ABSTRACT
Bacillus cereus is a soil-dwelling Gram-positive bacterium capable of forming structured multicellular communities, or biofilms. However, the regulatory pathways controlling biofilm formation are less well understood in B. cereus. In this work, we developed a method to study B. cereus biofilms formed at the air-liquid interface. We applied two genome-wide approaches, random transposon insertion mutagenesis to identify genes that are potentially important for biofilm formation, and transcriptome analyses by RNA sequencing (RNA-seq) to characterize genes that are differentially expressed in B. cereus when cells were grown in a biofilm-inducing medium. For the first approach, we identified 23 genes whose disruption by transposon insertion led to altered biofilm phenotypes. Based on the predicted function, they included genes involved in processes such as nucleotide biosynthesis, iron salvage, and antibiotic production, as well as genes encoding an ATP-dependent protease and transcription regulators. Transcriptome analyses identified about 500 genes that were differentially expressed in cells grown under biofilm-inducing conditions. One particular set of those genes may contribute to major metabolic shifts, leading to elevated production of small volatile molecules. Selected volatile molecules were shown to stimulate robust biofilm formation in B. cereus. Our studies represent a genome-wide investigation of B. cereus biofilm formation.
IMPORTANCE In this work, we established a robust method for B. cereus biofilm studies and applied two genome-wide approaches, transposon insertion mutagenesis and transcriptome analyses by RNA-seq, to identify genes and pathways that are potentially important for biofilm formation in B. cereus. We discovered dozens of genes and two major metabolic shifts that seem to be important for biofilm formation in B. cereus. Our study represents a genome-wide investigation on B. cereus biofilm formation.
KEYWORDS: Bacillus cereus, biofilm formation, transcriptome, transposon mutagenesis
INTRODUCTION
Bacteria are capable of forming multicellular communities, known as biofilms (1, 2). Biofilms are clinically significant, because biofilms formed by pathogenic species are often associated with hospital-acquired infections, both acute and chronic (3). Biofilms are also significant in industry and the environment, causing billions of dollars of loss every year in ship, water, and dairy industries; however, in some cases, biofilms can be beneficial. In the field of agriculture, some of the rhizosphere-associated bacteria are used as biological control agents for plant protection. Those bacteria are able to protect plants from infections by various bacterial pathogens, fungi, and even worms through different mechanisms (4).
Both Bacillus cereus and Bacillus subtilis belong to the rhizosphere-associated beneficial bacteria (4). Wild strains of B. subtilis are capable of forming robust pellicle biofilms at the air-liquid interface, colony biofilms on the solid surface, or plant root surface-associated biofilms (5, 6). Genes and regulatory pathways controlling biofilm formation have been well studied in B. subtilis (7, 8). At the center of the regulatory network is the biofilm master repressor SinR (9). SinR directly represses genes such as the epsA-to-epsO operon (epsA-O) and the tapA-sipW-tasA operon, which are responsible for making the sugar and protein moiety of the biofilm matrix, respectively (9, 10). SinR, acting together with other regulatory proteins, such as AbrB and DegU, also indirectly represses bslA, a gene encoding a small hydrophobin that was shown to form a layer of hydrophobic coating surrounding B. subtilis biofilms (11, 12). SinR is counteracted by a small antirepressor, SinI, whose expression is activated by a master regulator, Spo0A, under biofilm induction (13, 14). More recently, we showed that metabolic signals also play important roles in triggering biofilm formation in B. subtilis (15–17). In one study, we discovered a novel serine-sensing mechanism that triggers biofilm formation by altering the translation efficiency of the biofilm repressor SinR in response to a decrease in cellular serine levels, which act as a proxy for the nutrient status in the bacterium (15). In another study, acetic acid, a small volatile metabolite, was shown to stimulate biofilm formation in B. subtilis via air transmission (16).
Like B. subtilis, B. cereus is also known as a soilborne beneficial bacterium and has been frequently used as a biological control agent (4). However, some strains of B. cereus cause foodborne illness or even more severe infectious diseases, such as endophthalmitis and meningitis (18). The pathogenesis of B. cereus is related to several enterotoxins and hemolysins produced by some B. cereus strains, such as hemolysin BL (Hbl), nonhemolytic enterotoxin (Nhe), and cytotoxin K (CytK), whose genes are controlled by the transcriptional regulator PlcR (19). Previous studies suggested that certain B. cereus strains were able to form diverse biofilms, either submerged, bottom-surface-attached biofilms, floating pellicles, or pellicles attached to the side surfaces of the glass tubes. Different types of biofilms may require activities from different genetic determinants (20–22). For example, in one tested B. cereus strain, the tasA homologous genes were shown not to be required for submerged biofilms but were needed for the formation of floating pellicles (20). Nevertheless, compared to B. subtilis, much less is known about the genes in B. cereus that are important for biofilm formation and the functions of those genes. One previous study showed that the global regulator CodY plays an important role in biofilm formation in B. cereus (23). A recently published study also suggested that Spo0A acts as a key regulator for biofilm formation in B. cereus (20). Genes that are homologous to the epsA-O and the tapA biofilm matrix operons, and the dedicated regulatory genes, including sinI and sinR of B. subtilis, have been identified in several B. cereus strains, including AR156. Interestingly, in one recent study, the homologous operon to epsA-O of B. subtilis was shown not to be important for pellicle biofilm formation in a B. cereus strain when tested under laboratory conditions (20). This is very different from the case in B. subtilis, in which the epsA-O operon is essential for biofilm formation under all tested conditions. On the other hand, the results from another recent study demonstrated that protein components made from TasA- and SipW-like proteins in B. cereus seem to play a structural role in matrix assembly (21). However, even with recent progress, current knowledge about the genetics of B. cereus biofilm formation is still largely lacking.
In this study, we showed that an environmental isolate of B. cereus (AR156) is capable of forming robust pellicle biofilms in a newly formulated biofilm-inducing medium (LB-glycerol-MnSO4 [LBGM]) (24). AR156 is one of the environmental isolates of B. cereus that we obtained previously and has an excellent biological control efficacy against various plant fungal pathogens (25). We next took two global approaches, random transposon insertion mutagenesis to identify genes and genetic pathways that may play important roles in biofilm formation and genome-wide transcriptome analysis by RNA sequencing (RNA-seq) to identify genes that are differentially expressed when AR156 cells were grown in a biofilm-inducing medium. Our study represents a genome-wide investigation of B. cereus biofilm formation.
RESULTS AND DISCUSSION
An environmental isolate of B. cereus (AR156) formed robust pellicle biofilms in LBGM.
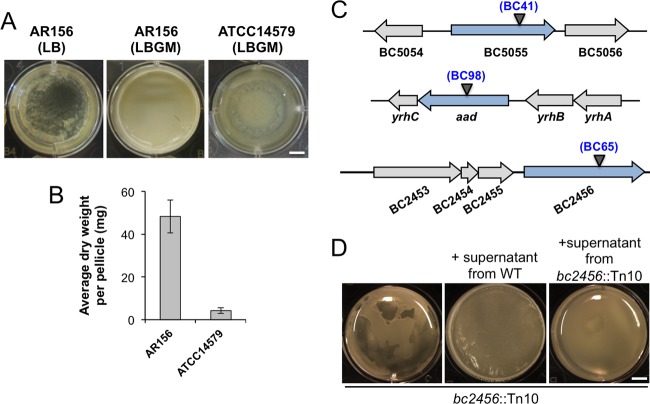
Biofilms formed by various B. cereus strains, as shown in previous studies, although diverse, were in general less robust and demonstrated fewer morphologically distinct features than those in B. subtilis (20, 21, 24, 26). We hoped to develop a robust method to study biofilm formation in B. cereus, similar to what we have in B. subtilis. In a previous study, we described LBGM (LB medium supplemented with 1% glycerol and 100 μM MnSO4) as a biofilm-inducing medium for multiple Bacillus species, including strains of B. subtilis, B. licheniformis, and B. cereus (24). The combination of glycerol and manganese converts LB, a biofilm-inert medium, into a strong biofilm-inducing medium for those bacteria. Colonies of B. subtilis, B. licheniformis, and B. cereus formed on LBGM agar plates also demonstrated complex surface morphology, distinct from those on LB agar plates, thus allowing for a quick screen for an altered biofilm phenotype (24). In this study, we modified LBGM by increasing the concentration of manganese from 100 to 200 μM. We also selected an environmental isolate of B. cereus (AR156), which is highly competent in forming floating pellicles compared to the commonly used laboratory strain ATCC 14579, which formed only a thin layer of floating pellicles under the same conditions (Fig. 1A) (27). To compare the biofilm robustness in a more quantitative fashion, we collected pellicle biofilms from AR156 and ATCC 14579 and measured the dry weight of the pellicles using a method previously developed in B. subtilis (28). Our result showed an almost 10-fold difference in biofilm biomass between AR156 and ATCC 14579 (Fig. 1B). We picked AR156 for further investigation in this study. The genome sequence of AR156 was recently determined (GenBank accession no. CP015589.1), allowing detailed genetic analysis.
FIG 1.
