ABSTRACT
We have previously demonstrated that inoculation of tomato plants with 2,4-diacetylphloroglucinol (DAPG)- and hydrogen cyanide (HCN)-producing Pseudomonas brassicacearum LBUM300 could significantly reduce bacterial canker symptoms caused by Clavibacter michiganensis subsp. michiganensis. In this study, in order to better characterize the population dynamics of LBUM300 in the rhizosphere of tomato plants, we characterized the role played by DAPG and HCN production by LBUM300 on rhizosphere colonization of healthy and C. michiganensis subsp. michiganensis-infected tomato plants. The impact of C. michiganensis subsp. michiganensis presence on the expression of DAPG and HCN biosynthetic genes in the rhizosphere was also examined. In planta assays were performed using combinations of C. michiganensis subsp. michiganensis and wild-type LBUM300 or DAPG (LBUM300ΔphlD) or HCN (LBUM300ΔhcnC) isogenic mutant strains. Populations of LBUM300 and phlD and hcnC gene expression levels were quantified in rhizosphere soil at several time points up to 264 h postinoculation using culture-independent quantitative PCR (qPCR) and reverse transcriptase quantitative PCR (RT-qPCR) TaqMan assays, respectively. The presence of C. michiganensis subsp. michiganensis significantly increased rhizospheric populations of LBUM300. In C. michiganensis subsp. michiganensis-infected tomato rhizospheres, the populations of wild-type LBUM300 and strain LBUM300ΔhcnC, both producing DAPG, were significantly higher than the population of strain LBUM300ΔphlD. A significant upregulation of phlD expression was observed in the presence of C. michiganensis subsp. michiganensis, while hcnC expression was only slightly increased in the mutant strain LBUM300ΔphlD when C. michiganensis subsp. michiganensis was present. Additionally, biofilm production was found to be significantly reduced in strain LBUM300ΔphlD compared to the wild-type and LBUM300ΔhcnC strains.
IMPORTANCE The results of this study suggest that C. michiganensis subsp. michiganensis infection of tomato plants contributes to increasing rhizospheric populations of LBUM300, a biocontrol agent, as well as the overexpression of the DAPG biosynthetic operon in this bacterium. The increasing rhizospheric populations of LBUM300 represent one of the key factors in controlling C. michiganensis subsp. michiganensis in tomato plants, as DAPG-producing bacteria have shown the ability to decrease bacterial canker symptoms in tomato plants.
KEYWORDS: Clavibacter michiganensis, DAPG, HCN, Pseudomonas, gene expression, rhizosphere, tomato
INTRODUCTION
Pseudomonads are well-studied plant-growth-promoting rhizobacteria (PGPR) that have long been recognized for their role in biological control of different plant pathogens (1–5). The adaptability of certain strains to inhabit the rhizosphere, the root surface, and sometimes the roots themselves is indispensable to their disease suppression capacities (6, 7). Their ability to produce different antimicrobial compounds has also been identified as another key factor for controlling plant pathogens (1, 4). Some beneficial pseudomonad strains are able to produce antimicrobial metabolites such as 2,4-diacetylphloroglucinol (DAPG) and hydrogen cyanide (HCN), which have been demonstrated to play a central role in pseudomonads' biocontrol capacities in different agrosystems (1, 4). DAPG, for instance, has broad-spectrum activity against a variety of fungi, bacteria, and nematodes (8). It can trigger an induced systemic resistance response in plants (9–11), stimulate root exudation (12), enhance root branching (13), and even stimulate plant-beneficial activities in other non-Pseudomonas PGPR (14). While HCN is rarely the sole antagonistic compound responsible for biocontrol in Pseudomonas strains and its effectiveness is not as important or as broad spectrum as DAPG, Pseudomonas strains producing DAPG generally also produce HCN (15). HCN has been studied for its antifungal properties and its involvement in disease suppression such as black root rot of tobacco (15).
Pseudomonas spp. have been extensively studied for their ability to protect plants from pathogenic fungi and oomycetes (16), but fewer studies have focused on their ability to suppress bacterial pathogens. Clavibacter michiganensis subsp. michiganensis is a Gram-positive actinomycete causing bacterial canker, the most destructive bacterial disease in tomato, for which no truly efficient chemical or biological control method exists. C. michiganensis subsp. michiganensis spreads through the xylem vessels, eventually colonizing the whole plant and causing systemic infection, which leads to unilateral wilting of leaves, development of canker lesions, tissue disintegration, and plant death (17). We previously reported (18) that under in planta conditions, development of bacterial canker disease in tomato was reduced by inoculation with Pseudomonas brassicacearum LBUM300, which is able to produce both DAPG and HCN (19), but not by inoculation with its phlD- or hcnC-deficient isogenic mutant counterparts. This suggests that the production of both antimicrobial metabolites contributes to reducing disease development and that P. brassicacearum LBUM300 may have the potential to be used as a biocontrol agent for bacterial canker.
In addition to biocontrol traits, the selection of rhizosphere-competent strains is essential to achieve efficient biological control of pathogens. Biofilms are communities of microbes living together in a self-produced extracellular matrix binding the cells to one another and adhering them to a surface (20). Biofilms are considered to be an important colonization strategy in various environments, including the rhizosphere, as the biofilm matrix is able to help protect the cells from desiccation and other stresses (21, 22). Most Pseudomonas isolates are able to form biofilms (21).
While pseudomonads are well known for their ability to protect plants from pathogens, the impact of pseudomonads' antimicrobial metabolite production on their own rhizosphere competency in the absence and especially in the presence of plant pathogens is not well documented. As most biocontrol agent-pathogen interaction studies generally focus on the ability of a potential biocontrol strain to suppress the growth of a specific plant pathogen and reduce disease symptom development, the biocontrol agent's behavior in the system is often overlooked. Previous studies have focused on the impact of soil factors, host genotype, and specific microbial genetic traits on rhizosphere colonization by Pseudomonas spp. (4, 23, 24). However, the role played by antimicrobial metabolite production in rhizosphere colonization remains controversial (25–28).
While it was demonstrated in our laboratory that P. brassicacearum LBUM300 is capable of significantly reducing disease symptoms of bacterial canker caused by C. michiganensis subsp. michiganensis (18), it remains unknown how the capacity to produce DAPG and HCN in the absence/presence of C. michiganensis subsp. michiganensis might impact rhizosphere colonization by P. brassicacearum LBUM300. Furthermore, it is unclear how the presence of C. michiganensis subsp. michiganensis might impact regulation of gene expression responsible for DAPG and HCN production.
In this study, the effects of inoculation with wild-type P. brassicacearum LBUM300 as well as two isogenic mutant strains incapable of producing DAPG or HCN (P. brassicacearum LBUM300ΔphlD and LBUM300ΔhcnC, respectively) on the population dynamics of P. brassicacearum LBUM300 and DAPG and HCN gene expression in the rhizosphere of healthy and C. michiganensis subsp. michiganensis-infected tomato plants were studied using quantitative PCR (qPCR) assays. Finally, the biofilm production of each Pseudomonas strain under study was measured in vitro and the plant-growth-promoting capacity of the three strains was also evaluated by measuring tomato root and shoot weight in the presence and absence of C. michiganensis subsp. michiganensis.
RESULTS
Primer design and evaluation.
