer using their own conjugative machinery, and to integrate in the chromosome of a recipient cell (4, 5). Moreover, various ICEs can promote the transfer of large fragments of the bacterial chromosome by an Hfr-like mechanism (8). They can also mobilize nonautonomous integrated transferable elements such as (i) integrative and mobilizable elements (IMEs), genetic elements unrelated to ICEs that can excise and integrate from the chromosome but need to hijack the transfer machinery of a conjugative element, and (ii) cis mobilizable elements (CIMEs), elements that derive from ICEs and IMEs by deletion of the conjugation/mobilization and recombination modules but retain recombination sites att (4). In the latter case, an ICE that integrates in a recombination site of a CIME can mobilize the nonautonomous element by mediating the excision of the whole composite element (process of accretion-mobilization) (9, 10). The increase of bacterial genome sequencing projects in the last few years provides a remarkable opportunity to explore the pool of bacterial genetic mobile elements (“mobilome”). These in silico analyses revealed the high abundance of ICEs in bacteria (11–13). In addition to the genes involved in or controlling their mobility, ICEs also carry cargo genes, which can provide new properties (virulence and antibiotic resistance, for example) to the recipient cell (7).
Streptococcus salivarius is a member of the Firmicutes that is a major constituent of the human oral cavity microbiota (14) and is commonly detected in the human gastrointestinal tract in healthy individuals (15, 16). A few strains have also been associated with opportunistic infections, in particular, in cases of meningitis (17), endocarditis (18) and bacteremia in immunocompromised patients (19, 20). Analyses of S. salivarius genomes pointed out the considerable variability of gene content and the differences in adaptive traits (21). Evidence of widespread HGT was obtained in association with the presence of diverse MGEs (21) and the competence for natural transformation of the species (22).
We recently screened a collection of 138 strains of S. salivarius for the presence of MGEs (23). This led to the identification of 60 strains (belonging to 39 multilocus sequence type [MLST] groups) with a positive PCR signal for the relaxase gene of ICESt3, an ICE previously characterized in the closely related species Streptococcus thermophilus (24–26). This indicated the presence in these strains of putative ICEs belonging to the ICESt3 subfamily. However, not all these strains showed a positive PCR signal for the ICESt3 integrase gene, which catalyzes the integration into the fda site. This suggested the presence of other recombination modules associated with the ICESt3 conjugation module in S. salivarius ICEs.
In this work, we selected 13 strains with different MLST patterns (including 6 strains with a negative PCR signal for the ICESt3 integrase gene) and sequenced their genomes in order to gain access to their ICE sequences. ICEs with a conjugation module closely related to the one in ICESt3 were searched in other NCBI database-available genomes of S. salivarius and in other genomes of Firmicutes. ICE sequences were compared and the putative functions encoded by their genes were analyzed in silico. Two putative ICEs from S. salivarius were analyzed experimentally to test their excision and their autonomous intraspecies and interspecies conjugative transfer.
RESULTS AND DISCUSSION
Diversity of integrative and conjugative elements of the ICESt3 subfamily identified in S. salivarius and in other streptococci. (i) Conjugation modules.
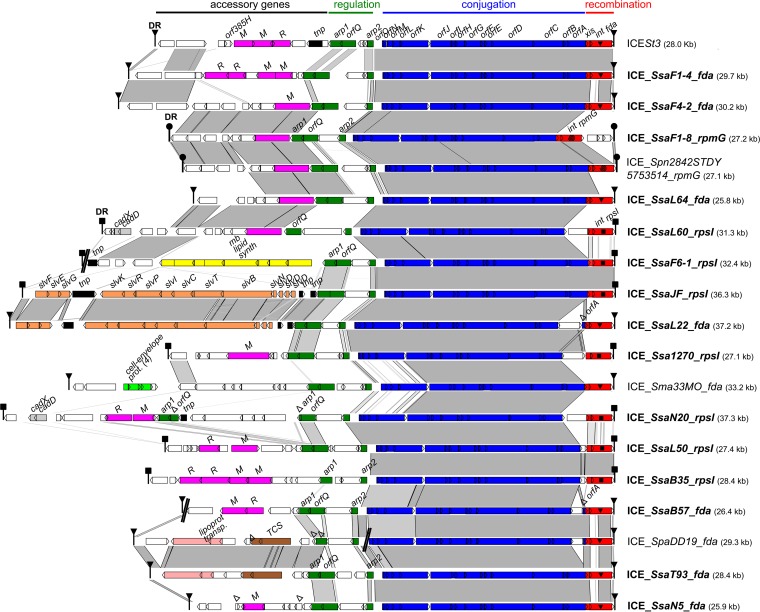
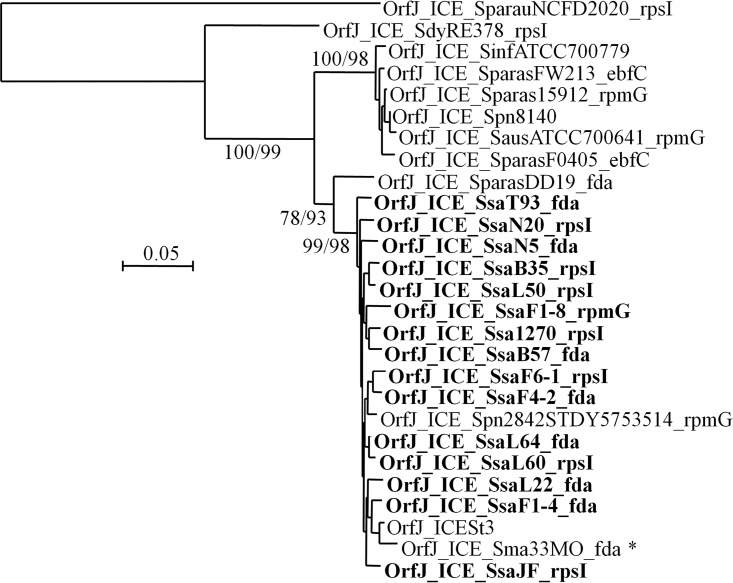
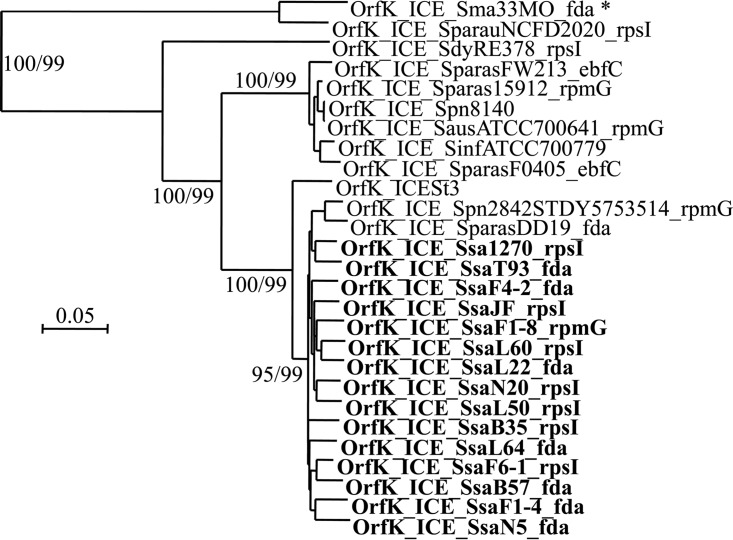
To explore the diversity of the ICEs of the ICESt3 subfamily found in S. salivarius, 13 strains (F1-4, F1-8, F4-2, F6-1, B35, B57, L22, L50, L60, L64, N5, N20, and T93) with different MLST patterns were selected from a previous work (23) (Table 1). These strains likely carry an ICE of the ICESt3 subfamily, as suggested by a positive PCR signal for the ICESt3 relaxase gene (23). Their genomes were sequenced and assembled in order to gain access to their ICE sequences. Except for 2 ICEs (ICE_SsaF6-1_rpsI and ICE_SsaB57_fda) split into 2 contigs, the sequences of the ICEs appear on a single contig (Fig. 1). As shown in Fig. 1, ICEs found in these 13 S. salivarius strains all display a conjugation module that is closely related to that of ICESt3 from S. thermophilus (>90% nucleic sequence identity). ICEs with a closely related full-conjugation module were also detected in 2 genomes of S. salivarius available in the NCBI database (those of strains JF and 1270) (Fig. 1). The element of S. salivarius NCTC 8618 (GenBank accession number CP009913.1) also has a closely related full-conjugation module but is not shown in the figure since it harbors a truncated integrase gene and is therefore defective. Elements with a closely related conjugation module (partial in most of the genomes due to gaps in the assembly) were also detected in the genomes of strains GED7778A (GenBank accession number LRQS00000000), 140 (GenBank accession number JVSQ01000000), 20-02 S1 (GenBank accession number LXMB00000000), 20-12 S2 (GenBank accession number LXMC00000000), and UC3162 (GenBank accession number JYOY01000000). These elements are not shown in Fig. 1 because their sequences are incomplete. ICEs with a closely related conjugation module (92 to 94% nucleic sequence identity with that of ICESt3) were also identified in the genome of Streptococcus pneumoniae 2842STDY5753514 (mitis group) and Streptococcus parasanguinis DD19 (sanguinis group) (Fig. 1). Phylogenetic analysis of the relaxase OrfJ (Fig. 2) and the coupling protein OrfK (Fig. 3) of these ICEs indicates that they group with OrfJ and OrfK from S. salivarius ICEs. Similar results were obtained for the other proteins of the conjugation module (data not shown). Taken as a whole, these phylogenetic analyses and sequence comparisons clearly showed that transfers of ICEs closely related to ICESt3 have occurred between distinct species from different streptococcal groups.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotype | Source or reference |
|---|---|---|
| Strains | ||
| S. salivarius | ||
| B35 | WTa strain carrying a putative ICE of the ICESt3 subfamily | 23 |
| B57 | WT strain carrying a putative ICE of the ICESt3 subfamily | 23 |
| F1-4 | WT strain carrying a putative ICE of the ICESt3 subfamily (ICE_SsaF1-4_fda) | 23 |
| F1-4(ICE_SsaF1-4_fda) Cmr | F1-4 carrying ICE_SsaF1-4_fda tagged with a Cmr cassette | This work |
| F1-8 | WT strain carrying a putative ICE in accretion with an IME integrated in the 3′ end of rpmG; no element in fda | 23 |
| F1-8(pMG36e) | F1-8 carrying pMG36e, a plasmid conferring erythromycin resistance | This work |
| F4-2 | WT strain carrying a putative ICE of the ICESt3 subfamily | 23 |
| F6-1 | WT strain carrying a putative ICE of the ICESt3 subfamily | 23 |
| JIM8777 | WT strain carrying a putative IME in the 3′ end of rpmG and a putative CIME in the 3′ end of rpsI; no element integrated in the fda gene | 57 |
| JIM8777(pMG36e) | JIM8777 carrying pMG36e, a plasmid conferring erythromycin resistance | This work |
| L22 | WT strain carrying a putative ICE of the ICESt3 subfamily | 23 |
| L50 | WT strain carrying a putative ICE of the ICESt3 subfamily | 23 |