B. cereus AR156 formed robust pellicle biofilms in the biofilm-inducing medium LBGM. (A) Pellicle biofilm formation by B. cereus AR156 in LB and LBGM, and by B. cereus ATCC 14579 in LBGM. Scale bar, 5 mm. (B) The average dry weights of the individual pellicle biofilms formed by AR156 and ATCC 14579 in LBGM were assayed by using the method described in Materials and Methods. The dry weight was calculated as milligrams per pellicle biofilm and was averaged from at least 3 independent samples. Error bars represent standard deviations. (C) The transposon insertion sites within specific genes on the chromosome of AR156 on selected mutants (those discussed in the Results) are indicated by triangles. The corresponding transposon insertion mutant was also indicated. Annotation of the genes is based on either the NCBI database entry for the sequenced B. cereus ATCC 14579 genome, or in some cases, on a BLAST search against strong homologous genes in the closely related B. subtilis. (D) Pellicle biofilm formation by the transposon insertion mutant BC65 (bc2456::Tn10) in LBGM, and in LBGM supplemented with the concentrated cell-free supernatant from the wild-type AR156 or that from the transposon insertion mutant BC65. Scale bar, 5 mm.
Screen of a transposon insertion library of AR156 with altered biofilm phenotypes.
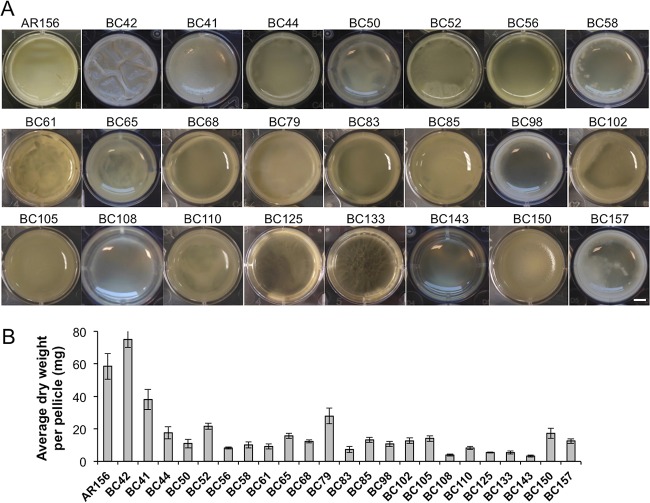
Our next goal was to identify genes that are potentially important for biofilm formation in B. cereus AR156. To do so, we applied two genome-wide approaches, random transposon insertion mutagenesis and transcriptome analysis by RNA-seq. In the first approach, we used the transposon mutagenesis system based on a mini-Tn10 transposable element (pIC333) (29, 30). Random transposon insertion mutagenesis was carried out in AR156. A total of ∼10,000 transposon insertion mutants were picked. These insertion mutants were first spotted on the biofilm-inducing LBGM plates (1.5% [wt/vol] agar) and screened for alterations in the colony phenotype. About 4% of the insertion mutants were found to be interesting to us because they showed altered colony morphology on LBGM plates. Those 400 mutants were further applied to pellicle biofilm formation. Based on the results from the pellicle biofilm assays, we subsequently picked 23 transposon insertion mutants, all of which showed an altered pellicle biofilm phenotype (Fig. 2A and Table 1). Quantitative analysis for the biomass of the pellicle biofilms by those mutants was performed in parallel by comparing the dry weights of the pellicle biofilms (Fig. 2B). Most insertion mutants showed reduced pellicle biofilm formation, and a number of them had >5-fold lower total biofilm biomass than that of AR156 (Fig. 2B). Next, we mapped the transposon insertion sites in those mutants. A description of the transposon mutants, including the mapped transposon insertion sites, is provided in Table 1. Based on the predicted function, genes disrupted by transposon insertion include those involved in nucleotide biosynthesis, iron salvage, antibiotic production, and sporulation, as well as genes encoding an ATP-dependent protease and transcription regulators (Table 1). One puzzle we had from the compilation of all the mapped insertion loci onto the AR156 chromosome is that the insertion loci are distributed in a way that is strongly biased toward the right arm of the chromosome (from 0° to 180°, see Fig. S1 in the supplemental material). It is unclear to us why this happened. Importantly, this indicates that the transposon mutagenesis we performed is not likely saturated. We also note that the genes homologous to the epsA-O and tapA matrix operons and the key biofilm regulatory genes sinI and sinR of B. subtilis are all located in regions at the left arm of the AR156 chromosome (the genetic loci for those homologous genes are as follows: open reading frame 3370 [ORF3370] [epsA], ORF3371 [epsB], ORF3376 [epsD], ORF3374 [epsG], ORF3373 [epsK], ORF3372 [epsM], ORF3430 [sipW], ORF3431 and ORF3433 [tasA; B. cereus genome contains two tasA homologs], ORF3434 [sinR], and ORF3435 [sinI]).
FIG 2.
Pellicle biofilm formation by 23 transposon insertion mutants. (A) Twenty-three individual transposon insertion mutants and the wild-type AR156 were assayed for pellicle biofilm formation in LBGM. Scale bar, 3 mm. (B) The average dry weights of the individual pellicle biofilms formed by the mutants and AR156 were assayed. The dry weight was calculated as milligram per pellicle biofilm and was averaged from at least 3 independent samples. Error bars represent standard deviations.
TABLE 2.
Upregulation of sigB and its regulon during biofilm induction in B. cereus AR156
| Locus tag | Predicted function | Fold induction | E value |
|---|---|---|---|
| BC1004 | SigB, stress response sigma factor | 5.1 | 0 |
| BC1003 | RsbW, serine protease, regulating SigB activity | 5.3 | 0 |
| BC1006 | RsbY, phosphatase, regulating SigB activity | 3.6 | 1.85E−05 |
| BC862 | Protease I, putative SigB regulon in B. cereus | 4.5 | 6.82E−14 |
| BC863 | Catalase, putative SigB regulon in B. cereus | 4.6 | 1.43E−13 |
| BC1012 | Heavy-metal binding protein, putative SigB regulon | 3.3 | 3.87E−12 |
| BC3130 | Hypothetical, putative SigB regulon | 6.4 | 3.40E−14 |
| BC3131 | Hypothetical, putative SigB regulon | 5.9 | 4.63E−10 |
TABLE 1.
Twenty-three transposon insertion mutants of B. cereus AR156
| Strain | Insertion site (AR156) (nt) | Gene locus (ATCC 14579) | Description (ATCC 14579)a |
|---|---|---|---|
| BC41 | 1283251 | BC_5055 | Possible wall-associated protein/M60 family peptidase |
| BC42 | 132817 | BC_3827 | ATP-dependent hsl protease ATP-binding subunit HslU (clpY) |
| BC44 | 3887313 | BC_2307 | Bacillibactin synthetase component F/adenylation domain of nonribosomal polyketide synthase |
| BC50 | 473010 | BC_4190 | Stage III sporulation protein AD (spoIIIAD) |
| BC52 | 591920 | BC_4325 | Putative pyrroline-5-carboxylate reductase (comER) |
| BC56 | 4475863 | Intergenic region between BC_4741 and BC_4742 | Downstream of TrkA domain site-specific recombinase (BC4171), upstream of ABC transporter NapA (BC4172) |
| BC58 | 1089845 | BC_4842 | Transcriptional regulator, GntR family |
| BC61 | 2003883 | BC_0334 | Phosphoribosylamine-glycine ligase (purD) |
| BC65 | 2386938 | BC_2456 | NRPS |
| BC68 | 1667177 | BC_5481 | Stage 0 sporulation protein J (soj) |
| BC79 | 1122960 | BC_4867 | 1,4-Alpha-glucan branching enzyme (glgB) |
| BC83 | 789947 | BC_4540 | DUF2196 superfamily (ywbE, in the putative operon of holin-antiholin genes cidAB in B. subtilis) |
| BC85 | 2002949 | BC_0333 | Phosphoribosyl-aminoimidazole carboxamide formyltransferase/IMP cyclohydrolase (purH) |
| BC98 | 626575 | BC_4365 | Aldehyde-alcohol dehydrogenase (aad) |
| BC102 | 1003735 | BC_4755 | PAP2 family protein, membrane-associated phospholipid phosphatase |
| BC105 | 2059639 | BC_0382 | Ferric-chrome ABC transporter (fluB) |
| BC108 | 1875050 | BC_0221 | Molybdenum ABC transporter (modA) |
| BC110 | 183948 | BC_3875 | Xaa-Pro dipeptidase (pepP) |
| BC125 | 2374525 | BC_0685 | Branched-chain amino acid transport system II carrier protein (brnQ) |
| BC133 | 1298318 | BC_5070 | Small function-unknown gene (BC_5070), downstream of multidrug efflux protein (BC5071) |
| BC143 | 1475960 | Intergenic region between BC_5292 and BC_5291 | Immediately downstream of NADH-ubiquinone dehydrogenase, M subunit (BC_5292), and upstream of N subunit (BC_5291) |
| BC150 | 2081676 | BC_0407 | OTCace |
| BC157 | 1817740 | BC_0167 | N-Acetylmuramoyl–l-alanine amidase (cwlD) |
NRPS, nonribosomal peptide synthetase; PAP2, type 2 phosphatidic acid phosphatase; OTCace, ornithine carbamoyltransferase.
Genes for the ClpYQ protease are important for biofilm formation in B. cereus.