The phlD primers and the probe designed in this study (Table 1) targeted a 73-bp region that remained uninterrupted during mutagenesis in wild-type and mutant strains of P. brassicacearum LBUM300 in order to enable absolute quantification of gene copy number in all strains, which was used as an indication of bacterial population numbers. The qPCR amplification of DNA extracted from nonsterile soil inoculated with P. brassicacearum LBUM300 (phlD and hcnC targets), LBUM300ΔphlD (phlD and hcnC targets), or LBUM300ΔhcnC (phlD target only) as well as DNA from pure cultures generated a single band of the appropriate size when DNA was run by conventional agarose gel electrophoresis. No amplification product of phlD or hcnC was obtained from DNA extracted from uninoculated soil (data not shown). For standard curves, a linear relation was observed between log copy numbers of cloned phlD and hcnC fragments and qPCR crossing points (R2 = 0.998 to 1.000) over 8 orders of magnitude ranging from 108 to 101 copies per qPCR, with amplification efficiencies ranging from 98% to 102%. The results obtained showed that in the soil system used in this study, 1:10 soil DNA dilutions were able to overcome the inhibition due to the presence of inhibitory compounds in soil while obtaining copy numbers representative of the amount of bacterial cells that were added to the soil (data not shown).
TABLE 1.
Nucleotide sequences of the primers and TaqMan probes designed and used in this study
| Target gene | Name of primer or probea | Sequence (5′ → 3′) | Product size (bp) | Reference |
|---|---|---|---|---|
| phlD | 300phlD28 (F) | GAGCGAAGCCGGGAACAT | 73 | This study |
| 300phlD100 (R) | TACAGGCCCGCTGTCGAA | |||
| 300phlD52 (P) | CGTGGTGGTCTTCGACGT | |||
| hcnC | hcnCfwd423 (F) | CCTGCCCCAGTCGTTCTTT | 60 | 52 |
| hcnCrev482 (R) | TGCAACTGCGGATACATTGC | |||
| hcnC443 (P) | ATTTCGCCTTGCAGTCC |
F, forward; R, reverse; P, probe.
Population of Pseudomonas spp. in the rhizosphere of tomato plants.
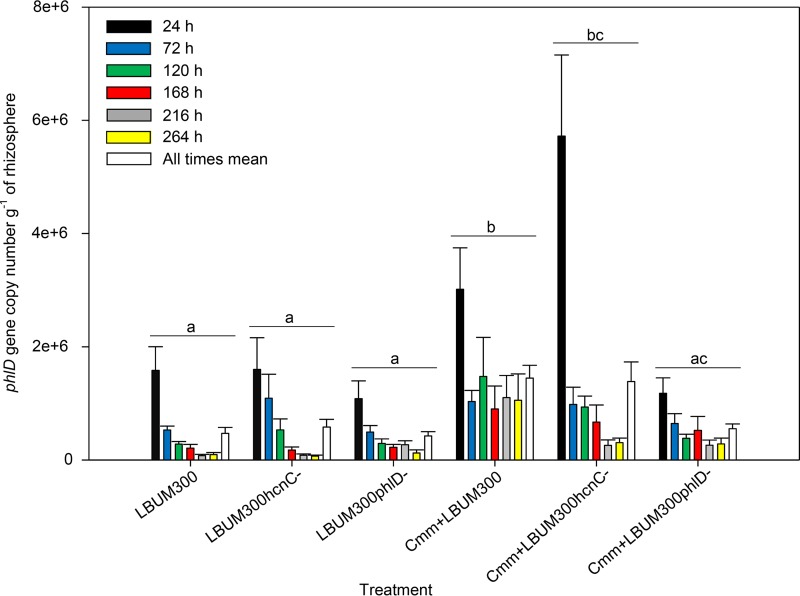
Absolute gene copy numbers were obtained by qPCR amplification of the phlD gene fragment and used to represent cell populations of P. brassicacearum LBUM300, LBUM300ΔphlD, or LBUM300ΔhcnC per gram of rhizosphere soil, as phlD is a single-copy gene in this Pseudomonas strain (29). In all treatments, populations generally decreased over time following inoculation (P < 0.0001). At the last harvest point, Pseudomonas species populations in treatments coinoculated with C. michiganensis subsp. michiganensis were maintained around 105 cells per g soil or higher, while treatments without the pathogen showed generally lower populations. Inoculation of the plant with the pathogen had an overall significant effect on Pseudomonas species populations in the rhizosphere (P < 0.0001). As shown in Fig. 1, analyses performed to evaluate the effects of different inoculation treatments indicated that when wild-type LBUM300 or LBUM300ΔhcnC was coinoculated with C. michiganensis subsp. michiganensis, their populations were significantly increased (P < 0.05) in comparison to treatments inoculated with Pseudomonas spp. alone. Populations of LBUM300ΔphlD when coinoculated with C. michiganensis subsp. michiganensis were not significantly different from those of LBUM300ΔphlD inoculated alone but were significantly lower than the wild-type LBUM300 coinoculated with C. michiganensis subsp. michiganensis (P < 0.05).
FIG 1.
Population of wild-type P. brassicacearum LBUM300 and isogenic mutant strains LBUM300ΔphlD and LBUM300ΔhcnC in the rhizosphere of tomato plants, represented by the phlD gene copy number detected per gram of soil at 24 h, 72 h, 120 h, 168 h, 216 h, and 264 h postinoculation using qPCR (n = 8 for each treatment). The mean of all times within each treatment is indicated by a white bar, and treatments showing different letters are significantly different (P < 0.05). Error bars are standard errors of the means.
Expression of phlD and hcnC genes in the rhizosphere of tomato plants.
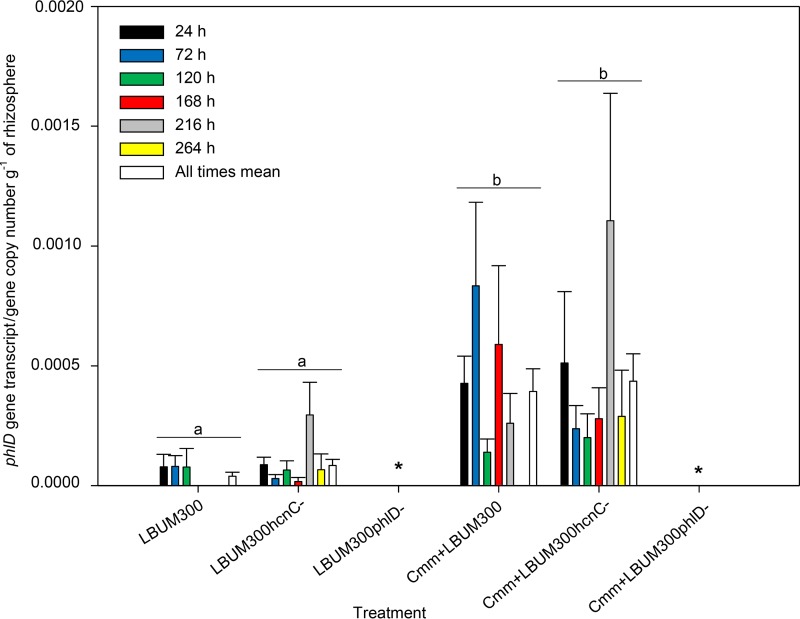
Transcripts of both phlD and hcnC genes were successfully detected from rhizosphere soil samples by reverse transcriptase quantitative PCR (RT-qPCR). Gene expression of phlD and hcnC was calculated as the ratio of transcript copy number to phlD DNA copy number as a way of describing transcription activity independently of potential cell number fluctuations.
As shown in Fig. 2, inoculation of tomato plants with only wild-type P. brassicacearum LBUM300 or strain LBUM300ΔhcnC led to similar levels of phlD gene expression. However, pathogen inoculation had a significant effect on phlD gene expression (P < 0.05). When plants were coinoculated with the pathogen and wild-type P. brassicacearum LBUM300 or LBUM300ΔhcnC, expression of phlD was significantly increased in comparison to treatments where C. michiganensis subsp. michiganensis was absent. Time postinoculation also had a significant effect on gene expression (P < 0.05), although no interaction between time and treatment was observed. No data are presented for plants inoculated with LBUM300ΔphlD, since phlD was disrupted and this rendered the organism incapable of gene expression and DAPG production.
FIG 2.