| L60 | WT strain carrying a putative ICE of the ICESt3 subfamily | 23 |
| L64 | WT strain carrying a putative ICE of the ICESt3 subfamily | 23 |
| N5 | WT strain carrying a putative ICE of the ICESt3 subfamily | 23 |
| N20 | WT strain carrying a putative ICE of the ICESt3 subfamily | 23 |
| T93 | WT strain carrying a putative ICE of the ICESt3 subfamily | 23 |
| S. thermophilus | ||
| CNRZ385(ICESt3cat) | CNRZ385 carrying ICESt3 with cat inserted in orf385J pseudogene; Cmr | 24 |
| LMG18311 | WT strain with no element in fda | BCCM/LMG |
| LMG18311(pMG36e) | LMG18311 carrying pMG36e, a plasmid conferring erythromycin resistance | 24 |
| E. faecalis | ||
| JH2-2 | WT strain with no element in fda | 58 |
| JH2-2(pMG36e) | JH2-2 carrying the plasmid pMG36e, conferring erythromycin resistance | 24 |
| Plasmids | ||
| pSET5s | pWV01-type thermosensitive replication origin from pVE6002; LacZ Cmr | 59 |
| pMG36e | 3.6 kb, replication origin from pWV01; Eryr | 49 |
WT, wild type.
FIG 1.
Comparison of the integrative and conjugative elements (ICEs) found in S. salivarius and in other streptococci. ICEs are named according to their host strains and integration sites. ICEs of S. salivarius are indicated in bold. For more clarity, elements in accretion with ICEs are not shown. Nucleic acid sequence identity higher than 80% between sequences is indicated in light gray and that higher than 90% in dark gray. Direct repeats (DR) delimiting ICEs are shown as triangles, circles, or squares depending on their sequences. Open reading frames (ORFs) appear as arrows (truncated genes are indicated by capital deltas). Modules of recombination (integrase [int] and excisionase [xis] genes), conjugation (orfO to orfA genes), and regulation (including the arp1, arp2, and orfQ genes) appear in red, blue, and green, respectively. The three different integration genes (fda, rpsI, or rpmG) targeted by the integrase are indicated by distinct symbols in the integrase gene and are part of the ICE name. Genes from the adaptation module encoding proteins with putative function inferred from in silico analysis are indicated in pink for RM systems and orphan methyltransferase genes, in dark gray for cadmium resistance genes (cadD and cadX), in yellow for the membrane lipid synthesis cluster, in orange for the bacteriocin synthesis cluster, in light green for genes encoding cell envelope proteins, in light pink for the cluster of genes for putative lipoprotein transport, in brown for the two-component system (TCS), and in black for the tnp transposase gene(s). Sequences used for this analysis were ICESt3 of S. thermophilus (AJ586568) and sequenced genomes of S. pneumoniae 2842STDY5753514 ( FDNK01000013), S. macedonicus 33MO (JNCV01000015), and S. parasanguinis DD19 (LQNY01000339, LQNY01000340). Gaps in the assembly found in ICE_SsaF6-1_rpsI, ICE_SsaB57_fda, and ICE_SpaDD19_fda are indicated by a double slash.
FIG 2.
Phylogenetic BioNJ tree obtained for relaxases (OrfJ) of ICEs belonging to the ICESt3 subfamily. The relaxase protein sequence of 27 ICEs (the 19 ICEs of the ICESt3 subfamily with closely related conjugation modules and 8 additional ones previously reported to belong to the same ICE subfamily but showing more distantly related conjugation modules) were included in the analysis. ICEs of S. salivarius are indicated in bold, and ICE_Sma33MO_fda is indicated by an asterisk. Bootstrap values supporting main branches are given for BioNJ and ML, respectively.
FIG 3.
Phylogenetic BioNJ tree obtained for coupling proteins (OrfK) of ICEs of the ICESt3 subfamily. The sequence of the coupling protein of 27 ICEs (the 19 ICEs of the ICESt3 subfamily with closely related conjugation modules and 8 additional ones previously reported to belong to the same ICE subfamily but showing more distantly related conjugation modules) were included in the analysis. ICEs of S. salivarius are indicated in bold, and ICE_Sma33MO_fda is indicated by an asterisk. Bootstrap values supporting main branches are given for BioNJ and ML, respectively.
The comparison of the clustering groups obtained for the relaxase (OrfJ) and the coupling protein (OrfK) indicated that OrfJ of ICE_Sma33MO_fda is closely related to the relaxases of ICESt3 and S. salivarius ICEs, whereas OrfK is clearly different from the coupling proteins of these ICEs but is closer to that of ICE_SparauNCFD2020_rpsI (see phylogenetic trees in Fig. 2 and 3). Nucleic sequence alignments indicated that the left extremity of the conjugation module (including the orfN to orfK genes) of ICE_Sma33MO_fda is closely related to the corresponding sequence of ICE_SparauNCFD2020_rpsI (86% nucleic sequence identity compared to 64% nucleic sequence identity with the corresponding genes of ICESt3), whereas the right extremity of the conjugation module of ICE_Sma33MO_fda (corresponding to the orfJ to orfA genes) is closely related to those of ICESt3 (96% nucleic sequence identity compared to 64% nucleic sequence identity with the corresponding genes of ICE_SparauNCFD2020_rpsI) and S. salivarius ICEs. The putative nic site of the transfer origin of ICEs of the ICESt3 subfamily, predicted by comparison with the oriT characterized for ICEBs1 of Bacillus subtilis (27), is located between the orfK and orfJ genes (Fig. S1 in the supplemental material). Sequence alignment of this region indicated that the drop in nucleic sequence identity between ICE_Sma33MO_fda and ICE_SparauNCFD2020_rpsI occurs at the nic site (Fig. S1 in the supplemental material). This suggests that a recombination event occurred at oriT in ICE_Sma33MO_fda. This oriT recombination was likely mediated by the relaxase of the ICE, which is able to recognize and nick this sequence. Such oriT site-specific recombination has already been demonstrated for several canonical relaxases of plasmids (belonging to the MobF, MobP, MobC, and MobM families) (4, 28, 29) and has been suggested to occur in ICEs of the SXT/R391 family and for ICEclc (both encoding canonical relaxases of the MobH family) (30, 31). This mechanism that enables creation of hybrid ICEs could thus also be mediated by relaxases of the noncanonical MobT family, which are related to rolling circle replication initiators and are found in ICEs of the Tn916/ICESt3/ICEBs1 family, a family of ICEs widespread in streptococci (11) and other Firmicutes (5).
(ii) Integration modules.
ICESt3 conjugation modules found in S. salivarius ICEs are associated with 3 different recombination modules (see Fig. S2 in the supplemental material) enabling integration of the corresponding ICEs in the 3′ ends of three different genes: fda (encoding the fructose-1-6-diphosphate aldolase, as for ICESt3), rpsI (encoding the S9 ribosomal protein), and rpmG (encoding the L33 ribosomal protein). This confirms our previous results showing a positive PCR signal for the ICESt3 relaxase gene but not for the ICESt3 integrase gene for 22 strains (23). Closely related integrases were found in ICEs of S. salivarius strains that are distant in the phylogenetic tree built on the basis of MLST data (for example, in strains F1-4, L22, and N5 or in strains B35 and L50) (23) but also in ICEs found in other species (for example, in S. pneumoniae 2842STDY5753514 or in S. parasanguinis DD19 and S. macedonicus 33MO) (Fig. S2 in the supplemental material). This is in agreement with the previously reported exchanges of recombination modules in the ICESt3 subfamily (11, 26).