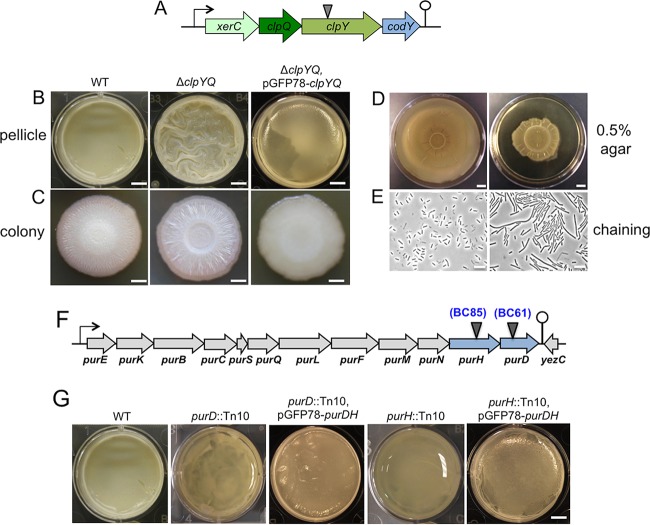
Most transposon insertion mutants we picked showed reduced biofilm formation, except for BC42, which formed a more robust pellicle biofilm (Fig. 2A). BC42 contained the transposon insertion in the clpY gene, which encodes the ATPase substrate-binding subunit for the ClpY-ClpQ (also known as HslVU) protease (Fig. 3A). In both B. cereus and B. subtilis, the clpY gene resides in an operon with clpQ, which encodes the catalytic subunit of the protease, and two other genes, codY and xerC (31). codY encodes a global regulator for stationary-phase metabolism and growth in B. subtilis (31–34), while xerC encodes a site-specific recombinase possibly involved in cell division (31). Although in Gram-negative bacteria, such as Escherichia coli, the biological function of ClpYQ has been relatively well studied under planktonic conditions, little is known about the role of ClpYQ in Gram-positive species.
FIG 3.
(A to E) The ΔclpYQ deletion mutant formed more robust biofilms while impaired for swarming motility and cell separation. (A) A schematic drawing of the presumptive operon of xerC-clpY-clpQ-codY in B. cereus. The transposon insertion site within the clpY gene in BC42 is indicated by the triangle. (B and C) Pellicle (B) and colony (C) biofilms formed by wild-type AR156 and the clpYQ in-frame deletion mutant (FY178), and the clpYQ complementation strain (YY250) in LBGM. Scale bars, 5 mm (B) and 2 mm (C). (D) Swarming motility of AR156 and FY178 on LB plates solidified with 0.5% agar. Plates were incubated at 30°C for 12 h before images were taken. Scale bars represent 1 cm. (E) Cells of AR156 and FY178 grown to late-exponential phase (OD600, 1) and observed under bright-field microscopy. Scale bars, 10 μm. (F and G) Genes involved in purine biosynthesis are important for biofilm formation in B. cereus. (F) A schematic drawing of the putative purine biosynthesis gene cluster in B. cereus AR156. The transposon insertion sites within the purH (BC85) and purD (BC61) genes are indicated by triangles. (G) Pellicle biofilm formation in LBGM by AR156, the two transposon insertion mutants, BC61 (purD::Tn) and BC85 (ΔpurH::Tn), and the two complementation strains, YY251(purD::Tn, pGFP78-purDH) and YY252(ΔpurH::Tn, pGFP78-purDH). Scale bars, 5 mm.
To avoid potential polar effect by the transposon insertion in clpY on the downstream codY gene, we constructed an in-frame deletion mutation in the clpYQ genes in B. cereus AR156. The deletion mutant (FY178) was then tested for pellicle biofilm formation. Similar to the transposon insertion mutant, the deletion mutant also formed more robust pellicle biofilms; the pellicle biofilm formed by the mutant was highly wrinkled within 48 h of incubation, while the wild-type pellicles at 48 h lacked surface features (Fig. 3B). A similar difference in colony biofilm morphology was also seen between the wild type and the clpYQ deletion mutant on LBGM plates (Fig. 3C). In addition, the biofilm phenotype of the mutant was fully suppressed when wild-type copies of the clpYQ genes were provided from a plasmid that was able to replicate in B. cereus AR156 (Fig. 3B and C). These results confirmed the role of clpYQ in biofilm formation in B. cereus. Further characterization revealed that the clpYQ deletion mutant also had a defect in swarming motility. On semisolid LB plates (+0.5% agar), the mutant showed reduced swarming zone size compared to that of the wild type (Fig. 3D). The clpYQ deletion mutant also seems to have a cell separation defect, since the mutant formed cell chains under shaking conditions (in LB to an optical density at 600 nm [OD600] of 1), while the wild type did not (Fig. 3E).
In summary, our evidence suggests that ClpYQ regulates multicellular behaviors, such as biofilm formation and swarming motility in B. cereus. If true, this will be a novel function for this family of ATP-dependent proteases. Interestingly, the clpYQ nonpolar deletion mutant of B. subtilis also largely phenocopies what was seen in the B. cereus mutant, implying a conserved function of ClpYQ in multicellularity (our unpublished data). In future studies, it will be interesting to identify the protein targets of ClpYQ and characterize how ClpYQ is involved in bacterial multicellularity.
Genes involved in purine biosynthesis are important for biofilm formation in B. cereus.
Two transposon insertion mutants (BC61 and BC85) had insertions in either the purD or purH gene (Fig. 3F). Both genes are highly conserved in different bacteria and are believed to be involved in purine biosynthesis (Fig. 4). Both mutants showed little floating pellicles; most cells were at the bottom of the well (Fig. 3G). Quantitatively, there were decreases of about 5- and 4-fold, respectively, in the total biomass of the floating pellicles of the two mutants compared to that of the wild type (Fig. 2B). Both mutants had no growth defect in LBGM (data not shown). More importantly, the biofilm defect in both transposon insertion mutants was largely rescued when wild-type copies of the purDH genes were provided from a plasmid that was able to replicate in B. cereus AR156 (Fig. 3G), indicating that the biofilm defect was due to the insertional mutation in purD or purH.
FIG 4.
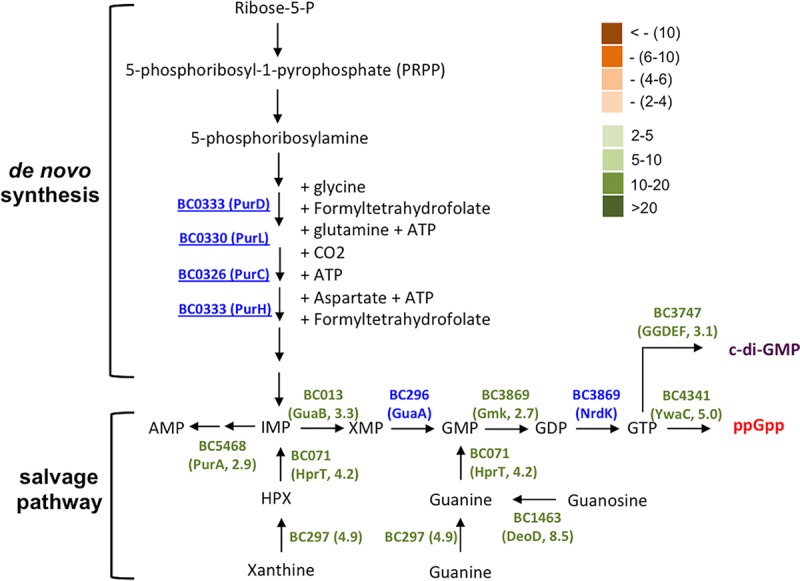
Genes in the proposed pathway for purine biosynthesis and GTP homeostasis were strongly upregulated during biofilm induction in B. cereus. Genes which are involved in the proposed pathway and whose expression was seen to be significantly altered under biofilm induction (in the RNA-seq experiment) are annotated. The fold change in gene expression was also indicated. Significantly increased expression (from 2- to >20-fold) under biofilm induction was seen for genes in green. Significantly decreased expression (from 2- to >10-fold) was seen for genes in orange. Genes in blue indicate insignificant change (<2-fold) in gene expression. Numbers next to the gene name in parentheses represent the fold changes in gene expression.
This result is interesting since in a previous study, a similar transposon mutagenesis was performed in the B. cereus laboratory strain ATCC 14579, and the authors obtained three insertion mutants that were impaired in biofilm formation and contained a transposon insertion in either the purA, purC, or purL gene, all of which are predicted to be involved in purine biosynthesis (Fig. 4) (26). Evidence from that study suggested that the reduced presence of extracellular DNA (eDNA) might be the reason for the impaired biofilm phenotype of the mutants and that eDNA plays an important role in biofilm formation in B. cereus ATCC 14579, likely by acting as an important structural component of the biofilm matrix (26).
We were curious about whether the biofilm defect of the insertion mutants in purD or purH in AR156 was due to similar reasons. To test that, we performed two experiments. First, we measured eDNA presence in the pellicle biofilms by the two mutants and the wild-type strain by quantitative PCR (qPCR). eDNA was collected from pellicle biofilms according to a method applied in our previous study (see Materials and Methods) (16). Three pairs of primers were used to amplify three short DNA fragments in the B. cereus genes clpY, sinR, and calY1, with the eDNA as the template. As shown in Fig. S2A, our results confirmed that eDNA was present in the pellicle biofilms of both the mutants and the wild-type strain. However, the differences in the eDNA levels in the pellicle biofilms from the two mutants and the wild-type strain were rather insignificant (P > 0.1, with the exception that for the abundance of the clpY transcript in the purD mutant versus that in the wild-type strain, P = 0.06; Fig. S2A). In Staphylococcus aureus, it was shown in some studies that biofilm matrix-associated eDNA was also important for biofilm formation, and the release of eDNA was caused by programmed cell death (35, 36). We thus further tested localized cell death within biofilms by performing LIVE/DEAD cell staining. A large number of dead cells were found present in all three pellicle biofilms (cells in red, Fig. S2B). The ratio of dead cells was also quantified by counting red cells versus total cells using MicrobeJ (37) and was calculated at 31.8%, 35.4%, and 32.9% for the wild type, the purD mutant, and the purH mutant, respectively (Fig. S2C). Again, our results suggest that there is no significant difference in the ratio of dead cells in the pellicle biofilms formed by the two mutants and the wild-type strain, since the P values for the variations between the wild type and the mutants were greater than 0.1. We thus suspected that the impaired biofilm phenotypes by the purD and purH insertion mutants of AR156 seen in this study could be due to an as-yet-unknown mechanism. Concerning the findings from the previous study (26), we believe that it will be very difficult to compare results from assays that used different B. cereus strains with different biofilm capacities and very different biofilm settings.