Transcriptional activity of the phlD gene in wild-type P. brassicacearum LBUM300 and strain LBUM300ΔhcnC in the rhizosphere of tomato plants detected at 24 h, 72 h, 120 h, 168 h, 216 h, and 264 h postinoculation using RT-qPCR (n = 8 for each treatment). phlD gene transcripts were normalized to phlD gene copy number per gram of rhizosphere soil. Strain LBUM300ΔphlD, carrying a disrupted phlD gene and incapable of phlD expression and DAPG production, is represented by an asterisk. The mean of all times within each treatment is indicated by a white bar, and treatments showing different letters are significantly different (P < 0.05). Error bars are standard errors of the means.
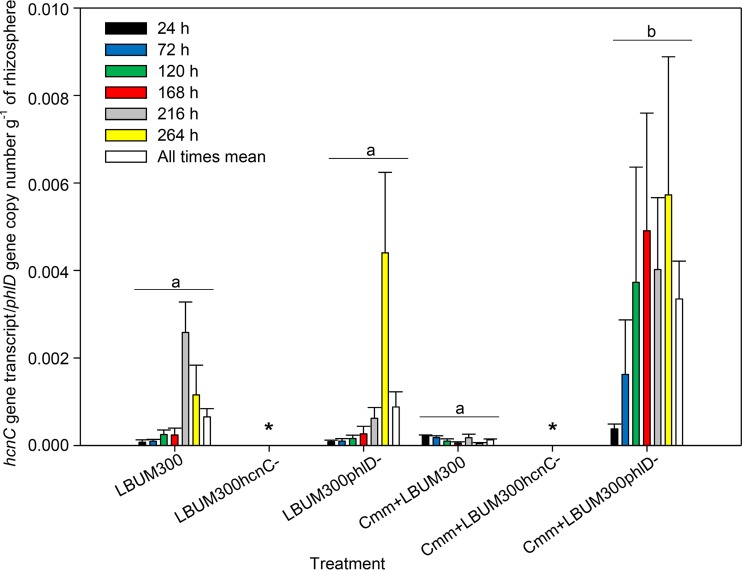
While statistical analysis also revealed a significant effect of pathogen inoculation on hcnC gene expression, it was demonstrated that hcnC expression was significantly upregulated only when the ΔphlD strain of P. brassicacearum LBUM300 was coinoculated with C. michiganensis subsp. michiganensis (Fig. 3). No statistical difference was observed between treatments when wild-type LBUM300 or LBUM300ΔphlD was inoculated alone or when C. michiganensis subsp. michiganensis was inoculated in combination with the wild-type P. brassicacearum LBUM300, capable of DAPG and HCN production. Time postinoculation had a significant effect (P < 0.05), with a general increase in hcnC expression over time in most treatments, but no interaction between time and treatment was detected. No data are presented for plants inoculated with strain LBUM300ΔhcnC, since hcnC was disrupted and this rendered the organism incapable of gene expression and HCN production.
FIG 3.
Transcription activity of the hcnC gene in wild-type P. brassicacearum LBUM300 and strain LBUM300ΔphlD in the rhizosphere of tomato plants detected at 24 h, 72 h, 120 h, 168 h, 216 h, and 264 h postinoculation using RT-qPCR (n = 8 for each treatment). hcnC gene transcripts were normalized to phlD gene copy number per gram of rhizosphere soil. Strain LBUM300ΔhcnC, carrying a disrupted hcnC gene and incapable of hcnC expression and HCN production, is represented by an asterisk. The mean of all times within each treatment is indicated by a white bar, and treatments showing different letters are significantly different (P < 0.05). Error bars are standard errors of the means.
Biofilm production by the three P. brassicacearum strains under study.
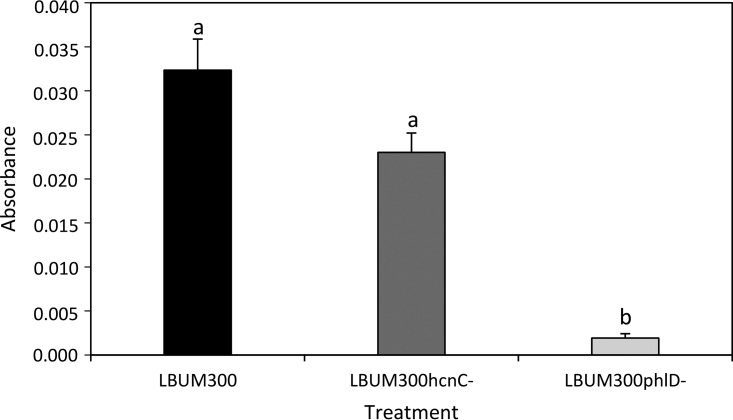
As shown in Fig. 4, the measured absorbance of wild-type LBUM300 and strain LBUM300ΔhcnC, both able to produce DAPG, was significantly higher than the absorbance of strain LBUM300ΔphlD, indicating a greater biofilm production. These results suggest that LBUM300's ability to produce DAPG leads to a greater biofilm production. HCN did not show an effect on biofilm formation.
FIG 4.
Biofilm production by wild-type P. brassicacearum LBUM300 and strains LBUM300ΔhcnC and LBUM300ΔphlD after 48 h of growth (n = 16 for each treatment). All treatments showing different letters are significantly different (P < 0.05). Error bars are standard errors of the means.
Influence of C. michiganensis subsp. michiganensis and P. brassicacearum inoculations on tomato root and shoot weights.
As shown in Tables 2 and 3, the presence or absence of P. brassicacearum LBUM300 or its mutant strains alone did not significantly alter tomato root or shoot weight in comparison to the control, with values ranging between 0.81 g and 0.97 g and between 2.37 g and 2.66 g, respectively. However, infection with C. michiganensis subsp. michiganensis had a significantly negative impact (P < 0.05) on these two growth parameters in comparison to the control or to plants inoculated with only Pseudomonas spp., with root and shoot weights dropping to 0.24 g and 0.76 g, respectively. The symptoms observed on the plants inoculated with C. michiganensis subsp. michiganensis were similar to those reported previously (18).
TABLE 2.
Root and shoot weight of tomato plants grown in nonsterile agricultural soil under growth chamber conditions at 4 weeks postinoculation treated with combinations of P. brassicacearum strains and C. michiganensis subsp. michiganensis
| Treatmenta | Wt (g) at wk 4 |
|
|---|---|---|
| Roots | Shoots | |
| None (control) | 0.97 | 2.54 |
| C. michiganensis subsp. michiganensis | 0.24 | 0.76 |
| P. brassicacearum LBUM300 | 0.89 | 2.66 |
| P. brassicacearum LBUM300ΔhcnC | 0.87 | 2.37 |
| P. brassicacearum LBUM300ΔphlD | 0.92 | 2.42 |
| C. michiganensis subsp. michiganensis + LBUM300 | 0.67 | 1.94 |
| C. michiganensis subsp. michiganensis + LBUM300ΔhcnC | 0.76 | 1.68 |
| C. michiganensis subsp. michiganensis + LBUM300ΔphlD | 0.89 | 2.39 |
n = 8 for each treatment.
TABLE 3.