(iii) Regulation modules.
Comparison of the regulation modules of all ICEs of the ICESt3 subfamily showed their high diversity (Fig. 1). The only common characteristic is the presence, in most of the ICEs, of homologs of the arp1, orfQ, and arp2 genes. This strongly suggests that these genes participate in the same regulation cascade and likely interact or interfere with each other. Exceptions are ICE_SsaL60_rpsI, devoid of the arp1 gene; ICE_SsaB35_rpsI, lacking the orfQ gene; and ICE_SpaDD19_fda, carrying truncated arp1 and orfQ genes. ICE_SsaN20_rpsI also displays a peculiar regulation organization, with full copies of the arp1 and orfQ genes separated by truncated copies of the orfQ and arp1 genes. This is due to an insertion of genes (including one encoding a transposase) in this region of the ICE (Fig. 1). The arp1, orfQ, and arp2 genes encode, respectively, a cI-related repressor, an ImmA-related putative protease (32), and a putative transcriptional regulator (33) that are related to those of ICESt3 from S. thermophilus (26, 34). Arp1 and Arp2 homologs all share an N-terminal helix-turn helix PF01381 domain that is found in several regulatory proteins, including Cro and cI regulators of the λ phage. Arp1 proteins also display a C-terminal PF00717 peptidase S24-like domain involved in the autocleavage of the cI repressor induced by DNA damage. OrfQ homologs are characterized by a COG2856 Zn-dependent peptidase ImmA domain, also found in the ImmA protease of ICEBs1 of B. subtilis that cleaves the ImmR repressor in response to DNA damage (32). A high diversity is observed in the nucleotide (Fig. 1) and amino acid primary sequences of the encoded regulators. Divergences of up to 45%, 58%, and 53% were observed between the sequences of the Arp1, OrfQ, and Arp2 proteins encoded by ICEs of S. salivarius. If considering only the ICEs that harbor the arp1, orfQ, and arp2 genes, the sequence divergence goes from 5 to 37% for Arp1, 7 to 58% for OrfQ, and 7 to 37% for Arp2.
In support of a modular evolution of ICEs that was already noticeable from the exchanges in the recombination modules mentioned previously, some ICEs harbor regulators which are distantly related to those found in the other ICEs of S. salivarius despite having a closely related conjugation module (deduced from the phylogenetic trees of relaxases and coupling proteins). This is the case for ICE_SsaF4-2_fda, which has a conjugation module closely related to the one of ICESt3 (94% nucleic sequence identity over the whole conjugation module) but encodes more distant regulators (with 63, 46, and 73% amino acid identity with those encoded by ICESt3 for Arp1, OrfQ, and Arp2, respectively).
Furthermore, all the regulation modules include additional genes (1 to 3 depending on the ICE) between the arp2 and orfQ genes. These genes that encode proteins of unknown function contribute to the diversity of the regulation modules.
(iv) Accretions.
Accretion of ICEs with other genetic elements was observed: ICEs of strains F1-8 and L60 were in accretion with a putative IME, whereas those of strains F6-1 and L50 were adjacent to 1 or 2 CIMEs (data not shown).
Evidence of accretion or recombination events was also found in the sequences of several ICEs of S. salivarius. A truncated supplementary copy of the orfA gene was found between orfA and the excisionase genes in the ICEs of S. salivarius L22 and B57 and in the closely related ICE found in S. parasanguinis DD19. The presence of supplementary truncated arp1 and orfQ genes and of a transposase gene was also detected in the regulation module of S. salivarius N20 ICE. These structures could result (i) from the integration of an ICE within an att site flanking a resident element (accretion) or (ii) from recombination between ICEs, followed by internal deletion(s) in both cases. These mechanisms contribute to the plasticity and evolution of ICEs and potentiate gene transfer mediated by these MGEs.
Cargo genes found on ICEs of the ICESt3 subfamily in S. salivarius and other streptococci.
A diversity of cargo genes was found in the left part of the ICEs of the ICESt3 subfamily, explaining the variation of their size ranges: from 25.8 kb for the smallest one (ICE_SsaL64_fda) to 37.3 kb for the largest one (ICE_SsaN20_rpsI) (Fig. 1). The 2 closely related strains included in the analysis (L22 and L64) harbor ICEs with unrelated cargo genes (Fig. 1). In addition, ICEs found in unrelated strains of S. salivarius (for example, in strains JF and L22 or F4-2 and F1-8) or in different species (in S. salivarius T93 and S. parasanguinis DD19) harbor the same cargo genes (Fig. 1). This distribution is due to the horizontal transfer of these genes (exchange of module between ICEs or transfer of the whole ICE).
Two-thirds (13/19 if including ICESt3 [35]) of the ICEs analyzed in this work carry a restriction-modification (RM) system (n = 6) or an orphan methyltransferase (n = 7) (Fig. 1). This is in accordance with a previous study indicating that the abundance of RM systems correlates with the presence of MGEs in small genomes (36). RM systems were first described as bacterial innate immune systems allowing protection against foreign unmethylated DNA (37). Unmethylated incoming DNA is degraded by restriction enzymes produced by the cell, while the genome of the host remains protected due to methylation by the cognate methyltransferase. RM systems carried by ICEs of S. salivarius could protect their host from invasion by other genetic elements such as phages, thus playing a role in “cellular defense” as described for other bacteria (38–40). RM systems also turn out to be selfish mobile elements themselves (41). RM systems that carry the restriction enzyme activity and modification enzyme activity on separate proteins (as observed for the ICEs of the ICESt3 subfamily) can have an impact on the maintenance of the MGEs carrying them (mechanism of genetic addiction). The MGE encodes the poison endonuclease activity and its “antidote,” the methyltransferase (37). Postsegregational killing would occur if the whole RM system was lost. Dilution of the modification enzyme by cell division will lead to the exposure of unmethylated recognition sites on newly replicated chromosomes that will be targeted by the restriction enzyme. Only a few remaining molecules of restriction enzymes are sufficient to kill the cell. Concerning orphan methyltransferases (encoded by 7 of the ICEs of the ICESt3 subfamily analyzed in this work), if these enzymes target the same DNA sequence as a resident RM system, they could protect the host from postsegregational killing, thus participating in the displacement of the resident MGE (42).
Two ICEs found in S. salivarius L60 and N20 carry a cadmium resistance cluster (cadD-cadX) (Fig. 1). Cadmium is a widespread heavy metal air pollutant which is commonly released into the environment from industrial processes (in particular, glass manufacturing) and urban activities, as well as from the widespread application of fertilizers, manures, and sewage sludge (43). The cadD-cadX genes also appear in a CIME in accretion with the ICE from strains F6-1 and L50 (data not shown). In the latter ones, full recombination sites are still present and thus the CIME and the ICE are considered separately. In these strains, the whole CIME-ICE composite element could excise and transfer by a process of accretion-mobilization already demonstrated with other ICEs of this family (9, 10). Sequence comparison revealed 98% nucleic acid identity over 2,683 bp between the CIME located upstream of the ICE in strain L50 and the region that includes cadD-cadX genes in ICE_SsaL60_rpsI. This suggests that in strain L60 the cadmium resistance cluster was acquired by the ICE after deletion of the recombination site delimiting the ICE and the adjacent CIME. This mechanism contributes to the plasticity and evolution of ICEs as previously suggested (4).
ICE_SsaF6-1_rpsI carries a cluster of genes (10.8 kb) that could be involved in membrane lipid synthesis. It includes, in particular, a fabF-like gene encoding a beta-ketoacyl (acyl carrier protein) synthase II (KASI/IIcd00834 domain, with an E value of 1.4e−144) and a KBL_like gene encoding a serine palmitoyltransferase involved in sphingolipid synthesis (cd06454 domain, with an E value of 3.7e−151).
Two ICEs (found in the unrelated strains S. salivarius JF and L22) harbor a closely related cluster of genes (slv cluster; 15.7 to 15.9 kb) involved in the biosynthesis of a bacteriocin (salivaricin D). These clusters differ from the one previously described for the commensal strain 5M6c of S. salivarius, isolated from a healthy infant (44), by the number of copies of the slvD structural gene (2 and 3 identical copies, respectively, compared to 1 in strain 5M6c). Salivaricin D is a nisin-like lantibiotic with a broad spectrum that includes a large array of Gram-positive bacteria, in particular, the important pathogens S. pneumoniae and S. pyogenes (44). Thus, it could be used as a potential weapon, for commensal species of the oral cavity such as S. salivarius, to compete with oronasopharynx-colonizing streptococci, including pathogens (44–46). Bacteriocins could act as an addiction system by killing neighboring cells that do not encode resistance to the bacteriocin and thus contribute to the maintenance of the MGE in the population (47).