Interestingly, our results from the genome-wide transcriptome analyses (discussed below) showed that many genes involved in GTP homeostasis were upregulated when cells were grown in the biofilm-inducing medium (Fig. 4 and Table S1). GTP homeostasis involves both GTP biosynthesis pathway and pathways making GTP derivative molecules, such as guanosine tetraphosphate (ppGpp) and cyclic-di-GMP (Fig. 4). In the previous study (26), upregulation of purA, purC, and purL was also seen under biofilm-promoting conditions. These results could provide new clues in terms of why those insertion mutants presumably deficient in GTP biosynthesis have a defective biofilm phenotype. GTP homeostasis was shown to be critically important in cell physiology, especially in stationary-phase cells of B. subtilis and Enterococcus faecalis through different mechanisms (e.g., ppGpp synthesis, cyclic-di-GMP [c-di-GMP] signaling, and toxicity of abnormal GTP accumulation) (38, 39). It was also shown that blocking activities of ppGpp-producing enzymes (RelA and RelA-like) had a negative impact on biofilm formation in different bacteria (40). It is possible that GTP homeostasis also plays an important role in biofilm induction in B. cereus and that altered GTP homeostasis may be another reason for the biofilm defect observed in the above-mentioned transposon insertion mutants. It will be interesting to test this hypothesis in future studies.
Other biofilm-defective transposon insertion mutants of B. cereus AR156.
BC41 (bc5055::Tn), in which the transposon insertion site was mapped to a gene encoding a M60 family peptidase (Fig. 1C and Table 1), showed an intermediate defect in pellicle biofilm robustness (Fig. 2A and B). This putative peptidase contains an HEXXH motif and conserved glutamic acid residues seen in zinc-dependent metallopeptidases (our unpublished data). Similar peptidases in the M60-like family were shown to play roles in bacterium-host interactions (41). For example, SslE from E. coli, an outer-surface-associated M60 family peptidase, was reported to be important for the colonization of E. coli cells onto the mucosal surfaces (42). More interestingly, in B. cereus, next to the peptidase gene is a gene (bc5056) that encodes a collagen-like adhesion protein (Fig. 1C). We speculate that this putative M60-like peptidase may play a role in bacterial attachment to the surface during biofilm formation or function as a matrix protein facilitating biofilm matrix assembly.
Another mutant of interest is BC65 (bc2456::Tn). The transposon insertion site in this mutant was mapped to a gene that resembles the ppsB gene in B. subtilis for nonribosomal antibiotic peptide synthesis (Fig. 1C and Table 1). This mutant showed a defective biofilm phenotype (Fig. 1D). In addition to those phenotypes, this mutant was found to completely lack any antagonistic activities against several pathogenic fungal species, such as Fusarium graminearum, Phytophthora capsici, and Botrytis cinema (data not shown), while the wild-type AR156 was known for its strong antifungal activities (25). This confirms that this gene is involved in nonribosomal synthesis of peptides with strong antifungal activities. Such peptides of nonribosomal sources have been extensively studied, and many of them showed antagonistic activities against bacteria, fungi, and even worms (43). What is interesting to us is why the mutant also demonstrated a defective pellicle biofilm phenotype. One possibility is that this peptide not only functions as antimicrobial agent but also as a signal to trigger biofilm formation. The homologous gene cluster is also present in closely related Bacillus thuringiensis strains. In B. thuringiensis, this homologous gene cluster was shown to encode enzymes involved in biosynthesis of an antifungal compound, kurstakin (44). Very interestingly, the B. thuringiensis mutant deficient in kurstakin production also had a strong defect in biofilm formation (45). To test the hypothesis that this putative peptide may be a signal to trigger biofilm formation, we prepared concentrated cell-free supernatants from both the wild-type AR156 and the BC65(bc2456::Tn) cells and applied both supernatants to the pellicle biofilm development by BC65(bc2456::Tn) cells. We observed a complete rescue of the biofilm defect by BC65 when the wild-type supernatants were added (Fig. 1D). Interestingly, a modest increase in pellicle biofilm formation was also observed when the supernatants from the BC65 cells were applied, indicating that other unknown factors in the supernatants also contributed to the pellicle-stimulating activity.
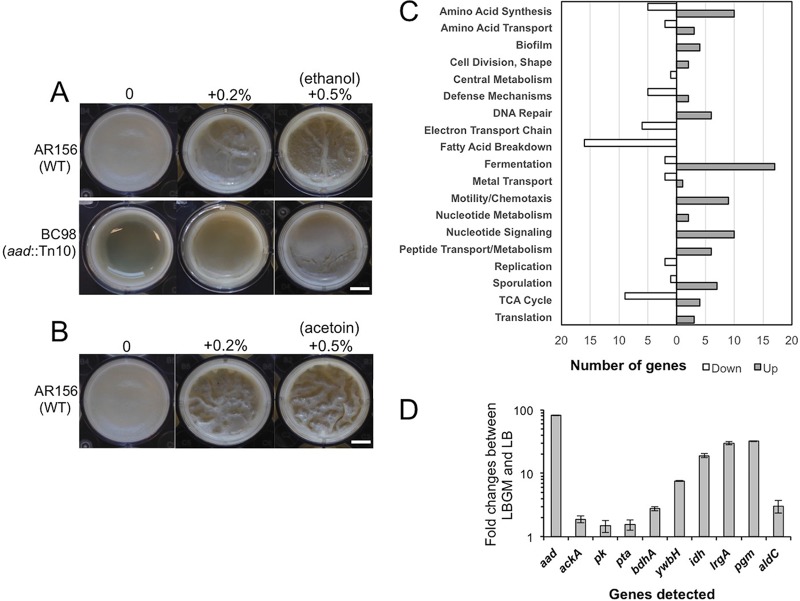
Another interesting transposon insertion mutant is BC98(aad::Tn). This mutant contained the insertional disruption in the aad gene (Table 1 and Fig. 1C). In B. cereus, the aad gene encodes a bifunctional alcohol/aldehyde dehydrogenase which carries out reactions that convert acetyl-coenzyme A (acetyl-CoA) to ethanol. This mutant showed a defective biofilm phenotype (Fig. 5A), possibly due to the lack of ethanol production in the mutant. If so, adding exogenous ethanol may be able to rescue the defective phenotype of the mutant. Indeed, when we applied increasing amounts of ethanol to the transposon insertion mutant of aad, we saw significantly improved biofilm formation (Fig. 5A). The addition of ethanol also seemed to stimulate biofilm robustness in the wild-type cells (Fig. 5A). Interestingly, our transcriptome analyses (discussed below) showed that aad is among the most highly induced genes when cells were grown under biofilm-inducing conditions (59.9-fold, Fig. 5D and 6).
FIG 5.
(A and B) Ethanol- and acetoin-stimulated biofilm formation in B. cereus. (A) Pellicle biofilm formation by AR156 and BC98(aad::Tn) in LBGM in the presence or absence of ethanol. (B) Pellicle biofilm formation by AR156 in LBGM in the presence or absence of acetoin. Scale bars, 2.5 μm. WT, wild type. (C and D) Genome-wide transcriptome analyses of B. cereus AR156 cells during biofilm induction. (C) Summary of the genome-wide transcriptome analysis by RNA-seq. A total of about 500 genes whose expression was significantly altered under biofilm induction (in LBGM) compared to non-biofilm induction (in LB), are categorized into differential functional groups. The number of genes in each functional group was calculated based on the predicted function of the genes. Numbers on the x axis represent number of genes showing differential expression. (D) Validation of upregulation of 10 selected genes under biofilm induction (in LBGM) compared to non-biofilm induction (in LB) by reverse transcription-quantitative PCR (RT-qPCR). Down, downregulated; Up, upregulated.
FIG 6.
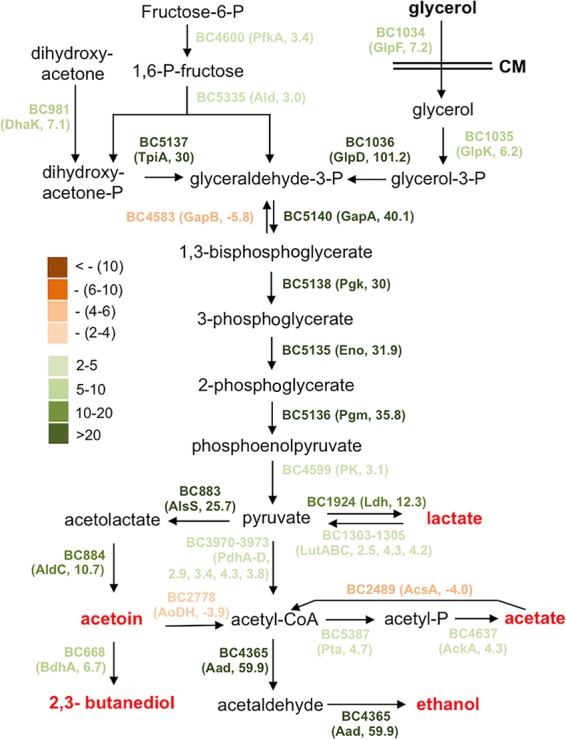
The proposed metabolic shift during biofilm induction in B. cereus. Most genes involved in this predicted metabolic pathway were found to be highly upregulated under biofilm induction (in LBGM) compared to non-biofilm induction (in LB) based on the results of the genome-wide transcriptome analysis by RNA-seq. The genes with different colors represent the difference in fold change in differential expression (LBGM versus LB). Dark green stands for highest induction, while dark brown stands for strongest repression. Numbers next to the gene name in the parentheses represent the fold changes in gene expression.