Resulting P values obtained by mixed ANOVAs using a priori comparisons between treatments of P. brassicacearum strains and C. michiganensis subsp. michiganensis at 4 weeks postinoculation on root and shoot weight of tomato plantsa
| Treatments compared |
P value |
|
|---|---|---|
| Roots | Shoots | |
| Control versus C. michiganensis subsp. michiganensis | 0.001 | 0.003 |
| Control versus LBUM300, LBUM300ΔhcnC, and LBUM300ΔphlD | 0.656 | 0.541 |
| C. michiganensis subsp. michiganensis versus LBUM300, LBUM300ΔhcnC, and LBUM300ΔphlD | 0.000 | 0.002 |
| Control versus C. michiganensis subsp. michiganensis + LBUM300, C. michiganensis subsp. michiganensis + LBUM300ΔhcnC, and C. michiganensis subsp. michiganensis + LBUM300ΔphlD | 0.229 | 0.159 |
| C. michiganensis subsp. michiganensis versus C. michiganensis subsp. michiganensis + LBUM300, C. michiganensis subsp. michiganensis + LBUM300ΔhcnC, and C. michiganensis subsp. michiganensis + LBUM300ΔphlD | 0.003 | 0.016 |
| C. michiganensis subsp. michiganensis + LBUM300 versus C. michiganensis subsp. michiganensis + LBUM300ΔhcnC and C. michiganensis subsp. michiganensis + LBUM300ΔphlD | 0.358 | 0.533 |
| C. michiganensis subsp. michiganensis + LBUM300ΔhcnC versus C. michiganensis subsp. michiganensis + LBUM300ΔphlD | 0.491 | 0.857 |
Significant values (P < 0.05) are in bold; n = 8 for each treatment.
A priori contrasts also revealed that shoot and root weights of plants coinoculated with the pathogen were able to regain levels similar to those of the uninfected control plants and did not differ significantly regardless of whether C. michiganensis subsp. michiganensis was coinoculated with wild-type P. brassicacearum LBUM300 or with strain LBUM300ΔphlD or LBUM300ΔhcnC.
DISCUSSION
Very few studies thus far have specifically looked at the impact of antimicrobial metabolite production on rhizosphere competency, and little is known about the contribution that these metabolites may have on the colonization and survival of the producing strains in the rhizosphere. In this study, we demonstrated that the inability to produce DAPG or HCN by P. brassicacearum LBUM300 did not influence its colonization of the tomato rhizosphere in the absence of the bacterial pathogen C. michiganensis subsp. michiganensis. However, this was not the case in the presence of C. michiganensis subsp. michiganensis, where the population dynamic of P. brassicacearum LBUM300 was clearly influenced by its capacity to produce DAPG. A higher abundance of strains able to produce DAPG was found in the rhizosphere of tomato when C. michiganensis subsp. michiganensis was present, and this effect was not seen for the non-DAPG-producing strain LBUM300ΔphlD, whose population did not differ significantly from those seen in treatments without the pathogen. These results strongly suggest that DAPG antibiotic production contributes to the ecological fitness of P. brassicacearum LBUM300 in the rhizosphere of tomato and that this is somehow related to the presence of C. michiganensis subsp. michiganensis.
Previous works have shown conflicting results as to the effect of DAPG production on a strain's rhizocompetence. Production of DAPG in Pseudomonas fluorescens P32 increased colonization capacity on wheat roots in natural soils in comparison to the parental strain, which produced HCN and siderophores but not DAPG (30). Additionally, a DAPG overproducer (due to a phlF gene mutation) was able to colonize the rhizosphere of tomato plants at a higher rate than the wild type (31). In contrast, the loss of DAPG production did not reduce the ability of P. fluorescens F113 to colonize and persist in the rhizosphere of sugar beets in short-term (27 days) and long-term (270 days) experiments (26, 32). Another study demonstrated that the inability to synthesize DAPG or HCN metabolites did not reduce the persistence of P. fluorescens CHA0 in the rhizosphere of wheat (33).
It has been previously noted that the presence of root pathogens can support larger populations of bacteria, presumably attributed to excess substrate availability due to root leakage of nutrients as a result of disease (28, 34). This alone does not explain why introduced P. brassicacearum strains specifically colonized the C. michiganensis subsp. michiganensis-infested rhizosphere to a higher extent only if these strains were capable of DAPG production. With respect to pathogen presence, Mazzola and colleagues observed that in the presence of Gaeumannomyces graminis var. tritici, a fungal pathogen responsible for take-all disease in wheat, phenazine-producing and -nonproducing strains maintained similar populations (28). In contrast to our results with DAPG producers, the population of the phenazine-producing strain was significantly higher than its nonproducing counterpart in the absence of the pathogen. Others have noted that the presence of root pathogens can lead to larger populations of phlD-positive Pseudomonas spp. (DAPG producers), for instance, in maize and cucumber infected with Pythium ultimum (35, 36), in bean infected with Rhizoctonia solani (37), and in wheat infected with G. graminis var. tritici (34, 38), although none of these studies clearly demonstrated the specific role of DAPG production in root colonization. Clearly, the results of this study and those available in the existing literature suggest that different responses could be expected depending on the antimicrobial metabolite produced, the host plant, and the pathogen present. Our results indicate that the capacity to produce DAPG allows a better establishment of P. brassicacearum LBUM300 in the rhizosphere of tomato in the presence of the bacterial pathogen C. michiganensis subsp. michiganensis.
We found that the presence of the tomato pathogen C. michiganensis subsp. michiganensis influenced expression of the phlD and hcnC genes, involved in the synthesis of DAPG and HCN, respectively. This clearly suggests that the bacterial pathogen C. michiganensis subsp. michiganensis has an effect on DAPG and HCN biosynthesis. Previous reports have shown that the expression of phlA in P. fluorescens CHA0 was significantly increased in the rhizosphere of bean in the presence of the fungal pathogen Rhizoctonia solani (37) and in the rhizosphere of maize and cucumber infected with Pythium ultimum (35). Other fungal pathogens, such as Fusarium oxysporum, have been found to repress phlA gene expression and DAPG production in the rhizosphere of wheat (39). While most studies have focused on the effects of fungal plant pathogens, we demonstrate here that the presence of a bacterial plant pathogen caused increases in phlD gene expression. The ability or inability to produce HCN did not have an effect on phlD gene expression.
In contrast to DAPG, very little information regarding expression of HCN biosynthetic genes in the plant rhizosphere is available, even though most DAPG producers also produce HCN. We report that expression of the hcnC gene was increased in the presence of C. michiganensis subsp. michiganensis in comparison to treatments without the pathogen, but only in the non-DAPG-producing strain LBUM300ΔphlD. These results indicate that the inability to produce DAPG clearly had an effect on hcnC expression. We demonstrated in an earlier study that the presence of the fungal pathogen Verticillium dahliae had a significant stimulatory effect on hcnC expression in the rhizosphere of strawberry compared to treatments in which the pathogen was absent (40). However, the increased expression was observed only in wild-type P. brassicacearum LBUM300, as isogenic mutants were not used. Jamali and colleagues monitored the expression of the hcnA gene in P. fluorescens CHA0 in the rhizosphere of bean infected with the pathogen R. solani under gnotobiotic conditions (37). An increase in hcnA gene expression per gram of rhizosphere was observed; however, this was attributed to an increase in root colonization by CHA0, since hcnA expression in individual cells was not significantly enhanced by the presence of R. solani (37).
We can further conclude that the upregulation of the phlD and hcnC genes was not attributable to an increase in colonization since all gene expression data for both genes were normalized to the phlD gene copy number in the rhizosphere. Hence, the results obtained here may be an indication that the presence of C. michiganensis subsp. michiganensis stimulates production of DAPG by P. brassicacearum LBUM300 in the rhizosphere of tomato and that when the strain is unable to produce DAPG, it may try to compensate through production of HCN. However, it is known that production of both metabolites by P. brassicacearum LBUM300 is necessary to reduce populations of C. michiganensis subsp. michiganensis in the rhizosphere of tomato and to reduce the development of bacterial canker (18).