The ICE found in S. macedonicus 33MO carries genes that encode cell envelope proteins: one with a PF03780 domain with cell envelope-related function (E = 7.93e−23) and three membrane proteins. Lastly, ICEs of S. salivarius T93 and S. parasanguinis DD19 harbor the same cargo genes, in particular, (i) a two-component system with a signal transduction histidine kinase (nitrate/nitrite-specific COG3850 NarQ domain, E = 5.3e−13) and a NarL DNA-binding response regulator (COG2197 domain, E = 6.1e−48) and (ii) a cluster of genes encoding a putative LolCDE complex which catalyzes the release of lipoproteins from the cytoplasmic membrane (LolC PF13521 AAA_28 domain, E = 1.2e−09; LolD cd03255 ABC_MJ0796_LolCDE_FtsE domain, E = 1.2e−108; and LolE COG4591 domain, E = 4.4e−14) (Fig. 1).
Excision testing of 2 putative ICEs: ICE_SsaF1-4_fda and ICE_SsaF4-2_fda.
Experiments were carried out to test the functionality of 2 putative ICEs of S. salivarius. The first one, ICE_SsaF1-4_fda, has a conjugation module showing the highest percentage of identity with that of ICESt3 of S. thermophilus, an ICE whose transfer was previously demonstrated (24) (Fig. 1). Arp1, OrfQ, and Arp2 regulators encoded by this ICE are also closely related to those of ICESt3 (85, 71, and 84% amino acid identities, respectively). As mentioned before, the second one, ICE_SsaF4-2_fda, has a conjugation module closely related to ICE_SsaF1-4_fda and ICESt3 but harbors a distantly related regulation module.
Excision of these 2 ICEs was tested by PCR. Amplifications were obtained for attI and attB, corresponding to the circular excised form and the empty chromosomal site, respectively, indicating the functionality of their recombination module. Recombination sites (attL and attR) flanking the ICEs were also amplified (data are shown only for ICE_SsaF1-4_fda [Fig. 4; Table 2]).
FIG 4.
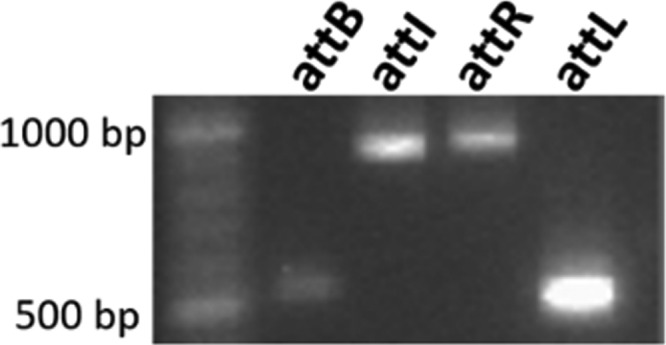
PCR detection of integrated and excised forms of ICE_SsaF1-4_fda. The sizes of the PCR fragments obtained for the amplification of the attB, attI, attR, and attL sites (586 bp, 887 bp, 900 bp, and 523 bp, respectively) were confirmed by parallel migration of a DNA ladder. The primer pairs used for these amplifications are listed in Table 2.
TABLE 2.
Primers used in this work
| Primer use | Primer name | Sequence (5′–3′) | Reference |
|---|---|---|---|
| ICE labeling | This study | ||
| F1-4 CDS7 F | GAGATTGAGCATATCCTTCC | ||
| F1-4 CDS8 R_Bis | GGTGACTAGTTATCTACACGCGAGATTCGTGGACTAACTT | ||
| F1-4 CDS9 F_Bis | CCATATCCTTCTTTTTCTGCTCACTATCTTGTTCGTTTTGT | ||
| F1-4 CDS9 R | GGAGAGTTTAGCTGGGAGG | ||
| ICE F4-2 fragment I_F | GGAAATATCCTGTTGTCATC | ||
| ICE F4-2 fragtI_R-catbis | GGTGACTAGTTATCTACACGCCTATAAAGTTGTTAAGTTCACT | ||
| ICE F4-2 fragtII_F-catbis | CCATATCCTTCTTTTTCTGGCGTGTAATTGAAGAGTGA | ||
| ICE F4-2 fragment II_R | GTCTAAACTGAGCCAAGAAG | ||
| Cat_F | GCCTCCTAAATTCACTTTAG | ||
| Cat_R | GTAAAAAGTACAGTCGGCAT | ||
| Detection of ICE integrated and excised forms | |||
| attB amplification | 23 | ||
| attBfdaSsal | GCCCAACCAAATAACACTAAA | ||
| attB ST3 Rev | CTCTTCGACCCACGTAAATTC | ||
| attI amplification | 23 | ||
| intST3 For | AGGGCTTTCTGACGAATTAG | ||
| attI ST3 Rev | CGGTGTAATGGGAAGTATGG | ||
| attL amplification | |||
| attL Rv CDS gwlG | CGGTGTAATGGGAAGTATGG | This study | |
| attBfdaSsal | GCCCAACCAAATAACACTAAA | 23 | |
| attR amplification | |||
| intST3 for | AGGGCTTTCTGACGAATTAG | 23 | |
| intICESt3-fdaRev | ACCAGGTTTCGATGCTATTACAG | 35 | |
| Integrase gene | 23 | ||
| intST3 for | AGGGCTTTCTGACGAATTAG | ||
| intST3 Rev | GAGTTCTAATAACTGAGGCTA | ||
| Semiquantitative PCR | |||
| attI amplification | This study | ||
| intICEF1-4 For | AGGTCTTTCTGACGAATTAG | ||
| attIICEF1-4 Rev | CGGCGTAATGGGAAGTATGG | ||
| attB amplification | 23 | ||
| attBfdaSsal | GCCCAACCAAATAACACTAAA | ||
| attB ST3 Rev | CTCTTCGACCCACGTAAATTC | ||
| fda amplification | |||
| Fda1 | TTCAAGAATTTACGTGGG | This study | |
| Fda2 | AGATGCTAAAGCTATGGTTG | 56 | |
| Donor/recipient discrimination | |||
| ITS16S/23S | 24 | ||
| 16SITS | TTGTACACACCGCCCGTCA | ||
| 23SITS | GGTACCTTAGATGTTTCAGTTC | ||
| ddlA gene | 60 | ||
| ddlA up | TCAAGTGTGGCTATGGA | ||
| ddlA dn | GTAGATGGCTCCATCCTC | ||
| E. faecalis fda-specific amplification | 24 | ||
| Efa fba1b | ATGTGTTCTTCTGCATCTTT | ||
| Efa fba2 | CCCATTGATTACGATTTTT | ||
| RAPD | 56 | ||
| XD9 | GAAGTCGTCC | ||
| pMG36e | |||
| pMG36e Fwd | GCCTCCTCATCCTCTTCAT | 24 | |
| pMG36e_R | ACAGAACCGTTTCTACTCAATGAAC | This study |
Test of intraspecies conjugative transfer of 2 putative ICEs of S. salivarius.
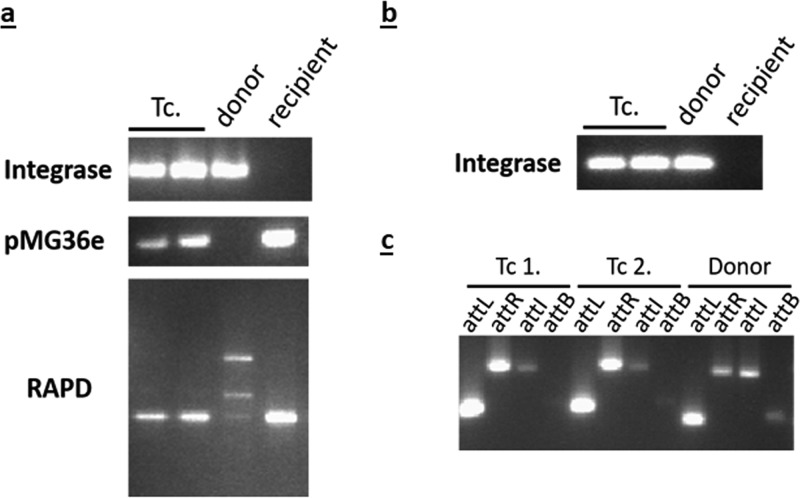
Both ICE_SsaF1-4_fda and ICE_SsaF4-2_fda were first tagged by a chloramphenicol resistance cassette (Table 1). Donor strains were then used in filter mating experiments with two S. salivarius recipient strains: JIM8777 and F1-8, displaying different genotypes (Table 1 and 3). Both strains carry an empty fda integration site, and strain F1-8 carries an ICE belonging to the ICESt3 subfamily (Fig. 1) in accretion with an IME integrated in the 3′ end of rpmG. Some putative transconjugants were recovered when S. salivarius JIM8777 or F1-8 was used as the recipient (Table 3). After subculturing, transconjugants were confirmed by detection of the integrase gene (data are shown only for ICE_SsaF1-4_fda [Fig. 5a and b]). For JIM8777 derivative clones, this screen was combined with detection of the pMG36e plasmid and random amplification of polymorphic DNA (RAPD) (Fig. 5a). F1-8 transconjugants were confirmed by sequencing of an MLST locus (ddlA gene). The site-specific insertion in the fda gene and the excision of the newly acquired ICEs were also confirmed by the PCR detection of attL, attR, attI, and attB sites for both F1-8 (data are shown only for ICE_SsaF1-4_fda [Fig. 5c]) and JIM8777 (data not shown). This suggests that the elements are still active in these transconjugants (at least for excision).
