ABSTRACT
Species in the extremely thermophilic genus Caldicellulosiruptor can degrade unpretreated plant biomass through the action of multimodular glycoside hydrolases. To date, most focus with these bacteria has been on hydrolysis of glucans and xylans, while the biodegradation mechanism for arabinose-based polysaccharides remains unclear. Here, putative α-l-arabinofuranosidases (AbFs) were identified in Caldicellulosiruptor species by homology to less-thermophilic versions of these enzymes. From this screen, an extracellular XynF was determined to be a key factor in hydrolyzing α-1,2-, α-1,3-, and α-1,5-l-arabinofuranosyl residues of arabinose-based polysaccharides. Combined with a GH11 xylanase (XynA), XynF increased arabinoxylan hydrolysis more than 6-fold compared to the level seen with XynA alone, likely the result of XynF removing arabinofuranosyl side chains to generate linear xylans that were readily degraded. A second AbF, the intracellular AbF51, preferentially cleaved the α-1,5-l-arabinofuranosyl glycoside bonds within sugar beet arabinan. β-Xylosidases, such as GH39 Xyl39B, facilitated the hydrolysis of arabinofuranosyl residues at the nonreducing terminus of the arabinose-branched xylo-oligosaccharides by AbF51. These results demonstrate the separate but complementary contributions of extracellular XynF and cytosolic AbF51 in processing the bioconversion of arabinose-containing oligosaccharides to fermentable monosaccharides.
IMPORTANCE Degradation of hemicellulose, due to its complex chemical structure, presents a major challenge during bioconversion of lignocellulosic biomass to biobased fuels and chemicals. Degradation of arabinose-containing polysaccharides, in particular, can be a key bottleneck in this process. Among Caldicellulosiruptor species, the multimodular arabinofuranosidase XynF is present in only selected members of this genus. This enzyme exhibited high hydrolysis activity, broad specificity, and strong synergism with other hemicellulases acting on arabino-polysaccharides. An intracellular arabinofuranosidase, AbF51, occurs in all Caldicellulosiruptor species and, in conjunction with xylosidases, processes the bioconversion of arabinose-branched oligosaccharides to fermentable monosaccharides. Taken together, the data suggest that plant biomass degradation in Caldicellulosiruptor species involves extracellular XynF that acts synergistically with other hemicellulases to digest arabino-polysaccharides that are subsequently transported and degraded further by intracellular AbF51 to produce short-chain arabino sugars.
KEYWORDS: Arabinofuranosidase, bioenergy, glycoside hydrolase, hyperthermophiles, synergism
INTRODUCTION
The processing of agricultural cereals generates large amounts of by-products, such as straw, stover, husks, and bagasse, which can be important sources of material for biorefineries and animal feeds. Arabinose-based polysaccharides constitute a major component of hemicellulose and occur as arabinoxylan (AX), arabinan, and arabinogalactan (1). For example, AXs constitute 64% to 69% of the nonstarch polysaccharides (NSPs) present in wheat bran and 88% of the NSPs identified in wheat endosperm (2). Heteropolysaccharides from deciduous hardwoods consist of a backbone of β-1,4-linked d-xylopyranosyl sugar units with random α-1,2- or α-1,3-l-arabinofuranosyl residues (i.e., in which it is random whether each is the α-1,2 form or the α-1,3 form) (Fig. 1a) (3). Side chain modifications containing arabinose or other monosaccharides restrict enzymatic hydrolysis of these heteropolymers (3, 4) such that endo-xylanases (EC 3.2.1.8), β-xylosidases (EC 3.2.1.37), and α-l-arabinofuranosidases (AbFs) (EC 3.2.1.55) are needed to degrade arabinoxylans (3). AbF catalyzes the hydrolysis of nonreducing α-1,2-, α-1,3-, and α-1,5-l-arabinofuranosyl residues from arabinose-based polysaccharides, such as AX and arabinan (3, 5). l-Arabinose is commercially used as a low-calorie sweetener whose use enables avoidance of the elevation of blood glucose levels otherwise induced by sucrose intake (6–8).
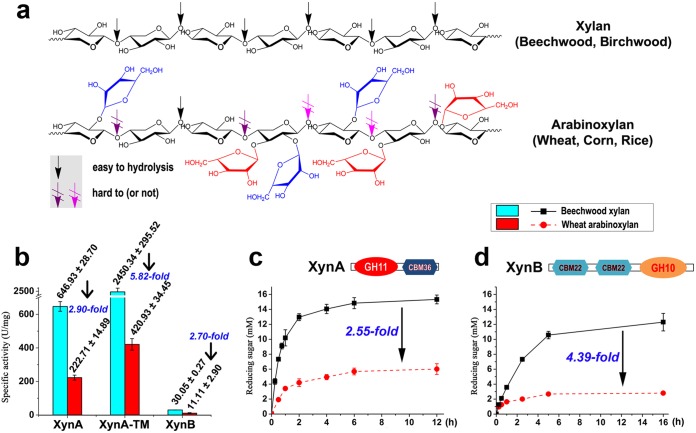
FIG 1.
Activities of endo-xylanases against beechwood xylan and wheat arabinoxylan. (a) Chemical structures of beechwood xylan and wheat arabinoxylan. (b) Specific activities of XynA, XynA-TM, and XynB on beechwood xylan and wheat arabinoxylan. (c) Time course of hydrolysis of beechwood xylan and wheat arabinoxylan by an endoxylanase XynA. (d) Time course of hydrolysis of beechwood xylan and wheat arabinoxylan by XynB. All the reactions were performed at pH 6 and 70°C in citrate buffer.