The biofilm results showed that the ability to produce DAPG by P. brassicacearum LBUM300 and P. brassicacearum LBUM300ΔhcnC leads to higher biofilm production than that of the mutant deficient in DAPG production. The ability to produce HCN did not influence biofilm production. Phenazines are known to significantly contribute to biofilm maturation in vitro and on plants (22); however, very few studies have shown an effect of DAPG on biofilm formation and the results have been contradictory to date. A study by Combes-Meynet et al. (41) showed that when adding DAPG to a culture of Azospirillum sp. strain Sp245-Rif, the biofilm production increased with increasing DAPG concentrations. They also observed that the expression of the ppk gene, which contributes to cell motility and biofilm formation (42, 43), was upregulated by DAPG (41). However, Powers et al. (20) demonstrated that subinhibitory concentrations of DAPG were able to inhibit Bacillus subtilis biofilm formation. These results indicate that DAPG can affect biofilm formation; however, the mechanism of action and the reasons leading to a positive or negative effect are clearly not known.
With regard to plant growth promotion, conflicting results on the potential implication of antimicrobial metabolite production are reported in the literature. In our experiments, the presence of the wild-type or mutant strains of P. brassicacearum LBUM300 did not significantly increase root or shoot weight or the general growth or vigor of tomato plants at 4 weeks postinoculation. Therefore, we can conclude that P. brassicacearum LBUM300 does not display general growth-promoting activities on tomato. Similarly, no significant difference in shoot or root weight was observed when pea plants were inoculated with the wild type and with non-DAPG producers (44, 45). However, in other studies, the inoculation of healthy pea plants with the DAPG producer P. fluorescens F113 or a DAPG-overproducing strain resulted in greater root weight than in plants inoculated with a non-DAPG-producing mutant strain (46).
Results following the coinoculation of plants with C. michiganensis subsp. michiganensis and any of the wild-type or mutant strains of P. brassicacearum LBUM300 indicated that the plants had significantly greater root and shoot weights than plants inoculated with the pathogen alone. Similar positive effects on shoot and/or root weight were observed when pea plants were inoculated with the pathogen Pythium and DAPG-producing strains of P. fluorescens (strain F113, CHA0, or Q2-87), but no plant growth promotion by DAPG producers was detected in healthy plants (47). In another study (48), similar results were obtained in wheat inoculated with the DAPG producer P. fluorescens Pf29 and/or G. graminis var. tritici, but in the absence of the pathogen, the presence of the DAPG-producing pseudomonads significantly increased root weight compared to the control plants. Our results suggest that regardless of the capacity to produce DAPG or HCN, inoculation of tomato plants with P. brassicacearum LBUM300 reduced the negative impact caused by the pathogen on root and shoot weight. The mechanisms involved, however, remain unknown.
In conclusion, our results demonstrated that the tomato bacterial pathogen C. michiganensis subsp. michiganensis had an effect on rhizosphere colonization by P. brassicacearum LBUM300 and that the capacity to produce DAPG by P. brassicacearum LBUM300 contributed to rhizosphere competency whereas this was not the case for HCN. While expression of both phlD and hcnC genes was detected in all treatments, upregulation of phlD was noted only in the presence of the pathogen and that of hcnC occurred only when the gene for production of DAPG was disrupted. Although P. brassicacearum LBUM300 cannot be considered a true tomato plant growth promoter, its presence in the rhizosphere attenuates the host plant's root and shoot weight decrease due to the presence of C. michiganensis subsp. michiganensis. This effect is, however, not associated with the inherent capacity to produce DAPG or HCN and remains to be characterized.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
P. brassicacearum LBUM300 carrying functional phl and hcn operons was previously isolated from the rhizosphere of strawberry plants in Bouctouche, NB, Canada (19). Isogenic mutant strains of P. brassicacearum LBUM300 deficient in DAPG production (P. brassicacearum LBUM300ΔphlD) or HCN production (P. brassicacearum LBUM300ΔhcnC) were previously developed by site-directed mutagenesis of phlD and hcnC genes using the suicide plasmid pKNOCK-Gmr (18). The pathogenic C. michiganensis subsp. michiganensis strain LMG 5644 was obtained from the Microbiology Laboratorium of the University of Ghent (Belgium). All bacterial strains were maintained and grown routinely in tryptic soy broth (TSB) or agar (TSA) at 25°C. Prior to in planta assays, strains were grown at 25°C in TSB with continuous shaking at 200 rpm for approximately 24 h and 48 h for P. brassicacearum strains and C. michiganensis subsp. michiganensis, respectively.
Biofilm assay.
Biofilm formation was assessed in the three P. brassicacearum strains according to the methods described by Arseneault et al. (49). Briefly, P. brassicacearum strains (wild-type LBUM300, LBUM300ΔhcnC, and LBUM300ΔphlD) were grown at 25°C for 48 h in TSB. An optical density at 600 nm (OD600) reading was performed to ensure equal concentrations among all samples. Cultures were diluted 1:10 in M9 minimal salts medium with Casamino Acids (BD), supplemented with 0.2% (wt/vol) glucose and 1 mM MgSO4, of which 100 μl was plated in each well of a 96-well polystyrene cell culture plate (Thermo Scientific, Waltham, MA). Plates were incubated without shaking at 25°C for 48 h, after which biofilm formation was assessed. The medium was removed, and wells were washed once with water. Each well was stained with 125 μl of 0.1% crystal violet for 10 min, washed 5 times with water, air dried, and destained for 10 min using 200 μl of 95% ethanol. The OD600 was quantified using a plate reader (Varioskan; Thermo Scientific) after pipetting 125 μl from each destained well into a new well. For each culture, in addition to a control with medium only (blank), 16 replicate wells were used.
In planta assays.
Seeds of Solanum lycopersicum cv. Scotia (Vesey's Seeds, York, Canada) were first germinated on watered compressed peat disks (Vesey's Seeds) and incubated for 24 days in a growth chamber (Conviron, Winnipeg, Canada) under controlled conditions (12-h light/12-h dark photoperiod, 25°C, 80% humidity). Plantlets were then transplanted into 4-inch-diameter pots and filled with sieved nonsterile agricultural field soil collected in Bouctouche, NB, Canada. The soil was stored at 4°C until use and characterized as a gleyed podzolic gray luvisol, a subgroup of the Canadian System of Soil Classification (50), pH 5.2, consisting of 62% sand, 25% silt, 13% clay, and 2.6% organic matter.
To prepare for root and soil inoculation, P. brassicacearum and C. michiganensis subsp. michiganensis cells were grown to late stationary phase as described above in TSB and harvested by centrifugation at 5,000 × g for 5 min at 4°C. Cells were washed twice, resuspended in 1× phosphate-buffered saline (PBS; 25 mM phosphate buffer, 125 mM NaCl, pH 7.4), and adjusted to 109 CFU ml−1 and 2 × 109 CFU ml−1 for the three P. brassicacearum strains and C. michiganensis subsp. michiganensis, respectively. At transplantation, root systems of plantlets were briefly dipped in the following P. brassicacearum suspensions: LBUM300, LBUM300ΔphlD, LBUM300ΔhcnC, or 1× PBS solution alone (“none-control”). An additional 5 ml of the respective P. brassicacearum solution and 5 ml of the C. michiganensis subsp. michiganensis solution (or 5 ml of 1× PBS for the none-control) were inoculated in the soil/root area of each plantlet. The plantlets were grown in a growth chamber (Conviron) (16-h light/8-h dark photoperiod with alternating 25°C/20°C thermoperiod, 100% humidity) and watered daily or when needed.
The experimental setup consisted of 6 harvesting dates (24 h, 72 h, 120 h, 168 h, 216 h, and 264 h), 8 treatments (LBUM300, LBUM300ΔphlD, LBUM300ΔhcnC, C. michiganensis subsp. michiganensis, C. michiganensis subsp. michiganensis plus LBUM300, C. michiganensis subsp. michiganensis plus LBUM300ΔphlD, C. michiganensis subsp. michiganensis plus LBUM300ΔhcnC, and control), and 4 biological replicates per time and treatment, for a total of 192 samples. An additional identical set of plants comprising all treatments and biological replicates were kept until 4 weeks postinoculation in order to weigh roots and shoots once symptoms of disease had developed. The whole experiment was repeated twice.