TABLE 3.
Mating pairs tested in filter experiments
| Type of transfer | Donor cells with ABra | Recipient cells with ABra |
|---|---|---|
| Intraspecies transfer | F1-4(ICE_SsaF1-4_fda) Cmr | JIM8777(pMG36e) Eryr |
| F1-8(pMG36e) Eryr | ||
| F4-2(ICE_SsaF4-2_fda) Cmr | JIM8777(pMG36e) Eryr | |
| F1-8(pMG36e) Eryr | ||
| Interspecies transfer | F1-4(ICE_SsaF1-4_fda) Cmr | LMG18311(pMG36e) Eryr |
| JH2-2(pMG36e) Eryr | ||
| F4-2(ICE_SsaF4-2_fda) Cmr | LMG18311(pMG36e) Eryr | |
| JH2-2(pMG36e) Eryr | ||
| ICE retransfer from transconjugants | F1-8(ICE_SsaF1-4_fda) Cmr | F1-8(pMG36e) Eryr |
| LMG18311(pMG36e) Eryr | ||
| LMG18311(ICE_SsaF1-4_fda) Cmr | LMG18311(pMG36e) Eryr | |
| JH2-2(ICE_SsaF1-4_fda) Cmr | JH2-2(pMG36e) Eryr |
ABr, antimicrobial resistance of the strain.
FIG 5.
Characterization of transconjugants carrying ICE_SsaF1-4_fda after intraspecies transfer. F1-4(ICE_SsaF1-4_fda) was used as the donor in mating experiments with JIM8777(pMG36e) (a) and F1-8(pMG36e) (b and c) as recipients. Tc, transconjugants; integrase, amplification of the integrase gene of ICE_SsaF1-4_fda; pMG36e, amplification of an internal fragment of the pMG36e plasmid; RAPD, random amplification of polymorphic DNA; attL, attR, attI, and attB, amplification of fragments carrying these attachments sites in F1-8 transconjugants. The primer pairs used for these amplifications are listed in Table 2.
Mating experiments were repeated with at least 3 different cultures of donor and recipient cells (biological repetitions) but very few transconjugants (2 clones for the whole experiment) were obtained, thus preventing any calculation of transfer frequency. Attempts to increase ICE_SsaF1-4_fda transfer frequency were made: (i) testing of different media (milk, brain heart infusion [BHI], mitis, and M17 broth with 1% final glucose instead of 0.5% final lactose); (ii) testing of different donor/recipient ratios ranging from 1:1, 2:1, 10:1, and 50:100 to 100:1; and (iii) mixing of donor and recipient at the end of exponential growth phase or stationary phase instead of mid-exponential phase. None of the tested conditions enabled an increase in transfer frequency, which remained inferior to 10−8 transconjugants per donor cells.
Testing of interspecies conjugative transfer of ICEs of S. salivarius.
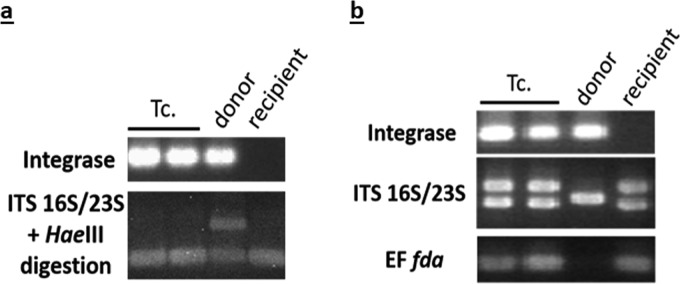
To evaluate ICE interspecies transfer, mating assays were carried out with two other Firmicutes as recipients: the closely related species S. thermophilus and Enterococcus faecalis. Recipient strains were S. thermophilus LMG18311 and E. faecalis JH2-2, both carrying pMG36e and described as recipients strains of ICESt3 from S. thermophilus (transfer frequency of 3.4 × 10−6 ± 0.5 × 10−6 and 3.9 × 10−7 ± 0.9 × 10−7, respectively [24]). Some putative transconjugants (2 clones for the whole experiment) were recovered when using strains F1-4 or F4-2 as donors in mating experiments (Table 3) and confirmed by PCR detection of the ICE integrase gene (data are shown only for ICE_SsaF1-4_fda [Fig. 6]). The genetic background of LMG18311 was confirmed by PCR amplification of the internal transcribed spacer (ITS) followed by HaeIII DNA digestion, which allows discrimination of S. salivarius strains and S. thermophilus LMG18311 (data are shown only for ICE_SsaF1-4_fda [Fig. 6a]). The genetic background of JH2-2 was confirmed by the PCR amplification of the ITS and by the amplification of fda gene with primers specific to E. faecalis (data are shown only for ICE_SsaF1-4_fda [Fig. 6b]).
FIG 6.
Characterization of transconjugants carrying ICE_SsaF1-4_fda after interspecies transfer. F1-4(ICE_SsaF1-4_fda) was used as the donor in mating experiments with LMG18311(pMG36e) (a) and JH2-2(pMG36e) (b) as recipients. Tc, transconjugants; integrase, amplification of the integrase gene of ICE_SsaF1-4_fda; ITS16S/23S, amplification of the internal transcribed spacer; ITS16S/23S+HaeIII digestion, amplification of the internal transcribed spacer followed by HaeIII digestion of the obtained fragment; EF fda, amplification of fda fragment using E. faecalis-specific primers.
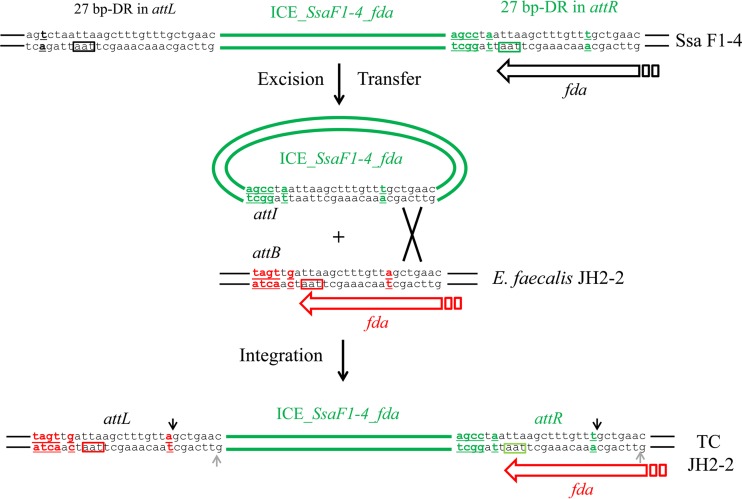
ICE_SsaF1-4_fda carries 27 bp showing identity with the 3′ end of the fda gene of S. salivarius (Fig. 7). This sequence (present in attI) enables specific recombination with the chromosomal integration site of the recipient cell (attB site) and generates a 27-bp imperfect direct repeat (DR) in intraspecies transconjugants (Fig. 7). Sequencing of the attR site obtained after integration of ICE_SsaF1-4_fda in E. faecalis JH2-2 recipient cells provided relevant information on the DNA localization of one cutting site of fda integrase. This attR site is a hybrid between the sequence found in the attI of ICE_SsaF1-4_fda and the attB of E. faecalis JH2-2 recipient cells. Since the attB sequence in the recipient strains is different from the attI of ICE_SsaF1-4_fda (Fig. 7), the presence of a T nucleotide at position 20 of the DR in the attR of the transconjugants, as in ICE_SsaF1-4_fda, indicates that integrase cuts downstream of this nucleotide to allow strand exchange during recombination. Previous work aimed at studying CIME-ICE accretion also identified the same cutting position as well as the position of a second staggered cutting site located 6 bp downstream (Fig. 7) (48).
FIG 7.
Localization of the DNA cutting site of integrase by sequencing of the attR site in E. faecalis JH2-2 transconjugants. ICE_SsaF1-4_fda is shown in green in its integrated form in donor S. salivarius strain F1-4 (top), its excised form (middle), and its integrated form in E. faecalis JH2-2 recipient strain after transfer and integration (bottom). Nucleotide differences in att sequences are indicated in green for ICE_SsaF1-4_fda and in red for E. faecalis JH2-2. The position of the DNA cutting site of integrase deduced from this analysis is indicated by a black arrow. Previous work aimed at studying CIME-ICE accretion also identified the same cutting position as well as the position of a second staggered cutting site located 6 bp from the first one (indicated in gray) (48).
Intra- and interspecies conjugative transfer of two different ICEs found in S. salivarius was observed under laboratory conditions. These ICEs could thus play a significant role in HGT occurring both in the oral cavity (as exemplified by the presence of a closely related ICE in S. parasanguinis) and in the digestive tract.
Impact of MMC on excision and transfer of ICE_SsaF1-4_fda.