Genome-wide transcriptome analyses of AR156 cells under biofilm-inducing conditions.
Genes that are differentially expressed under biofilm-inducing conditions are more likely to play a role in biofilm formation. This prompted us to perform genome-wide transcriptome analyses on AR156 cells collected under either non-biofilm- or biofilm-inducing conditions. We used LB shaking culture as a non-biofilm-inducing condition and LBGM shaking culture as a biofilm-inducing condition. This is supported by the idea that AR156 cells form robust pellicle biofilms in LBGM but not in LB (Fig. 1A), and that biofilm-associated gene expression program may be similar enough under shaking conditions in the biofilm-inducing medium versus in real biofilms, based on our previous studies in B. subtilis (10, 15). Among a total of approximately 5,200 genes in B. cereus AR156, we observed about 350 genes (∼6.7%) whose expression was increased by at least 2-fold under the biofilm-inducing conditions (Table S1) and about 140 genes (2.7%) whose expression was decreased by at least 2-fold under the same conditions (Table S2). We subsequently grouped those genes into a number of functional categories based on their predicted functions, which include genes involved in fermentation, respiration, metabolism of amino acids and fatty acids, biofilm, sporulation, and the stress response (Fig. 5C). Ten genes whose expression was significantly upregulated were picked and validated for the induction of these genes by real-time qPCR (Fig. 5D).
The most highly induced genes when cells are grown in LBGM include (i) those involved in carbon metabolism and fermentation, ranging from severalfold to 100-fold (Fig. 6). Activation of these fermentation pathways may lead to the production of small fermentation products, such as ethanol, acetate, lactate, acetoin, and 2,3-butanediol (Fig. 6). We suspect that those genes may not be directly involved in biofilm formation but rather trigger the biofilm induction pathway through altered metabolism activities. (ii) Another important group of induced genes includes those involved in purine biosynthesis, GTP homeostasis, and nucleotide signaling (e.g., ppGpp and cyclic-di-GMP) (Fig. 4). Consistent with this, some of the nucleotide biosynthesis genes were also reported to be upregulated during biofilm formation in B. cereus in other studies (23, 26). (iii) sigB and several genes in the putative SigB regulon were also highly upregulated (Table 2). The stress sigma factor SigB and SigB-activated genes were shown to be important in biofilm formation in several bacterial species, including Listeria monocytogenes, Staphylococcus epidermidis, and Staphylococcus aureus (46–48). (iv) Putative genes associated with regulated cell death were also strongly upregulated (Table 3). Some of those so-called holin antiholin-like genes were among the most highly induced ones when AR156 cells were grown in LBGM (up to 40-fold; Table 3). Those genes were previously shown to play important roles in biofilm formation in S. aureus and B. subtilis (16, 35). On the other hand, a number of genes involved in the tricarboxylic acid (TCA) cycle and oxidative respiration were downregulated when cells were grown in the biofilm-inducing medium (Fig. 5C and Table S2). Some of the fatty acid biosynthesis genes were also seen downregulated (Fig. 5C and Table S2). This may indicate changes in metabolic activities or cytoplasmic membrane structures. Finally, we also noted that a good number of putative toxin genes, including those that encode hemolysin, enterotoxin, and perfringolysin O, were modestly downregulated (Table S2). We have yet to understand the significance of this regulation.
TABLE 3.
Upregulation of genes possibly involved in regulated cell lysis in B. cereus AR156
| Locus tag | Predicted function | Fold induction | E value |
|---|---|---|---|
| BC5439 | LrgA, holin antiholin-like protein | 40 | 2.42E−12 |
| BC5438 | LrgB, holin antiholin-like protein | 12 | 0 |
| BC5133 | YwbH, holin antiholin-like protein | 7 | 2.14E−09 |
| BC813 | Cell wall hydrolase, similar to YocH in B. subtilis | 2.6 | 1.23E−12 |
| BC5239 | Cell wall hydrolase, similar to YocH in B. subtilis | 3.0 | 4.61E−13 |
In B. subtilis, there are dozens of matrix genes (e.g., the epsA-O and tapA operons) that are strongly induced during biofilm formation and play important roles for biofilm assembly (7). The B. cereus AR156 genome contains homologs of those matrix genes in B. subtilis (our unpublished data). However, we noticed that most of those genes were not among the differentially expressed ones (using the 2-fold threshold; Tables S1 and S2). A previous study showed that the eps-like operon was not needed for floating pellicle formation in a B. cereus strain (20). Thus, it is possible that AR156 has different genetic requirements from that of B. subtilis in floating pellicle formation.
Finally, we hope to point out that our approach here is still an indirect approach to study biofilm-associated genes for at least two reasons. First, although biofilm-associated gene expression program may be similar enough under shaking conditions versus in real biofilms, they likely still differ. Second, and more importantly, although the addition of glycerol and manganese (in LBGM) causes differential expression of about 500 genes, and although we believe that at least the activities from some of those genes should lead to biofilm assembly (which explains why LBGM is a biofilm-inducing medium), many of the 500 differentially expressed genes may have little to do with biofilm induction or are indirectly involved in biofilm formation. This has been commonly seen in similar transcriptome-based studies, since subsequent gene knockout studies showed that many of the differentially expressed genes under biofilm conditions did not play clear roles in biofilm formation (10, 49).
Small volatile chemicals stimulate biofilm formation in B. cereus.
Our transcriptome analyses showed that a number of glycolytic and fermentation genes were strongly and differentially upregulated, while TCA cycle and respiration genes were downregulated, when cells were grown in LBGM (Fig. 6 and Tables S1 and S2). This suggests that major metabolic shifts may occur, which may subsequently lead to strong accumulation of fermentation products, such as ethanol, acetate, lactate, acetoin, and 2,3-butanediol (Fig. 6). One could argue that these changes might simply be due to the addition of glycerol and have little relevance to biofilm formation. However, our previous studies in B. subtilis (16) and studies in S. aureus (50) suggest that metabolic changes may be important and necessary for biofilm induction. Interestingly, we showed earlier that the aad transposon insertion mutant failed to form a robust pellicle biofilm. The aad gene encodes a putative bifunctional alcohol/aldehyde dehydrogenase that converts acetyl-CoA to ethanol and is among the most highly induced genes when cells were grown in the biofilm-inducing medium (59.9-fold, Fig. 5D). The biofilm defect of the aad mutant is likely due to a lack of ethanol production, since the defect was rescued by the addition of ethanol to the medium (Fig. 5A). Indeed, even for the wild-type cells, when we added increasing amounts of ethanol (up to 0.5%) to the LBGM, it also stimulated formation of thick structured floating pellicles (Fig. 5A). We thus speculate that ethanol, a small volatile chemical derived from metabolic shifts, can act as a stimulating signal for biofilm formation in B. cereus. Interestingly, in Staphylococcus, ethanol is one of the environmental signals shown to trigger SigB activity and induce biofilm formation (46). Coincidently enough, the sigB gene and several genes in the sigB regulon were among the strongly induced genes when AR156 cells were grown in LBGM (Table 2).
Among other strongly upregulated fermentation pathways from our transcriptome analyses is the pathway for acetoin production. The alsS and aldC genes, whose enzymatic products are involved in the production of acetoin from pyruvate, were upregulated 25.7- and 10.7-fold, respectively (Fig. 6). Strong induction of aldC was also validated by qPCR (Fig. 5D). We similarly speculated that acetoin might be another biofilm-stimulating signal. We tested this accordingly. Increasing amounts of acetoin (from 0, to 0.2%, to 0.5%) were added to LBGM inoculated with AR156 cells for pellicle biofilm development. We saw increased pellicle robustness, evidenced by the formation of thick floating pellicles and hyperwrinkled surface features (Fig. 5B). This result indicates that, like ethanol, acetoin can also stimulate biofilm formation in B. cereus. Our result is also consistent with the findings of a previous study suggesting that acetoin may stimulate biofilm formation in B. subtilis (49).
Previously, it was not entirely clear how the addition of glycerol and manganese can convert LB, a biofilm-inert medium, into a biofilm-inducing medium, even though as part of the answer, we showed in our own study that glycerol or its derivatives were able to stimulate one of the sensory histidine kinases that mediate biofilm induction in B. subtilis (24). Nevertheless, our evidence in that study suggests that glycerol utilization is still necessary for full biofilm induction (24). In this study, our genome-wide transcriptome analyses and subsequent investigations of those volatile chemicals provide evidence for additional mechanisms for the biofilm-stimulating activities of glycerol. We propose that full biofilm induction in LBGM is also dependent on the expected metabolic shifts, which likely result in increased production of small fermentative products, such as acetate, lactate, acetoin, and ethanol (Fig. 6).
Another important group of genes whose expression was also significantly upregulated in the RNA-seq experiment include those involved in purine biosynthesis, GTP homeostasis, and nucleotide signaling (e.g., ppGpp and cyclic-di-GMP) (Fig. 4 and Table S1). This echoed our previous discussion on the insertion mutants for purine biosynthesis genes (e.g., purD and purH). We had hypothesized that GTP homeostasis and nucleotide signaling may play an important role in biofilm formation in B. cereus. Taken together, we propose based on our genome-wide transcriptome analyses that during biofilm formation by B. cereus in LBGM, there may be at least two major metabolic shifts, one leading to strong fermentation and increased production of small fermentative products, and the other possibly leading to the production of ppGpp and c-di-GMP, as well as active nucleotide-based signaling.