Plant harvest.
Plants and rhizosphere soil were destructively sampled at 24 h, 72 h, 120 h, 168 h, 216 h, and 264 h postinoculation, for a total of 32 plants per time point. Rhizosphere soil and plant roots were immediately frozen in liquid nitrogen and stored at −80°C until use. Rhizosphere soil was later lyophilized using a ModulyoD freeze dryer (Thermo Fisher Scientific, Waltham, MA, USA), and plant roots were crushed in liquid nitrogen and stored at −80°C until microbial DNA and RNA extractions. After 4 weeks of growth postinoculation, eight plants per treatment were harvested and plant roots and shoots were weighed.
Nucleic acid extraction.
Total DNA and RNA were coextracted from 0.5 g rhizosphere soil samples as previously described (51) with the following modification: 15% polyethylene glycol (PEG) 6000 was used instead of 30% PEG 6000. Samples were kept on ice during the whole procedure, and final extracts were resuspended in 50 μl of DNase- and RNase-free diethyl pyrocarbonate (DEPC)-treated water (Fisher Scientific, Mississauga, Canada) and stored at −80°C. Total DNA and RNA were also extracted from 0.1 g of crushed root samples using the DNeasy plant minikit (Qiagen, Mississauga, Canada) and the RNeasy plant minikit (Qiagen), respectively, by following the manufacturer's protocol. Final extracts were resuspended in DNase- and RNase-free DEPC-treated water (Fisher Scientific); RNA samples were stored at −80°C, and DNA samples were stored at −20°C. For all samples, DNA and RNA quantity and quality were evaluated using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
DNase treatment and reverse transcription.
To eliminate contaminating DNA, RNA samples extracted from rhizosphere soil were treated with 6 U and subsequently 3 U of Turbo DNA-free DNase I enzyme as suggested by the manufacturer (Life technologies, Burlington, Canada). RNA from plant roots were subjected to an on-column DNase digestion as indicated by the manufacturer of the RNeasy plant minikit (Qiagen) and subsequently to 6 U and 3 U of DNase I enzyme (Life Technologies). Immediately after DNase treatments, 7.9-μl subsamples of each DNase-treated extract were used as a template for gene-specific reverse transcription using 200 nM appropriate reverse primers (Table 1) and TaqMan reverse transcription reagents according to the manufacturer protocol (Life Technologies).
Primer and probe design for qPCR.
The design of RT-qPCR primers and a TaqMan probe based on the hcnC sequence of P. brassicacearum LBUM300 was previously described (Table 1) (52). The design of primers and a TaqMan probe for the phlD gene was performed using the PrimerExpress 3.0 software (Applied Biosystems, Foster City, CA) based on the phlD gene sequence of P. brassicacearum LBUM300 (GenBank accession number DQ788986). Because the designed primers and probe for the phlD gene were developed in a region that was not interrupted during mutagenesis in LBUM300, LBUM300ΔphlD, and LBUM300ΔhcnC, the same set could be used for DNA quantification of the three strains. Specificity was confirmed through a BLASTn search in the NCBI database and through qPCR amplification on DNA extracted from nonsterile field soil samples uninoculated and inoculated with each of the wild-type and mutant P. brassicacearum strains under study in addition to DNA extracted from pure cultures (data not shown). All TaqMan probes were labeled with a 6-carboxyfluorescein (6-FAM) reporter dye at the 5′ end and an MGBNFQ quencher dye at the 3′ end (Applied Biosystems). The primers were custom synthesized by Integrated DNA Technologies (Coralville, IA, USA). The sequences of the primers and probe are listed in Table 1.
Standard curves for absolute quantification of hcnC and phlD genes.
A standard curve for hcnC was prepared by cloning a 60-bp PCR amplicon into the TOPO-TA 3,956-bp plasmid (Invitrogen, Burlington, Canada). For phlD, a 73-bp PCR amplicon was cloned into the 2,976-bp pKRX plasmid (National Institute of Genetics, Mishima, Japan). Plasmid copy numbers were quantified using a spectrophotometer (NanoDrop Technologies), and the gene copy numbers were calculated according to the molar mass derived from the plasmid and amplicon lengths. Dilutions of the known concentrations of plasmid DNA containing the appropriate inserts were made to generate standard curves ranging from 108 to 101 gene copies per qPCR mixture obtained by 10-fold dilutions.
Because the phlD primers and probe were compatible with a region that was not interrupted during mutagenesis in LBUM300, LBUMΔ300phlD, and LBUM300ΔhcnC, the phlD standard curve was used for the absolute quantification of phlD gene copy numbers in the three strains. This standard curve was also used to quantify phlD mRNA transcripts, while hcnC mRNA transcripts were quantified using the hcnC standard curve. Gene expression data of phlD and hcnC were presented as ratios of mRNA transcripts normalized to the phlD DNA gene copy number. Standard curves were always run in triplicate, and amplification efficiency (E) was calculated from the slope of the standard curve using the equation E = 10(−1/slope) − 1.
Quantitative PCR.
qPCR targeting DNA and cDNA from hcnC and phlD genes (Table 1) was performed using a Bio-Rad CFX Connect real-time PCR detection system and the iTaq universal probe supermix kit (Bio-Rad Laboratories, Mississauga, Canada). Each qPCR mixture contained 6 μl of template DNA (diluted 1 in 10 in DEPC-treated water following extraction), cDNA, or DNase-treated RNA (no RT control) and was prepared with 10 μl (1×) of iTaq universal probe supermix, 0.8 μl (200 nmol/liter) of appropriate reverse and forward primers, 1.2 μl (100 nmol/liter) of appropriate probe, and 1.1 μl of sterile double-distilled water (ddH2O), for a total volume of 20 μl. The cycling conditions were 95°C for 2 min followed by 50 cycles of 95°C for 5 s and 60°C for 30 s. Negative-control reactions were performed during each qPCR run by replacing DNA with sterile ddH2O. All qPCRs were replicated 3 times.
Statistical analysis.
Statistical analyses were performed using the SAS Statistical Software version 9.2 (SAS institute Inc., 1992). Mixed analyses of variance (ANOVAs) were carried out on the qPCR data and were followed by a posteriori comparisons using Tukey's tests at a 5% significance level. An analysis was performed on all the data to examine the effects of time, the presence of the pathogen, and the presence of the P. brassicacearum strain. This was followed by a second analysis between different treatments grouping all sampling dates without factoring in the presence of the pathogen. The biofilm data were analyzed by ANOVA to examine the effect of the P. brassicacearum strain on biofilm formation, followed by a posteriori comparisons using Tukey's test at a 5% significance level. Mixed ANOVAs using a priori contrasts between appropriate treatments were carried out on the shoot and root weight data. To meet the requirements of the tests, the qPCR data used in the mixed ANOVA analysis were either log or rank transformed whereas the biofilm data and the week 4 shoot weight data were both rank transformed.
ACKNOWLEDGMENTS
We acknowledge Ivan Oresnik (University of Manitoba, Winnipeg, MB, Canada) and Niki Hironori (Genetic Strain Research Center, National Institute of Genetics, Mishima, Shizuoka, Japan) for providing the pKNOCK-Gmr plasmid vector and the E. coli strain S17-1/λpir, respectively.
This study was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) grant to Martin Filion.