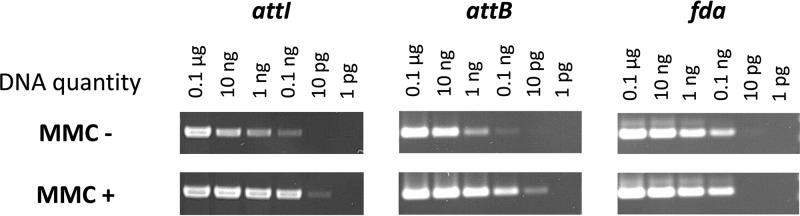
Previous work demonstrated that excision and transfer of ICESt3 of S. thermophilus can be increased by treating donor cells with mitomycin C (MMC) (34). Hence, we examined the impact of such treatment on excision and transfer of an ICE from S. salivarius (ICE_SsaF1-4_fda). Induction of ICE excision by MMC was tested by PCR amplification of attI and attB fragments using different amounts of genomic DNA (0.1 μg to 1 pg) after MMC treatment or not of the cells (Fig. 8). The minimal quantity of DNA producing a positive result for attI and attB was 0.1 ng for nontreated cells, whereas 10 pg of DNA was sufficient to detect a signal after MMC treatment of the cells (Fig. 8). Furthermore, when cells were treated with MMC, a higher intensity of the PCR signal was observed for attI and attB using 0.1, 1, and 10 ng of DNA than in cells without MMC treatment (Fig. 8). Such a difference was not observed for the fda control (Fig. 8). Thus, as for ICESt3 (34), MMC treatment leads to an increase of ICE_SsaF1-4_fda excision.
FIG 8.
Impact of mitomycin C on ICE_SsaF1-4_fda excision. Fragments corresponding to the recombination sites attB and attI of nontreated (MMC−) or treated (MMC+) cells were amplified by PCR using template DNA quantities ranging from 0.1 μg to 1 pg. The amplification of the fda gene was used as a control.
To evaluate the effect of MMC on ICE transfer, strain F1-4 was treated with MMC (at the concentration inducing the maximum level of ICE excision) and used in filter mating experiments with S. salivarius strain F1-8 as the recipient. No difference of ICE transfer was detectable between cells treated or not with MMC. This is in contrast to what was observed for ICESt3 (34). Thus, it appears that for ICE_SsaF1-4_fda excision is not a limiting step for transfer.
Retransfer of ICE_SsaF1-4_fda from transconjugants.
To test whether ICE_SsaF1-4_fda transfers autonomously, retransfer was tested by using transconjugants (obtained as described above) as donor cells. Plasmid pMG36e was removed by curing transconjugants prior to the retransfer assays. F1-8(ICE_SsaF1-4_fda) cells were used as donor cells in filter mating experiments with either F1-8(pMG36e) or LMG18311(pMG36e) cells as recipient cells. LMG18311(ICE_SsaF1-4_fda) and JH2-2(ICE_SsaF1-4_fda) cells were used as donor cells with LMG18311(pMG36e) and JH2-2(pMG36e), respectively (Table 3). For each mating assay, transconjugants were recovered and confirmed by PCR detection of both the ICE integrase gene and plasmid pMG36e present in the recipients (data not shown). A small number of transconjugant colonies (as for intraspecies transfer of ICE_SsaF1-4_fda, i.e., 2 clones for the whole experiment) was observed when using S. salivarius as the donor and strains with the same genetic background as the recipients (F1-8/F1-8 mating pairs) or S. thermophilus LMG18311 cells as recipient cells. In contrast, more transconjugant colonies (at least 20 clones for the whole experiment) were obtained using S. thermophilus LMG18311(ICE_SsaF1-4_fda) and E. faecalis JH2-2(ICE_SsaF1-4_fda) as donors and LMG18311(pMG36e) and JH2-2(pMG36e) cells as recipient cells. These results confirm that ICE_SsaF1-4_fda is able to transfer autonomously but also suggest that (i) host factors could impact ICE replication and/or assembly of the conjugation machinery in S. salivarius, as already suggested for some strains of S. thermophilus (26), or (ii) molecules displayed at the cell surface of S. salivarius could impact the donor/recipient cell contacts and thus the assembly of the conjugative machinery. These hypotheses are consistent with the fact that no transconjugant was recovered when S. thermophilus CNRZ368 donor cells carrying ICESt3 were mated with S. salivarius JIM8777 or F1-8 recipient cells (data not shown), whereas transconjugants were obtained using S. thermophilus LMG18311 as recipient cells as already reported (24).
It should be kept in mind that S. salivarius encounters different physiological conditions in the oral cavity and along the digestive tract. The impact on ICE regulation of these complex interactions between bacteria and of changing environment in these ecological niches should be explored in the near future.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used in the experimental part of this study are listed in Table 1.
Thirteen S. salivarius strains, including 4 commensal (F1-4, F1-8, F4-2, and F6-1) and 9 clinical (B35, B57, L22, L50, L60, L64, N5, N20, and T93) strains, were selected on the basis of a previous study (23). These strains were chosen according to 3 criteria. First, the selected strains likely harbor an ICE with a conjugation module related to ICESt3 (as suggested by a positive PCR signal for the relaxase and coupling protein genes of ICESt3). Second, to increase the diversity of ICEs included in the analysis, half of the strains were selected according to the absence of a PCR signal for the integrase gene of ICESt3, thus suggesting the presence of a different recombination module. Lastly, the selected strains are distributed all over the phylogenetic tree built from MLST data (23). They belong to different MLST groups and differ by at least 3 MLST alleles (out of 6 alleles analyzed) and can thus be considered unrelated, except two of them (strains L22 and L64) which differ by only 1 allele (23). These 13 strains are available upon request.
Modified strains were named according to the modification. For example, S. salivarius F1-8(pMG36e) corresponds to a derivative of S. salivarius F1-8 carrying plasmid pMG36e. pMG36e is a small plasmid (3,611 bp) that carries only genes required for its replication and an erythromycin resistance gene. It is a nonconjugative plasmid derived from pWV01, which is a broad-spectrum plasmid replicating in E. coli and in Firmicutes (49) and has been successfully used for labeling recipient strains in conjugation experiments with S. thermophilus (24). S. salivarius, E. faecalis, and S. thermophilus were grown in M17 broth supplemented with 0.5% lactose (LM17) at 37°C (S. salivarius and E. faecalis) or 42°C (S. thermophilus) without shaking. Selective mitis salivarius agar (MSA; Difco) containing a 1% (wt/vol) tellurite solution, brain heart infusion (BHI; Difco), and reconstituted skim milk broths were also used for mating assays. Solid cultures were made in an oxygen-free environment induced by GasPak utilization (bioMérieux).
When required, cultures were supplemented with the following antibiotics: chloramphenicol (5 μg ml−1 for S. thermophilus or 8 μg ml−1 for S. salivarius and E. faecalis) and erythromycin (5 μg ml−1 for S. thermophilus or 10 μg ml−1 for S. salivarius and E. faecalis).
DNA sequencing and sequence analysis.
The 13 selected S. salivarius strains were subjected to whole-genome sequencing. Sequencing was done with an Illumina HiSeq2000 sequencer by Beckman Coulter genomics (2 × 100 bp after paired-end library construction, with at least 60× coverage expected). Observed sequencing depth was higher than 200× for all the 13 genomes. De novo assembly was performed using CLC Genomics Workbench (CLC Bio) with default parameters. A scaffold of the genomes was built by using the Genome Finishing module of CLC Genomics Workbench with the S. salivarius JIM8777 genome as reference. Some assembly gaps were filled by PCR and sequencing. This enabled us to obtain genomes fragmented in less than 75 contigs (less than 30 contigs for two-thirds of the genomes; the median size of the contigs was higher than 40 kb for half of them). Contigs were first annotated using the RAST annotation server (http://rast.nmpdr.org/) (50, 51). Then, contigs containing genes closely related to conjugation genes of ICESt3 were identified by BLASTN analysis (using the orfO to orfA genes as a query by megaBlast analysis with default parameters and filter disabled), and annotation was completed manually.
In NCBI sequence data banks, ICEs closely related to ICESt3 were searched by using microbial nucleotide blast (using the orfO to orfA genes as a query by megaBlast analysis with default parameters and filter disabled) on complete (n = 816; last accessed 29 July 2016) and draft (n = 9,573; last accessed 29 July 2016) genomes of Firmicutes. Hits with more than 90% identity with the whole query sequence were further analyzed.
Pairwise comparisons of elements were performed with Artemis Comparison Tool provided by the Sanger Centre using comparison files generated by Double Act (available at http://www.hpa-bioinfotools.org.uk/pise/double_actv2.html) (52). Manual editing of comparison figures was performed using Inkscape.
Phylogenetic tree construction.
Proteins of 27 ICEs were included in the analysis. These ICEs correspond to (i) the 19 ICEs of the ICESt3 subfamily with closely related conjugation modules and (ii) 8 additional ones previously reported to belong to the same ICE subfamily but showing more distantly related conjugation modules (11, 26). The sequences of signature proteins were aligned using Clustal omega with default parameters (53). The trees were built with MEGA (54) using (i) maximum likelihood (ML) based on the Jones-Taylor-Thornton (JTT) model including amino acid empirical frequencies (partial deletion of gaps and missing data at 80% cutoff, gamma distribution in 5 categories, and allowance for invariant sites), and (ii) BioNJ methods with the Poisson model (55). The branch support of the groupings was estimated using bootstrap (100 replicates).
ICE tagging.