Conclusion.
Our current investigation represents a genome-wide study of B. cereus biofilm formation. In this study, we established a fairly robust method for biofilm studies in B. cereus by formulating a modified biofilm-inducing medium and by selecting AR156, an environmental isolate of B. cereus capable of forming robust pellicle biofilms. We also took two genome-wide approaches, transposon insertion mutagenesis and global transcriptome analyses by RNA-seq, in order to characterize the genes and pathways that may play important roles in biofilm formation in B. cereus. Transposon mutagenesis allowed us to identify 23 genes. The role of those genes in biofilm formation in B. cereus is manifested by altered biofilm robustness. Nevertheless, for many of those genes identified in transposon insertion mutagenesis, it is still unclear to us how they can be possibly involved in biofilm formation. This investigation provides a starting point for our future studies aiming to characterize the exact function of these genes and detailed molecular mechanisms in biofilm formation in B. cereus. Our genome-wide transcriptome analyses provided evidence that when cells were grown in the biofilm-inducing medium LBGM, major metabolic shifts caused by the addition of glycerol and manganese could be part of the biofilm-inducing mechanisms in B. cereus. We showed that genes involved in the various fermentation pathways were strongly upregulated (Fig. 6 and Table S1), while genes involved in the citric acid cycle and electron transfer chain were significantly downregulated in LBGM (Fig. 5C and Table S2), suggesting a metabolic shift. This metabolic shift is expected to result in increased production of small fermentative products. We then showed that some of the fermentation products, such as acetoin and ethanol, were able to stimulate strong biofilm formation in B. cereus. Some of these fermentation products may act as small ligands and directly interact with dedicated targets in the cells to alter the expression of genes, whose activities in turn contribute to biofilm formation.
MATERIALS AND METHODS
Strains and media.
Bacillus subtilis and Bacillus cereus strains were routinely cultured in lysogenic broth (LB) (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter of broth) or LB plates solidified with 1.5% agar at 37°C. All strains, plasmids, and primers used in this study are listed in Table 4. The genome sequence of B. cereus AR156 was deposited in GenBank (accession no. CP015589.1). For assays of biofilm formation, LBGM was used. LBGM is composed of LB broth (or solidified LB agar) supplemented with 1% of glycerol and 200 μM MnSO4, modified from a previous recipe for B. subtilis (24). Chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA). Restriction enzymes and other enzymes for molecular cloning were obtained from New England BioLabs (Ipswich, MA, USA). Primers were ordered from Integrated DNA Technologies (Coralville, IA, USA). DNA sequencing was performed at Genewiz (Cambridge, MA, USA).
TABLE 4.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primera | Description or sequence (5′ to 3′)b | Reference or source |
|---|---|---|
| Strains | ||
| NICB3610 | Undomesticated wild strain of B. subtilis | 6 |
| YC1025 | Mutant of B. subtilis 3610 with transposon insertion in yclQ | This study |
| YC1104 | Mutant of B. subtilis 3610 with transposon insertion in fhuD | This study |
| AR156 | Environmental isolate of B. cereus | 25 |
| ATCC 14579 | Domesticated laboratory strain of B. cereus | 27 |
| FY178 | clpYQ in-frame deletion mutant of AR156 | This study |
| YY250 | clpYQ in-frame deletion mutant (FY178), complemented by pGFP78-clpYQ | This study |
| YY251 | purD::Tn10 (BC61), complemented by pGFP78-purDH | This study |
| YY252 | purD::Tn10 (BC85), complemented by pGFP78-purDH | This study |
| Plasmids | ||
| pIC333 | Plasmid containing the mini-Tn10 transposition system, Spcr | 53 |
| pMAD | Suicide vector containing temp-sensitive origin, Mlsr Spcr | 51 |
| pGFP78 | Plasmid replicable in B. cereus with gfp under constitutive promoter | 20 |
| Primers | ||
| Tn10-(113–98) | GCCGCGTTGGCCGATTC | |
| Tn10-(2235–2249) | GATATTCACGGTTTA | |
| clpYQ(Bc)-P1 | GTACGGATCCACTTTGTACTGACACCACTGCT | |
| clpYQ(Bc)-P2 | GTACGTCGACACGTTGAGGAGAAATTAGCAACG | |
| clpYQ(Bc)-P3 | GTACGAATTC CGCCATTGTTTCGCCATCTC | |
| clpYQ(Bc)-P4 | GTACCCATGG ACGACGTTGCACGATGAAAA | |
| clpYQ(Bc)-ComF | GTACTCTAGAAGGAGGGATGAAACAATGGGAAATTTCCACGCT | |
| clpYQ(Bc)-ComR | GTACTCTAGATTACAAAATAAACTGGCT | |
| purDH(Bc)-ComF | GTACTCTAGAAGGAGGGATGAAACAATGAAAAAGCGTGCATTA | |
| purDH(Bc)-ComR | GTACTCTAGATTAACTTCTCGCCATCTC | |
| aad-1 | TAGGACATTCAGCAGTTATC | |
| aad-2 | CAAGCGTAAGTGATGGAA | |
| ackA-1 | ACAGTAGAAGAAGTGGTTA | |
| ackA-2 | TACGGCTTACGAATACAT | |
| aldC-1 | GAAGAAGTGGAAGCATTA | |
| aldC-2 | ATGAGAAGATTGGTTGTG | |
| bdhA-1 | CATTAGCAGGCGTTGATAT | |
| bdhA-2 | GTTATTCGGAGTAATTGTTG | |
| ldh-1 | TAGAGCACTTAGGATGGA | |
| ldh-2 | ATTAGCGAACTCAGAACA | |
| lrgA-1 | TGCTGTTCCAAGTGCTAA | |
| lrgA-2 | TGCGTTACCAATCTCTGAA | |
| pgm-1 | GCTATGGTGAATGGTGAA | |
| pgm-2 | GGCGTGTTATCTTCGTTA | |
| pk-1 | TTACAACAAGCACTGATA | |
| pk-2 | GGAATACCGATTGATACA | |
| pta-1 | CATTAGCAGGCGTTGATAT | |
| pta-2 | GAGCGTCTTCTTCAGTTG | |
| ywbH-1 | TGCTGTTCCAAGTGCTAA | |
| ywbH-2 | TGCGTTACCAATCTCTGAA | |
| clpY-Int(Bc)-F | GAAACGCTCCAGCTGCAACG | |
| clpY-Int(Bc)-R | GTCGAATAGCGTAGATGTATC | |
| sinR-Int(Bc)-F | GAAGTAGAGTATCAACTGGA | |
| sinR-Int(Bc)-R | GCTGGTGTTGCTAAATCTTAC | |
| calY1-Int(Bc)-F | AGGTGATTAGCAACGTAATCT | |
| calY1-Int(Bc)-R | GTTCCATCGATCCTGAAAGAA |
All transposon insertion mutants of B. cereus AR156 are described in Table 1.
Spcr, spectinomycin resistance; Mlsr, erythromycin resistance.
Strain construction.
To construct the nonpolar in-frame deletion mutation in clpYQ in B. cereus AR156 (FY175), the temperature-sensitive suicide vector pMAD was used (51). To explain briefly, about 1-kb regions both upstream and downstream of the clpYQ genes were amplified by PCR using primers clpYQ(Bc)-P1 and clpYQ(Bc)-P2, and clpYQ(Bc)-P3 and clpYQ(Bc)-P4, respectively. The two PCR products were sequentially cloned into the pMAD plasmid, resulting in the recombinant plasmid pMAD-ΔclpYQ(Bc). The resulting plasmid was then introduced into B. cereus AR156 by electroporation (52). Transformants with the plasmid integrated into the chromosomal locus via Campbell integration were selected at nonpermissive temperature (45°C) on LB agar plates supplemented with Mls (1 μg/ml erythromycin plus 24 μg/ml lincomycin) and 40 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl β-d-galactopyranoside). Mls-resistant blue colonies were picked and grown at permissive temperature (25°C) in LB broth to the stationary phase to allow the integrated plasmid to excise from the chromosome. Cells were then diluted 1,000-fold into fresh LB broth and grown at nonpermissive temperature (45°C) for 4 h. Cells were then diluted again and plated on LB agar plates (+X-Gal). The next day, white colonies were picked from the plates and checked for loss of Mls drug resistance. The presence of an in-frame deletion in clpYQ in B. cereus AR156 was confirmed by PCR amplification of the clpYQ locus and DNA sequencing.
To complement the ΔclpYQ mutation, the clpYQ gene was amplified by PCR using the AR156 genomic DNA as the template and primers clpYQ(Bc)-comF1 and clpYQ(Bc)-comR1 (Table 4). The PCR products were digested with XbaI and cloned into the XbaI site of pGFP78, a shuttle plasmid that contains a strong and constitutively expressed promoter, F78, and is able to replicate in both B. cereus and Escherichia coli (20). The correct orientation of the insertion of the PCR products was verified, and the recombinant plasmid was introduced into the ΔclpYQ mutant strain by electroporation (2.5 kV, MicroPulser electroporator; Bio-Rad, Hercules, CA, USA). The transformants were selected on LB agar plates supplemented with 10 μg/ml tetracycline. Complementation in purD::Tn10 (BC61) and purH::Tn10 (BC85) was performed similarly, except that the PCR products containing both purD and purH were applied in both complementation experiments. The purD-purH DNA fragments were amplified by PCR using the AR156 genomic DNA as the template and primers purDH(Bc)-comF1 and purDH(Bc)-comR1 (Table 4).