REFERENCES
- 1.Haas D, Défago G. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 2.Maurhofer M, Baehler E, Notz R, Martinez V, Keel C. 2004. Cross talk between 2,4-diacetylphloroglucinol-producing biocontrol pseudomonads on wheat roots. Appl Environ Microbiol 70:1990–1998. doi: 10.1128/AEM.70.4.1990-1998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raaijmakers JM, Vlami M, de Souza JT. 2002. Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek 81:537–547. doi: 10.1023/A:1020501420831. [DOI] [PubMed] [Google Scholar]
- 4.Weller DM. 2007. Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97:250–256. doi: 10.1094/PHYTO-97-2-0250. [DOI] [PubMed] [Google Scholar]
- 5.Weller DM, Raaijmakers JM, Gardener BBM, Thomashow LS. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu Rev Phytopathol 40:309–348. doi: 10.1146/annurev.phyto.40.030402.110010. [DOI] [PubMed] [Google Scholar]
- 6.Couillerot O, Pringent-Combaret C, Caballero-Mellado J, Moenne-Loccoz Y. 2009. Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett Appl Microbiol 48:505–512. doi: 10.1111/j.1472-765X.2009.02566.x. [DOI] [PubMed] [Google Scholar]
- 7.Lugtenberg BJJ, Dekkers LC. 1999. What makes Pseudomonas bacteria rhizosphere competent? Environ Microbiol 1:9–13. doi: 10.1046/j.1462-2920.1999.00005.x. [DOI] [PubMed] [Google Scholar]
- 8.Weller DM, Landa BB, Mavrodi OV, Schroeder KL, de la Fuente L, Bankhead SB, Molar RA, Bonsall RF, Mavrodi DV, Thomashow LS. 2007. Role of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant Biol 9:4–20. doi: 10.1055/s-2006-924473. [DOI] [PubMed] [Google Scholar]
- 9.Iavicoli A, Boutet E, Buchala A, Metraux JP. 2003. Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. Mol Plant Microbe Interact 16:851–858. doi: 10.1094/MPMI.2003.16.10.851. [DOI] [PubMed] [Google Scholar]
- 10.Siddiqui IA, Shahid Shaukat S. 2003. Suppression of root-knot disease by Pseudomonas fluorescens CHA0 in tomato: importance of bacterial secondary metabolite, 2,4-diacetylphloroglucinol. Soil Biol Biochem 35:1615–1623. doi: 10.1016/j.soilbio.2003.08.006. [DOI] [Google Scholar]
- 11.Weller DM, Mavrodi DV, van Pelt JA, Pieterse CM, van Loon LC, Bakker PAMH. 2012. Induced systemic resistance in Arabidopsis thaliana against Pseudomonas syringae pv. tomato by 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens. Phytopathology 102:403–412. doi: 10.1094/PHYTO-08-11-0222. [DOI] [PubMed] [Google Scholar]
- 12.Phillips DA, Fox TC, King MD, Bhuvaneswari TV, Teuber LR. 2004. Microbial products trigger amino acid exudation from plant roots. Plant Physiol 136:2887–2894. doi: 10.1104/pp.104.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brazelton JN, Pfeufer EE, Sewat TA, Gardener BBM, Coenen C. 2008. 2,4-Diacetylphloroglucinol alters plant root development. Mol Plant Microbe Interact 21:1349–1358. doi: 10.1094/MPMI-21-10-1349. [DOI] [PubMed] [Google Scholar]
- 14.Combes-Meynet E, Pothier JF, Moënne-Loccoz Y, Prigent-Combaret C. 2011. The Pseudomonas secondary metabolite 2,4-diacetylphloroglucinol is a signal inducing rhizoplane expression of Azospirillum genes involved in plant-growth promotion. Mol Plant Microbe Interact 24:271–284. doi: 10.1094/MPMI-07-10-0148. [DOI] [PubMed] [Google Scholar]
- 15.Rezzonico F, Zala M, Keel C, Duffy B, Moenne-Loccoz Y, Défago G. 2007. Is the ability of biocontrol fluorescent pseudomonads to produce the antifungal metabolite 2,4-diacetylphloroglucinol really synonymous with higher plant protection? New Phytol 173:861–872. doi: 10.1111/j.1469-8137.2006.01955.x. [DOI] [PubMed] [Google Scholar]
- 16.Sharifi-Tehrani A, Zala M, Natsch A, Moënne-Loccoz Y, Défago G. 1998. Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Eur J Plant Pathol 104:631–643. doi: 10.1023/A:1008672104562. [DOI] [Google Scholar]
- 17.Eichenlaub R, Gartemann KH. 2011. The Clavibacter michiganensis subspecies: molecular investigation of gram-positive bacterial plant pathogens. Annu Rev Phytopathol 49:445–464. doi: 10.1146/annurev-phyto-072910-095258. [DOI] [PubMed] [Google Scholar]
- 18.Lanteigne C, Gadkar VJ, Wallon T, Novinscak A, Filion M. 2012. Production of DAPG and HCN by Pseudomonas sp. LBUM300 contributes to the biological control of bacterial canker of tomato. Phytopathology 102:976–973. doi: 10.1094/PHYTO-11-11-0312. [DOI] [PubMed] [Google Scholar]
- 19.Paulin MM, Novinscak A, St-Arnaud M, Goyer C, DeCoste NJ, Privé J-P, Owen J, Filion M. 2009. Transcriptional activity of antifungal metabolite-encoding genes phlD and hcnBC in Pseudomonas spp. using qRT-PCR. FEMS Microbiol Ecol 68:212–222. doi: 10.1111/j.1574-6941.2009.00669.x. [DOI] [PubMed] [Google Scholar]
- 20.Powers MJ, Sanabria-Valentín E, Bowers AA, Shank EA. 2015. Inhibition of cell differentiation in Bacillus subtilis by Pseudomonas protegens. J Bacteriol 197:2129–2138. doi: 10.1128/JB.02535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ude S, Arnold DL, Moon CD, Timms-Wilson T, Spiers AJ. 2006. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ Microbiol 8:1997–2011. doi: 10.1111/j.1462-2920.2006.01080.x. [DOI] [PubMed] [Google Scholar]
- 22.Danhorn T, Fuqua C. 2007. Biofilm formation by plant-associated bacteria. Annu Rev Microbiol 61:401–422. doi: 10.1146/annurev.micro.61.080706.093316. [DOI] [PubMed] [Google Scholar]
- 23.Landa BB, Mavrodi OV, Raaijmakers JM, Gardener BBM, Thomashow LS, Weller DM. 2002. Differential ability of genotypes of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens strains to colonize the roots of pea plants. Appl Environ Microbiol 68:3226–3237. doi: 10.1128/AEM.68.7.3226-3237.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lugtenberg BJJ, Dekkers LC, Bloemberg GV. 2001. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu Rev Phytopathol 39:461–490. doi: 10.1146/annurev.phyto.39.1.461. [DOI] [PubMed] [Google Scholar]
- 25.Athukorala SNP, Fernando WGD, Rashid KY, de Kievit T. 2010. The role of volatile and non-volatile antibiotics produced by Pseudomonas chlororaphis strain PA23 in its root colonization and control of Sclerotinia sclerotiorum. Biocontrol Sci Technol 20:875–890. doi: 10.1080/09583157.2010.484484. [DOI] [Google Scholar]
- 26.Carroll H, Moenne-Loccoz Y, Dowlong DN, Ogara F. 1995. Mutational disruption of the biosynthesis genes-coding for the antifungal metabolite 2,4-diaceylphloroglucinol does not influence the ecological fitness of Pseudomonas fluorescens F113 in the rhizosphere of sugar beets. Appl Environ Microbiol 61:3002–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghirardi S, Dessaint F, Mazurier S, Corberand T, Raaijmakers JM, Meyer JM, Dessaux Y, Lemanceau P. 2012. Identification of traits shared by rhizosphere-competent strains of fluorescent pseudomonads. Microb Ecol 64:725–737. doi: 10.1007/s00248-012-0065-3. [DOI] [PubMed] [Google Scholar]