ICE_SsaF1-4_fda and ICE_SsaF4-2_fda were tagged by a chloramphenicol resistance cassette originating from the pSET5s plasmid. The resistance cassette was inserted in an intergenic region located between convergent coding sequences in the adaptive module to avoid impacting ICE functionality. Two DNA fragments of about 1,000 bp corresponding to the upstream and the downstream regions of the integration locus were amplified by PCR using specific primers that present an extended sequence matching with the 5′ and the 3′ ends of the chloramphenicol resistance cassette. A second PCR amplification was carried out using these two PCR fragments and the resistance gene as a template to synthesize a fragment carrying the antibiotic resistance cassette flanked by the upstream and downstream chromosomal regions of the gene. The natural competence of S. salivarius cells was induced by addition of the synthetic peptide (H2N-LPYFTGCL-COOH) (22), and the overlap PCR product was then added for transformation. The crossover events, upstream and downstream from the tagged region, were positively selected by the newly acquired antibiotic resistance of the transformed clones. The integrity of the regions flanking the antibiotic cassette was confirmed by PCR.
PCRs were done using the Phusion high-fidelity DNA polymerase (Thermo Scientific). PCRs were performed with 50 ng of genomic DNA, 200 μM each deoxynucleotide triphosphate (dNTP), 0.5 μM each primer (for primer sequences, see Table 2), and 0.02 U μl−1 of Phusion DNA polymerase in an appropriate buffer per 50-μl reaction volume. Cycling conditions for the overlap PCR were 3 min at 98°C, 30 s at annealing temperature (with 1°C of incrementation at each cycle), and 30 s/kb at 72°C, followed by 30 additional cycles with an annealing temperature of 55°C and a final extension of 10 min at 72°C.
Excision testing with or without mitomycin C (MMC).
PCR amplifications of attB, attI, attR, and attL fragments were carried out in a 25-μl volume containing 1 μl of overnight culture, 200 μM each dNTP (Thermo Scientific, France), 0.5 μM each primer (for primer sequences, see Table 2), and 0.025 U μl−1 of DreamTaq DNA polymerase in an appropriate buffer (Thermo Scientific, France). PCR amplifications were performed using the following cycling parameters: 10 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 50°C, and 1 min/kb at 72°C, with a final 5-min extension at 72°C. Amplified products were analyzed by electrophoresis on a 1.5% agarose gel.
To test the impact of MMC on ICE excision, PCR amplifications of the attachment sites were done after a 2.5-h treatment of the cells with MMC. To select the MMC concentration that gives the highest induction of excision, a range of MMC concentrations (0.025 to 0.4 mg ml−1) was first tested using 1 ng of genomic DNA as the template. Semiquantitative PCR (performed with 30 cycles) was then done at the selected concentration of MMC (0.05 mg ml−1) using different genomic DNA quantities (0.1 μg to 1 pg). Amplifications of the fda gene were done in parallel as controls.
Mating experiments.
Donor and recipient strains were grown overnight with an appropriate antibiotic. Fifteen milliliters of broth medium was inoculated with 150 μl of donor or recipient stationary-phase cultures. Cultures were grown until mid-exponential phase (optical density at 600 nm of 0.4) and then were mixed and centrifuged for 15 min in a prewarmed centrifuge at 4,500 × g to pellet cells. The pellet was resuspended in 1 ml of LM17 broth, and 150 μl was spread on 0.45-μm-pore-size cellulose nitrate filters (Sartorius Stedim Biotech) deposited on LM17 soft agar (0.8%) plates. Plates were then incubated at 37°C (for S. salivarius-S. salivarius and S. salivarius-E. faecalis mating pairs) or at 39°C (for S. salivarius-S. thermophilus mating pairs). After an overnight incubation, the filters were removed from the agar plates and placed into 10 ml of LM17 liquid medium. Bacteria were recovered by vortexing for 30 s. The suspension was then directly spread on agar plates supplemented with the appropriate antibiotics or concentrated 10 times by centrifugation at 4,500 × g for 15 min to enable counting of the CFU of the donor, the recipient, and the transconjugant cells after a 24-h incubation. Transconjugant clones obtained after S. salivarius intraspecies mating were typed by DNA sequencing of PCR products corresponding to the ddlA gene (for primer sequences, see Table 2).
Treatment of donor cells with mitomycin C was done as follows. Cells carrying ICE_SsaF1-4_fda were grown in LM17 liquid medium at 37°C to an optical density at 600 nm of 0.4. The culture was then diluted 10-fold in 15 ml of prewarmed LM17 medium containing MMC at the concentration that showed the maximum ICE excision level (0.05 mg ml−1). A 10-fold dilution without MMC was used as a control. After 1 h of culture, the cells were harvested by centrifugation in a prewarmed centrifuge and washed once with 15 ml of prewarmed LM17 medium. The donor cells treated with MMC were mixed with recipient cells grown at an optical density at 600 nm of 0.4 and centrifuged for 15 min in a prewarmed centrifuge at 4,500 × g to pellet cells. The pellet was resuspended in 1 ml of LM17 broth, and 150 μl was spread on nitrocellulose filters on LM17 soft agar plates before incubation for 4 h or overnight at appropriate temperatures. The filters were then treated as previously described.
Mating frequencies were calculated by dividing the number of transconjugants by the number of donor cells, except in the case of donor cells treated with MMC, where mating frequencies were calculated relative to recipients. At least three independent biological replicates were done.
Plasmid curing.
Transconjugants carrying plasmid pMG36e were cultured overnight without erythromycin and were then spread on LM17 plates at different dilutions. One hundred isolated clones were then streaked on LM17 plates with or without erythromycin. Erythromycin-sensitive clones were then confirmed by PCR for the absence of plasmid (see Table 2 for primer sequences).
RAPD.
Random amplification of polymorphic DNA (RAPD) (56) was carried out with DreamTaq enzyme (for primer sequences, see Table 2). Cycling conditions were 40 cycles consisting of 94°C for 1 min, 31°C for 1 min, and 72°C for 2 min; the final extension was continued for 7 min at 72°C. One microliter of liquid culture was used as the DNA template. The PCR products were separated by electrophoresis on a 1.5% agarose gel.
Accession number(s).
The sequences of ICEs have been deposited in the EMBL Nucleotide Sequence Database under accession numbers LT622825 to LT622837 .
Supplementary Material
ACKNOWLEDGMENTS
We thank Stéphane Bertin and Emilie Robert for their technical help.
N.D. is recipient of a scholarship funded by INRA and Région Grand Est (formerly Région Lorraine). This work received financial support from the Région Lorraine and Université de Lorraine (2011–2013) and from ANR (MATICE project ANR-15-CE21-0007).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00337-17.