Transposon mutagenesis.
The temperature-sensitive plasmid pIC333 was used for random transposon insertional mutagenesis. The pIC333 plasmid contains four key elements, the mini-Tn10 transposon element with the replication region of pBR322, and a spectinomycin resistance gene, an erythromycin resistance gene for counterselection, a transposase gene, and a thermosensitive origin of replication functional in Gram-positive species (53). Transposon mutagenesis was performed as described previously (54). The pIC333 plasmid was first transformed into B. cereus AR156 by electroporation. The resulting transformants containing pIC333 were selected based on resistance to both spectinomycin and erythromycin and grown at 25°C to mid-log phase in LB medium supplemented with spectinomycin and erythromycin. The culture was diluted 1:100 into fresh LB medium supplemented with spectinomycin only. The incubation temperature was then shifted to 45°C. This dilution-and-regrow step was repeated 6 to 8 times. Appropriate dilutions of final cultures were plated on LB agar plates supplemented with spectinomycin and incubated at 45°C overnight. All selected transposon mutants were verified for both resistance to spectinomycin and sensitivity to erythromycin. A total of ∼10,000 transposon insertion mutants were picked. These insertion mutants were first spotted on the biofilm-inducing LBGM plates (1.5% [wt/vol] agar) and screened for alteration of colony phenotype. On LBGM plates, about 400 colonies showed altered colony morphology. Those mutants were further selected and applied to pellicle biofilm formation, as described below.
To map the transposon insertion site, genomic DNA of the mutant was prepared by application of a cell lysis kit (lot number 0000195519; Promega, Madison, WI, USA). Five micrograms of DNA was digested with EcoRI or HindIII (the two restriction sites that are not present in pIC333). The digested DNA was purified and self-ligated with T4 DNA ligase overnight at 16°C, and the ligation product was transformed into E. coli DH5α. Plasmid DNA was prepared from the E. coli cells and sent for DNA sequencing by using the primers Tn10(113–98) and Tn10(2235–2249) listed in Table 4. The two primers allow sequence reading outward from the border sequences of the transposon insertion site. The obtained DNA sequences were used to map the transposon insertion sites by aligning the sequence with the genome sequences of both B. cereus ATCC 14579 and AR156.
Transcriptome analysis.
B. cereus AR156 cells were grown to mid-log phase in fresh LB medium and then diluted into LB and LBGM, with a starting OD600 of 0.005. Cells were grown with shaking at 37°C. After grown to late-log phase (OD600, 1.0), cells were sampled, and total RNAs were purified using the RNeasy miniprep kit (Qiagen, Hilden, Germany). The total RNAs were sent on dry ice to BGI Americas (Cambridge, MA, USA) for RNA-seq analysis. Sequencing and analysis were done using duplicate samples. Bioinformatics analyses were also performed by BGI Americas. The published genome sequence of B. cereus ATCC 14579 was used a reference for bioinformatics analysis (27).
Assays of biofilm formation.
To analyze pellicle biofilm formation, cells were first grown in 3 ml of LB broth to exponential phase. Four microliters of culture was then added into the 12-well microliter plates filled with 4 ml of LBGM broth. The plates were incubated at 30°C. Images were taken by a Nikon Coolpix camera after incubation for 2 days. For formation of colony biofilms, cells were similarly grown in 3 ml of LB broth to exponential phase. A 2-μl culture was then spotted onto the LBGM agar plates. The plates were incubated at 30°C for 3 days.
Pellicle dry-weight assay.
Pellicle biofilm formation was carried out in Costar 6-well polystyrene plates filled with Netwell insert with a polyester mesh bottom (opening size, 440 μm) (Corning, NY, USA). LBGM medium inoculated with B. cereus cells was added, and pellicles were allowed to develop for 48 h at 30°C. Individual wells were then removed and air-dried completely in a laminar flow hood for 24 h. Dried pellicles were carefully removed from the well and weighed using an analytic balance. Assays were done in triplicate.
Cell LIVE/DEAD staining.
Cells were grown to exponential-growth phase in 3 ml of fresh LB broth. A 2-μl culture was diluted into the 12-well microliter plates filled with 3 ml of LBGM medium. The plates were placed in incubation for 2 days at 30°C. The cultures were gently discarded, and the pellicle was washed twice with phosphate-buffered saline (PBS) buffer and resuspended in 100 μl of PBS buffer. The LIVE/DEAD staining was performed by using the LIVE/DEAD BacLight bacterial viability kits (catalog no. L7007; Life Technologies, Carlsbad, CA, USA). We first combined equal volumes of components A and B, mixed them thoroughly, and added 1 μl of dye mixture to 10-μl samples. The samples were mixed thoroughly and incubated at room temperature in the dark for 5 min. Five-microliter samples were spotted on the center of the glass slides and were observed using Leica AF6000 modular microsystems. The number of LIVE/DEAD cells was quantified and analyzed by using MicrobeJ (37).
Real-time qPCR.
Total RNAs was isolated from mid-log-phase cells by using the RNeasy miniprep kit (Qiagen, Hilden, Germany). One nanogram of total RNA was applied to the reverse transcription reaction using the AMV first-strand cDNA synthesis kit (New England BioLabs, Ipswich, MA, USA), as described in the provided protocol. Real-time PCR was performed to compare the level of expression in LBGM medium and LB medium with the Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA, USA). The cDNA samples were serially diluted and used as the template for real-time PCR. Serially dilutions of purified genomic DNAs of AR156 with known concentrations were used to generate a standard curve. PCR was performed in the StepOne Plus real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA) by using the following programs: one cycle of 95°C for 3 min and 40 cycles of 95°C for 3 s, 53°C for 20 s, and 60°C for 20 s. All quantitative PCR results were analyzed using the StepOne software.
Quantification of eDNA release by qPCR.
Quantification of pellicle biofilm-associated eDNA release was conducted by using a qPCR-based method, according to a published protocol (35), with some modifications. Pellicle biofilms by the wild-type cells and the mutants were collected after 48 h of incubation at 30°C by brief centrifugation at 4°C and 16,000 × g. Pellicle biofilms were resuspended in 50 mM Tris-HCl–10 mM EDTA–500 mM NaCl (pH 8.0), vigorously vortexed for 3 min, and centrifuged again for 5 min at 4°C and 16,000 × g. Five hundred microliters of the supernatant was transferred to a clean tube and extracted once with an equal volume of phenol-chloroform–isopropanol (25:24:1) and once with chloroform-isopropanol (24:1). The aqueous phase was then mixed with 3 volumes of ice-cold 100% (vol/vol) ethanol and a 1/10 volume of 3 M Na-acetate (pH 5.2) for DNA precipitation. DNA samples were stored at −20°C for 2 h. The ethanol-precipitated DNA was spun down by centrifugation for 25 min at 4°C and 16,000 × g, washed with ice-cold 70% (vol/vol) ethanol once, air-dried, and finally dissolved in 20 μl of Tris-EDTA (TE) buffer. The amount of eDNA was quantified by qPCR using three different primer pairs (for clpY, sinR, and calY1) listed in Table 4. The concentrations of the DNA templates and the primers were determined according to the manufacturer's instructions. PCR was performed in the StepOne Plus real-time PCR system by using the following programs: one cycle of 95°C for 3 min and 40 cycles of 95°C for 3 s, 53°C for 20 s, and 60°C for 20 s. All quantitative PCR results were analyzed using the StepOne software.
Microscopic analysis.
Cells of B. cereus AR156 and its derivatives were cultured to exponential-growth phase in 3 ml LB broth. Cells were then harvested and resuspended in 200 μl of PBS buffer. Five-microliter resuspensions were spotted on the center of the glass slides and covered by coverslips pretreated with poly-l-lysine. All samples were observed using Leica AF6000 modular microsystems.
Assays of swarming motility.
Cells were grown to mid-log phase in fresh LB medium. One milliliter of cultures was sampled, washed with PBS buffer twice, and resuspended in 100 μl of PBS buffer. Swarming plates (LB semisolidified with 0.5% agar) were poured and dried overnight at room temperature. A 5-μl sample was spotted at the center of the swarming plate, dried for 10 min in the laminar hood, and incubated at 37°C. After 12 h, the swarming plates were removed from incubation and dried for 1 h in the laminar hood, and the plates were left on the bench overnight. The next day, the diameter of the swarming zone was measured.
Assays of pellicle promotion by cell-free supernatants.
The wild-type AR156 and BC65 (bc2456::Tn10) cells were first applied to pellicle biofilm formation in LBGM. After 48 h of incubation at 30°C, supernatants were collected from both samples and filtered twice to remove cells and large debris by using the 0.2-μm-pore-size filter (VWR, Radnor, PA, USA). The supernatants were then concentrated 10 times by using a speed vacuum. Those concentrated supernatants were added to fresh LBGM liquid broth at a ratio of 1:4 (cell-free supernatants were 20% of the total volume) to allow pellicle biofilm development by the BC65(bc2456::Tn10) cells. Pellicle biofilms were recorded by camera after 48 h of incubation at 30°C.
Accession number(s).
The genome sequence of B. cereus AR156 was deposited in GenBank under accession no. CP015589.1.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Chai lab for helpful suggestions and comments during the study and manuscript preparation, particularly Yuxuan Qin for statistical analyses.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00561-17.