- 28.Mazzola M, Cook RJ, Thomashow LS, Weller DM, Pierson LS. 1992. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl Environ Microbiol 58:2616–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novinscak A, Gadkar VJ, Joly DL, Filion M. 2016. Complete genome sequence of Pseudomonas brassicacearum LBUM300, a disease-suppressive bacterium with antagonistic activity toward fungal, oomycete, and bacterial plant pathogens. Genome Announc 4(1):e01623-15. doi: 10.1128/genomeA.01623-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou HY, Wei HL, Liu XL, Wang Y, Zhang LQ, Tang WH. 2005. Improving biocontrol activity of Pseudomonas fluorescens through chromosomal integration of 2,4-diacetylphloroglucinol biosynthesis genes. Chin Sci Bull 50:775–781. [Google Scholar]
- 31.Zhou T-T, Li C-Y, Chen D, Wu K, Shen Q-R, Shen B. 2014. phlF− mutant of Pseudomonas fluorescens J2 improved 2,4-DAPG biosynthesis and biocontrol efficacy against tomato bacterial wilt. Biol Control 78:1–8. doi: 10.1016/j.biocontrol.2014.07.006. [DOI] [Google Scholar]
- 32.Fenton AM, Stephens PM, Crowley J, Ocallaghan M, Ogara F. 1992. Exploitation of gene(s) involved in 2,4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl Environ Microbiol 58:3873–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Natsch A, Keel C, Pfirter HA, Haas D, Défago G. 1994. Contribution of the global regulator gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 introduced into soil microcosms. Appl Environ Microbiol 60:2553–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raaijmakers JM, Weller DM. 1998. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol Plant Microbe Interact 11:144–152. doi: 10.1094/MPMI.1998.11.2.144. [DOI] [Google Scholar]
- 35.Notz R, Maurhofer M, Schnider-Keel U, Duffy B, Haas D, Défago G. 2001. Biotic factors affecting expression of the 2,4-diacetylphloroglucinol biosynthesis gene phlA in Pseudomonas fluorescens biocontrol strain CHA0 in the rhizosphere. Phytopathology 91:873–881. doi: 10.1094/PHYTO.2001.91.9.873. [DOI] [PubMed] [Google Scholar]
- 36.Rotenberg D, Joshi R, Benitez M-S, Chapin LG, Camp A, Zumpetta C, Osborne A, Dick WA, McSpadden Gardener BB. 2007. Farm management effects on rhizosphere colonization by native populations of 2,4-diacetylphloroglucinol-producing Pseudomonas spp. and their contributions to crop health. Phytopathology 97:756–766. doi: 10.1094/PHYTO-97-6-0756. [DOI] [PubMed] [Google Scholar]
- 37.Jamali F, Sharifi-Tehrani A, Lutz MP, Maurhofer M. 2009. Influence of host plant genotype, presence of a pathogen, and coinoculation with Pseudomonas fluorescens strains on the rhizosphere expression of hydrogen cyanide and 2,4-diacetylphloroglucinol biosynthetic genes in P. fluorescens biocontrol strain CHA0. Microb Ecol 57:267–275. doi: 10.1007/s00248-008-9471-y. [DOI] [PubMed] [Google Scholar]
- 38.McSpadden Gardener BB, Weller DM. 2001. Changes in populations of rhizosphere bacteria associated with take-all disease of wheat. Appl Environ Microbiol 67:4414–4425. doi: 10.1128/AEM.67.10.4414-4425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Notz R, Maurhofer M, Dubach H, Haas D, Défago G. 2002. Fusaric acid-producing strains of Fusarium oxysporum alter 2,4-diacetylphloroglucinol biosynthetic gene expression in Pseudomonas fluorescens CHA0 in vitro and in the rhizosphere of wheat. Appl Environ Microbiol 68:2229–2235. doi: 10.1128/AEM.68.5.2229-2235.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeCoste NJ, Gadkar VJ, Filion M. 2010. Verticillium dahlia alters Pseudomonas spp. populations and HCN gene expression in the rhizosphere of strawberry. Can J Microbiol 56:906–915. doi: 10.1139/W10-080. [DOI] [PubMed] [Google Scholar]
- 41.Combes-Meynet E, Pothier JF, Moënne-Loccoz Y, Prigent-Combaret C. 2011. The Pseudomonas secondary metabolite 2,4-diacetylphloroglucinol is a signal inducing rhizoplane expression of Azospirillum genes involved in plant-growth promotion. Mol Plant Microbe Interact 2:271–284. doi: 10.1094/MPMI-07-10-0148. [DOI] [PubMed] [Google Scholar]
- 42.Brown MRW, Kornberg A. 2008. The long and short of it—polyphosphate, PPK and bacterial survival. Trends Biochem Sci 33:284–290. doi: 10.1016/j.tibs.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Daniels R, Reynaert S, Hoekstra H, Verreth C, Janssens J, Braeken K, Fauvart M, Beullens S, Heusdens C, Lambrichts I, De Vos DE, Vanderleyden J, Vermant J, Michiels J. 2006. Quorum signal molecules as biosurfactants affecting swarming in Rhizobium etli. Proc Natl Acad Sci U S A 103:14965–14970. doi: 10.1073/pnas.0511037103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naseby DC, Lynch JM. 1998. Impact of wild-type and genetically modified Pseudomonas fluorescens on soil enzyme activities and microbial population structure in the rhizosphere of pea. Mol Ecol 7:617–625. doi: 10.1046/j.1365-294x.1998.00367.x. [DOI] [Google Scholar]
- 45.Naseby DC, Pascual JA, Lynch JM. 1999. Carbon fractions in the rhizosphere of pea inoculated with 2,4-diacetylphloroglucinol producing and non-producing Pseudomonas fluorescens F113. J Appl Microbiol 87:173–181. doi: 10.1046/j.1365-2672.1999.00809.x. [DOI] [Google Scholar]
- 46.Naseby DC, Lynch JM. 2001. Effect of 2,4-diacetylphloroglucinol producing, overproducing, and nonproduction Pseudomonas fluorescens F113 in the rhizosphere of pea. Microb Ecol 42:193–200. [DOI] [PubMed] [Google Scholar]
- 47.Naseby DC, Way JA, Bainton NJ, Lynch JM. 2001. Biocontrol of Pythium in the pea rhizosphere by antifungal metabolite producing and non-producing Pseudomonas strains. J Appl Microbiol 90:421–429. doi: 10.1046/j.1365-2672.2001.01260.x. [DOI] [PubMed] [Google Scholar]
- 48.Chapon A, Guillerm A-Y, Delalande L, Lebreton L, Sarniquet A. 2002. Dominant colonisation of wheat roots by Pseudomonas fluorescens Pf29A and selection of the indigenous microflora in the presence of the take-all fungus. Eur J Plant Pathol 108:449–459. doi: 10.1023/A:1016099707119. [DOI] [Google Scholar]
- 49.Arseneault T, Goyer C, Filion M. 2016. Biocontrol of potato common scab is associated with high Pseudomonas fluorescens LBUM223 populations and PCA biosynthetic transcripts accumulation. Phytopathology 106:963–970. doi: 10.1094/PHYTO-01-16-0019-R. [DOI] [PubMed] [Google Scholar]
- 50.Agriculture and Agri-Food Canada. 1998. The Canadian system of soil classification, 3rd ed. NRC Research Press, Ottawa, ON, Canada. [Google Scholar]
- 51.Paulin MM, Nicolaisen MH, Sørensen J. 2011. (R,S)-dichlorprop herbicide in agricultural soil induces proliferation and expression of multiple dioxygenase-encoding genes in the indigenous microbial community. Environ Microbiol 13:1513–1523. doi: 10.1111/j.1462-2920.2011.02456.x. [DOI] [PubMed] [Google Scholar]
- 52.DeCoste NJ, Gadkar VJ, Filion M. 2011. Relative and absolute quantitative real-time PCR-based quantifications of hcnC and phlD gene transcripts in natural soil spiked with Pseudomonas sp. strain LBUM300. Appl Environ Microbiol 77:41–47. doi: 10.1128/AEM.01387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]