REFERENCES
- 1.Abby SS, Tannier E, Gouy M, Daubin V. 2012. Lateral gene transfer as a support for the tree of life. Proc Natl Acad Sci U S A 109:4962–4967. doi: 10.1073/pnas.1116871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 3.Llosa M, Gomis-Ruth FX, Coll M, de la Cruz F. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol 45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 4.Bellanger X, Payot S, Leblond-Bourget N, Guédon G. 2014. Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev 38:720–760. doi: 10.1111/1574-6976.12058. [DOI] [PubMed] [Google Scholar]
- 5.Roberts AP, Mullany P. 2013. Bacterial integrative mobile genetic elements. Landes Bioscience, Austin, TX. [Google Scholar]
- 6.Toussaint A, Merlin C. 2002. Mobile elements as a combination of functional modules. Plasmid 47:26–35. doi: 10.1006/plas.2001.1552. [DOI] [PubMed] [Google Scholar]
- 7.Burrus V, Pavlovic G, Decaris B, Guédon G. 2002. Conjugative transposons: the tip of the iceberg. Mol Microbiol 46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 8.Brochet M, Rusniok C, Couvé E, Dramsi S, Poyart C, Trieu-Cuot P, Kunst F, Glaser P. 2008. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc Natl Acad Sci U S A 105:15961–15966. doi: 10.1073/pnas.0803654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellanger X, Morel C, Gonot F, Puymege A, Decaris B, Guédon G. 2011. Site-specific accretion of an integrative conjugative element and a related genomic island leads to cis-mobilization and gene capture. Mol Microbiol 81:912–925. doi: 10.1111/j.1365-2958.2011.07737.x. [DOI] [PubMed] [Google Scholar]
- 10.Puymège A, Bertin S, Chuzeville S, Guédon G, Payot S. 2013. Conjugative transfer and cis-mobilization of a genomic island by an integrative and conjugative element of Streptococcus agalactiae. J Bacteriol 195:1142–1151. doi: 10.1128/JB.02199-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambroset C, Coluzzi C, Guédon G, Devignes MD, Loux V, Lacroix T, Payot S, Leblond-Bourget N. 2015. New insights into the classification and integration specificity of Streptococcus integrative conjugative elements through extensive genome exploration. Front Microbiol 6:1483. doi: 10.3389/fmicb.2015.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. 2011. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puymège A, Bertin S, Guédon G, Payot S. 2015. Analysis of Streptococcus agalactiae pan-genome for prevalence, diversity and functionality of integrative and conjugative or mobilizable elements integrated in the tRNALys CTT gene. Mol Genet Genomics 290:1727–1740. doi: 10.1007/s00438-015-1031-9. [DOI] [PubMed] [Google Scholar]
- 14.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. 2005. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van den Bogert B, Boekhorst J, Herrmann R, Smid EJ, Zoetendal EG, Kleerebezem M. 2013. Comparative genomics analysis of Streptococcus isolates from the human small intestine reveals their adaptation to a highly dynamic ecosystem. PLoS One 8:e83418. doi: 10.1371/journal.pone.0083418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson M, Martin R, Walk ST, Young C, Grossman S, McKean EL, Aronoff DM. 2012. Clinical and laboratory features of Streptococcus salivarius meningitis: a case report and literature review. Clin Med Res 10:15–25. doi: 10.3121/cmr.2011.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitten T, Munro CL, Zollar NQ, Lee SP, Patel RD. 2012. Oral streptococcal bacteremia in hospitalized patients: taxonomic identification and clinical characterization. J Clin Microbiol 50:1039–1042. doi: 10.1128/JCM.06438-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corredoira JC, Alonso MP, Garcia JF, Casariego E, Coira A, Rodriguez A, Pita J, Louzao C, Pombo B, Lopez MJ, Varela J. 2005. Clinical characteristics and significance of Streptococcus salivarius bacteremia and Streptococcus bovis bacteremia: a prospective 16-year study. Eur J Clin Microbiol Infect Dis 24:250–255. doi: 10.1007/s10096-005-1314-x. [DOI] [PubMed] [Google Scholar]
- 20.Han XY, Kamana M, Rolston KV. 2006. Viridans streptococci isolated by culture from blood of cancer patients: clinical and microbiologic analysis of 50 cases. J Clin Microbiol 44:160–165. doi: 10.1128/JCM.44.1.160-165.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delorme C, Abraham AL, Renault P, Guedon E. 2015. Genomics of Streptococcus salivarius, a major human commensal. Infect Genet Evol 33:381–392. doi: 10.1016/j.meegid.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, Boyaval P, Hols P. 2010. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J Bacteriol 192:1444–1454. doi: 10.1128/JB.01251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaffanel F, Charron-Bourgoin F, Libante V, Leblond-Bourget N, Payot S. 2015. Resistance genes and genetic elements associated with antibiotic resistance in clinical and commensal isolates of Streptococcus salivarius. Appl Environ Microbiol 81:4155–4163. doi: 10.1128/AEM.00415-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellanger X, Roberts AP, Morel C, Choulet F, Pavlovic G, Mullany P, Decaris B, Guédon G. 2009. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J Bacteriol 191:2764–2775. doi: 10.1128/JB.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burrus V, Pavlovic G, Decaris B, Guédon G. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77–97. doi: 10.1016/S0147-619X(02)00102-6. [DOI] [PubMed] [Google Scholar]
- 26.Carraro N, Libante V, Morel C, Decaris B, Charron-Bourgoin F, Leblond P, Guédon G. 2011. Differential regulation of two closely related integrative and conjugative elements from Streptococcus thermophilus. BMC Microbiol 11:238. doi: 10.1186/1471-2180-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee CA, Grossman AD. 2007. Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol 189:7254–7261. doi: 10.1128/JB.00932-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kishida K, Inoue K, Ohtsubo Y, Nagata Y, Tsuda M. 2017. Host range of the conjugative transfer system of IncP-9 naphthalene-catabolic plasmid NAH7 and characterization of its oriT region and relaxase. Appl Environ Microbiol 83:e02359-16. doi: 10.1128/AEM.02359-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P, Zhang C, Zhu Y, Deng Y, Guo S, Peng D, Ruan L, Sun M. 2013. The resolution and regeneration of a cointegrate plasmid reveals a model for plasmid evolution mediated by conjugation and oriT site-specific recombination. Environ Microbiol 15:3305–3318. doi: 10.1111/1462-2920.12177. [DOI] [PubMed] [Google Scholar]
- 30.Ceccarelli D, Daccord A, Rene M, Burrus V. 2008. Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J Bacteriol 190:5328–5338. doi: 10.1128/JB.00150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyazaki R, van der Meer JR. 2011. A dual functional origin of transfer in the ICEclc genomic island of Pseudomonas knackmussii B13. Mol Microbiol 79:743–758. doi: 10.1111/j.1365-2958.2010.07484.x. [DOI] [PubMed] [Google Scholar]
- 32.Bose B, Auchtung JM, Lee CA, Grossman AD. 2008. A conserved anti-repressor controls horizontal gene transfer by proteolysis. Mol Microbiol 70:570–582. doi: 10.1111/j.1365-2958.2008.06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Auchtung JM, Lee CA, Garrison KL, Grossman AD. 2007. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol Microbiol 64:1515–1528. doi: 10.1111/j.1365-2958.2007.05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellanger X, Morel C, Decaris B, Guédon G. 2007. Derepression of excision of integrative and potentially conjugative elements from Streptococcus thermophilus by DNA damage response: implication of a cI-related repressor. J Bacteriol 189:1478–1481. doi: 10.1128/JB.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carraro N, Libante V, Morel C, Charron-Bourgoin F, Leblond P, Guédon G. 2016. Plasmid-like replication of a minimal streptococcal integrative and conjugative element. Microbiology 162:622–632. doi: 10.1099/mic.0.000219. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira PH, Touchon M, Rocha EP. 2014. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res 42:10618–10631. doi: 10.1093/nar/gku734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mruk I, Kobayashi I. 2014. To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res 42:70–86. doi: 10.1093/nar/gkt711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balado M, Lemos ML, Osorio CR. 2013. Integrating conjugative elements of the SXT/R391 family from fish-isolated vibrios encode restriction-modification systems that confer resistance to bacteriophages. FEMS Microbiol Ecol 83:457–467. doi: 10.1111/1574-6941.12007. [DOI] [PubMed] [Google Scholar]
- 39.Price VJ, Huo W, Sharifi A, Palmer KL. 2016. CRISPR-Cas and restriction-modification act additively against conjugative antibiotic resistance plasmid transfer in Enterococcus faecalis. mSphere 1(3):e00064-16. doi: 10.1128/mSphere.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasu K, Nagaraja V. 2013. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77:53–72. doi: 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocha EP, Danchin A, Viari A. 2001. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res 11:946–958. doi: 10.1101/gr.GR-1531RR. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi N, Naito Y, Handa N, Kobayashi I. 2002. A DNA methyltransferase can protect the genome from postdisturbance attack by a restriction-modification gene complex. J Bacteriol 184:6100–6108. doi: 10.1128/JB.184.22.6100-6108.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Järup L, Akesson A. 2009. Current status of cadmium as an environmental health problem. Toxicol Appl Pharmacol 238:201–208. doi: 10.1016/j.taap.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 44.Birri DJ, Brede DA, Nes IF. 2012. Salivaricin D, a novel intrinsically trypsin-resistant lantibiotic from Streptococcus salivarius 5M6c isolated from a healthy infant. Appl Environ Microbiol 78:402–410. doi: 10.1128/AEM.06588-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dobson A, Cotter PD, Ross RP, Hill C. 2012. Bacteriocin production: a probiotic trait? Appl Environ Microbiol 78:1–6. doi: 10.1128/AEM.05576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wescombe PA, Heng NC, Burton JP, Tagg JR. 2010. Something old and something new: an update on the amazing repertoire of bacteriocins produced by Streptococcus salivarius. Probiotics Antimicrob Proteins 2:37–45. doi: 10.1007/s12602-009-9026-7. [DOI] [PubMed] [Google Scholar]
- 47.Rankin DJ, Rocha EP, Brown SP. 2011. What traits are carried on mobile genetic elements, and why? Heredity 106:1–10. doi: 10.1038/hdy.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bellanger X. 2009. Transfert, accrétion et mobilisation des éléments intégratifs conjugatifs et des îlots génomiques apparentés de Streptococcus thermophilus: un mécanisme clef de l'évolution bactérienne? PhD thesis Nancy-Université, Nancy, France. [Google Scholar]
- 49.van de Guchte M, van der Vossen JM, Kok J, Venema G. 1989. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl Environ Microbiol 55:224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, Parkhill J, Rajandream MA. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 56.Blaiotta G, Sorrentino A, Ottombrino A, Aponte M. 2011. Short communication: technological and genotypic comparison between Streptococcus macedonicus and Streptococcus thermophilus strains coming from the same dairy environment. J Dairy Sci 94:5871–5877. doi: 10.3168/jds.2011-4630. [DOI] [PubMed] [Google Scholar]
- 57.Guédon E, Delorme C, Pons N, Cruaud C, Loux V, Couloux A, Gautier C, Sanchez N, Layec S, Galleron N, Almeida M, van de Guchte M, Kennedy SP, Ehrlich SD, Gibrat JF, Wincker P, Renault P. 2011. Complete genome sequence of the commensal Streptococcus salivarius strain JIM8777. J Bacteriol 193:5024–5025. doi: 10.1128/JB.05390-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacob AE, Hobbs SJ. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol 117:360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takamatsu D, Osaki M, Sekizaki T. 2001. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46:140–148. doi: 10.1006/plas.2001.1532. [DOI] [PubMed] [Google Scholar]
- 60.Delorme C, Poyart C, Ehrlich SD, Renault P. 2007. Extent of horizontal gene transfer in evolution of streptococci of the salivarius group. J Bacteriol 189:1330–1341. doi: 10.1128/JB.01058-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.