REFERENCES
- 1.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 2.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 3.Hall-Stoodley L, Stoodley P. 2009. Evolving concepts in biofilm infections. Cell Microbiol 11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 4.Emmert EAB, Handelsman J. 1999. Biocontrol of plant disease: a (Gram-) positive perspective. FEMS Microbiol Lett 171:1–9. doi: 10.1111/j.1574-6968.1999.tb13405.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Yan F, Chai Y, Liu H, Kolter R, Losick R, Guo J-h. 2013. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ Microbiol 15:848–864. doi: 10.1111/j.1462-2920.2012.02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mielich-Süss B, Lopez D. 2015. Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ Microbiol 17:555–565. doi: 10.1111/1462-2920.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 10.Chu F, Kearns DB, Branda SS, Kolter R, Losick R. 2006. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol 59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- 11.Hobley L, Ostrowski A, Rao FV, Bromley KM, Porter M, Prescott AR, MacPhee CE, van Aalten DM, Stanley-Wall NR. 2013. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci U S A 110:13600–13605. doi: 10.1073/pnas.1306390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi K, Iwano M. 2012. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol 85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 13.Newman JA, Rodrigues C, Lewis RJ. 2013. Molecular basis of the activity of SinR, the master regulator of biofilm formation in Bacillus subtilis. J Biol Chem 288:10766–10778. doi: 10.1074/jbc.M113.455592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai U, Mandic-Mulec I, Smith I. 1993. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev 7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- 15.Subramaniam AR, DeLoughery A, Bradshaw N, Chen Y, O'Shea E, Losick R, Chai Y. 2013. A serine sensor for multicellularity in a bacterium. Elife 2:e01501. doi: 10.7443/eLife.01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Gozzi K, Yan F, Chai Y. 2015. Acetic acid acts as a volatile signal to stimulate bacterial biofilm formation. mBio 6(3):e00392-15. doi: 10.1128/mBio.00392-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Gozzi K, Chai Y. 2015. A bacterial volatile signal for biofilm formation. Microb Cell 2:406–408. doi: 10.15698/mic2015.10.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotiranta A, Lounatmaa K, Haapasalo M. 2000. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect 2:189–198. doi: 10.1016/S1286-4579(00)00269-0. [DOI] [PubMed] [Google Scholar]
- 19.Gohar M, Faegri K, Perchat S, Ravnum S, Økstad OA, Gominet M, Kolsto AB, Lereclus D. 2008. The PlcR virulence regulon of Bacillus cereus. PLoS One 3:e2793. doi: 10.1371/journal.pone.0002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao T, Foulston L, Chai Y, Wang Q, Losick R. 2015. Alternative modes of biofilm formation by plant-associated Bacillus cereus. Microbiologyopen 4:452–464. doi: 10.1002/mbo3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caro-Astorga J, Pérez-García A, de Vicente A, Romero D. 2015. A genomic region involved in the formation of adhesin fibers in Bacillus cereus biofilms. Front Microbiol 5:745. doi: 10.3389/fmicb.2014.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wijman JGE, de Leeuw PPLA, Moezelaar R, Zwietering MH, Abee T. 2007. Air-liquid interface biofilms of Bacillus cereus: formation, sporulation, and dispersion. Appl Environ Microbiol 73:1481–1488. doi: 10.1128/AEM.01781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindbäck T, Mols M, Basset C, Granum PE, Kuipers OP, Kovács ÁT. 2012. CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus. Environ Microbiol 14:2233–2246. doi: 10.1111/j.1462-2920.2012.02766.x. [DOI] [PubMed] [Google Scholar]
- 24.Shemesh M, Chai Y. 2013. A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. J Bacteriol 195:2747–2754. doi: 10.1128/JB.00028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niu D-D, Liu H-X, Jiang C-H, Wang Y-P, Wang Q-Y, Jin H-L, Guo J-H. 2011. The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling. Mol Plant Microbe Interact 24:533–542. doi: 10.1094/MPMI-09-10-0213. [DOI] [PubMed] [Google Scholar]
- 26.Vilain S, Pretorius JM, Theron J, Brözel VS. 2009. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl Environ Microbiol 75:2861–2868. doi: 10.1128/AEM.01317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, Chu L, Mazur M, Goltsman E, Larsen N, D'Souza M, Walunas T, Grechkin Y, Pusch G, Haselkorn R, Fonstein M, Ehrlich SD, Overbeek R, Kyrpides N. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87–91. doi: 10.1038/nature01582. [DOI] [PubMed] [Google Scholar]
- 28.Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R. 2013. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci U S A 110:E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kearns DB, Chu F, Rudner R, Losick R. 2004. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol 52:357–369. doi: 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- 30.Dartois V, Djavakhishvili T, Hoch JA. 1996. Identification of a membrane protein involved in activation of the KinB pathway to sporulation in Bacillus subtilis. J Bacteriol 178:1178–1186. doi: 10.1128/jb.178.4.1178-1186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slack FJ, Serror P, Joyce E, Sonenshein AL. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol Microbiol 15:689–702. doi: 10.1111/j.1365-2958.1995.tb02378.x. [DOI] [PubMed] [Google Scholar]
- 32.Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol 191:2953–2963. doi: 10.1128/JB.01492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roux A, Todd DA, Velázquez JV, Cech NB, Sonenshein AL. 2014. CodY-mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J Bacteriol 196:1184–1196. doi: 10.1128/JB.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belitsky BR, Sonenshein AL. 2013. Genome-wide identification of Bacillus subtilis CodY-binding sites at single-nucleotide resolution. Proc Natl Acad Sci U S A 110:7026–7031. doi: 10.1073/pnas.1300428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A 104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma-Kuinkel BK, Mann EE, Ahn J-S, Kuechenmeister LJ, Dunman PM, Bayles KW. 2009. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J Bacteriol 191:4767–4775. doi: 10.1128/JB.00348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ducret A, Quardokus EM, Brun YV. 2016. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaca AO, Kajfasz JK, Miller JH, Liu K, Wang JD, Abranches J, Lemos JA. 2013. Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. mBio 4(5):e00646-13. doi: 10.1128/mBio.00646-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kriel A, Bittner Alycia N, Kim Sok H, Liu K, Tehranchi Ashley K, Zou WY, Rendon S, Chen R, Tu BP, Wang JD. 2012. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol Cell 48:231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakjang S, Ndeh DA, Wipat A, Bolam DN, Hirt RP. 2012. A novel extracellular metallopeptidase domain shared by animal host-associated mutualistic and pathogenic microbes. PLoS One 7:e30287. doi: 10.1371/journal.pone.0030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nesta B, Valeri M, Spagnuolo A, Rosini R, Mora M, Donato P, Alteri CJ, Del Vecchio M, Buccato S, Pezzicoli A, Bertoldi L, Tuscano G, Falduto M, Rippa V, Ashbab Y, Bensi G, Fontana MR, Seib KL, Mobley HLT, Pizza M, Soriani M, Serino L. 2014. SslE elicits functional antibodies that impair in vitro mucinase activity and in vivo colonization by both intestinal and extraintestinal Escherichia coli Strains. PLoS Pathog 10:e1004124. doi: 10.1371/journal.ppat.1004124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ongena M, Jacques P. 2008. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol 16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 44.Béchet M, Caradec T, Hussein W, Abderrahmani A, Chollet M, Leclère V, Dubois T, Lereclus D, Pupin M, Jacques P. 2012. Structure, biosynthesis, and properties of kurstakins, nonribosomal lipopeptides from Bacillus spp. Appl Microbiol Biotechnol 95:593–600. doi: 10.1007/s00253-012-4181-2. [DOI] [PubMed] [Google Scholar]
- 45.Dubois T, Faegri K, Perchat S, Lemy C, Buisson C, Nielsen-LeRoux C, Gohar M, Jacques P, Ramarao N, Kolsto A-B, Lereclus D. 2012. Necrotrophism is a quorum-sensing-regulated lifestyle in Bacillus thuringiensis. PLoS Pathog 8:e1002629. doi: 10.1371/journal.ppat.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knobloch JK-M, Bartscht K, Sabottke A, Rohde H, Feucht H-H, Mack D. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J Bacteriol 183:2624–2633. doi: 10.1128/JB.183.8.2624-2633.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Veen S, Abee T. 2010. Importance of SigB for Listeria monocytogenes static and continuous-flow biofilm formation and disinfectant resistance. Appl Environ Microbiol 76:7854–7860. doi: 10.1128/AEM.01519-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. 2009. Interconnections between sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun 77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanley NR, Britton RA, Grossman AD, Lazazzera BA. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J Bacteriol 185:1951–1957. doi: 10.1128/JB.185.6.1951-1957.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas VC, Sadykov MR, Chaudhari SS, Jones J, Endres JL, Widhelm TJ, Ahn J-S, Jawa RS, Zimmerman MC, Bayles KW. 2014. A central role for carbon-overflow pathways in the modulation of bacterial cell death. PLoS Pathog 10:e1004205. doi: 10.1371/journal.ppat.1004205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turgeon N, Laflamme C, Ho J, Duchaine C. 2006. Elaboration of an electroporation protocol for Bacillus cereus ATCC 14579. J Microbiol Methods 67:543–548. doi: 10.1016/j.mimet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 53.Steinmetz M, Richter R. 1994. Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J Bacteriol 176:1761–1763. doi: 10.1128/jb.176.6.1761-1763.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan F, Yu Y, Wang L, Luo Y, Guo J, Chai Y. 2016. The comER gene plays an important role in biofilm formation and sporulation in both Bacillus subtilis and Bacillus cereus. Front Microbiol 7:1025. doi: 10.3389/fmicb.2016.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.