Caldicellulosiruptor species are bacteria that are extremely thermophilic (range, approximately 70 to 85°C) and produce a wide range of glycosyl hydrolases to attack plant cell walls (9–12). These thermostable enzymes can have significant advantages in biotechnological processes due to higher reaction velocities, reduced risk of contamination, and enhanced substrate solubility (13). Therefore, thermostable hemicellulases, including AbFs, from Caldicellulosiruptor species are of considerable interest for industrial applications.
In the Carbohydrate-Active enZymes (CAZy) database (14), several AbFs, belonging to glycoside hydrolase (GH) families 3, 43, 51, 54, and 62, have been characterized biochemically (15–18). Though most GH3 and GH43 AbFs exhibit broad substrate specificities with respect to synthetic p-nitrophenyl (pNP)-linked or methylumbelliferyl (MUF)-linked monosaccharides, these hemicellulases can cleave arabinose only from natural substrates (3, 19, 20). Most GH51 AbFs release arabinose from AXs and arabino-containing saccharides, but their activity on the branched arabinan is much higher than on the debranched arabinan. This suggests that GH51 AbFs prefer the α-1,2 and α-1,3 linkages between arabinose substituents and backbone residues (3, 16). Finally, AbFs from the less common GH54 and GH62 families release arabinose from the branches but exhibit little or no activity on debranched arabinan (15, 18, 21).
Here, nine putative AbFs from the genus Caldicellulosiruptor were identified and two, an extracellular multimodular GH43-containing enzyme (XynF) and an intracellular GH51 hydrolase (AbF51), were characterized biochemically. AbF51 exhibited strong hydrolytic activity on debranched arabinan, while XynF exhibited broad substrate specificity and worked synergistically with other extracellular xylanases. These two enzymes likely play an essential role in converting arabino-based polysaccharides into ultimately intracellular fermentable sugars that can be used by Caldicellulosiruptor species as carbon and energy sources.
RESULTS
Genus-wide screening of the putative AbFs in Caldicellulosiruptor.
Previously, two thermostable xylanases (XynA and XynB) from Caldicellulosiruptor sp. strain F32 were shown to degrade linear beechwood xylans (BWX) (22) (Fig. 1a). XynA, with a GH-11 catalytic domain and a carbohydrate-binding module (CBM), namely, CBM36, was 3-fold less active on arabinose-branched hemicellulose, such as wheat arabinoxylan (WAX) (Fig. 1a), than on BWX (Fig. 1b). Also, XynA-TM (a truncated mutant of XynA without CBM36) and XynB (containing a GH10 domain and two CBM22 domains) had 5.82- and 2.70-fold-lower activities, respectively (Fig. 1b), yielding lower concentrations of reducing saccharides from WAX digestion than from BWX digestion (Fig. 1c and d; see also Fig. S1 in the supplemental material). To improve the hydrolysis of WAX, identification of an accessory debranching enzyme, arabinofuranosidase (AbF), was sought in Caldicellulosiruptor genomes (3).
According to the CAZy database, AbFs belong to GH families 3, 43, 51, 54, and 62 (3). In all 12 sequenced genomes of Caldicellulosiruptor strains, putative AbFs from GH3, GH43, and GH51, but not GH54 or GH62, could be identified (Table 1 and Fig. 2a) (12). Recombinant forms of two GH3 enzymes, five GH43 enzymes, and one GH51 (i.e., AbF51) enzyme from Caldicellulosiruptor sp. strain F32 were produced in Escherichia coli (Fig. 2b). Initial attempts to produce a multimodular GH43-containing enzyme (XynF) from Caldicellulosiruptor saccharolyticus in E. coli strains were unsuccessful (data not shown) (10). However, hydrolytic activity on WAX and pNP-α-l-arabinofuranoside (pNP-AraF) was detected in supernatant fractions of E. coli expressing this gene (data not shown), confirming functional expression of XynF.
TABLE 1.
Putative α-l-arabinofuranosidases in Caldicellulosiruptor speciesa
The protein loci and identities of different AbFs are present in this table.
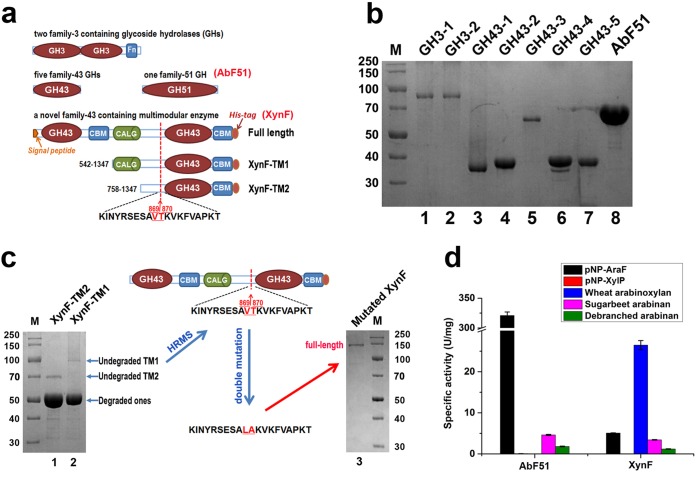
FIG 2.
Schematic structures and genus-wide screening of the putative AbFs in Caldicellulosiruptor species. (a) Schematic diagram of two GH3s, five GH43s, one AbF51, a multimodular XynF enzyme, and two truncated mutants of XynF. (b) SDS-PAGE analysis of two GH3s, five GH43s, and one AbF51 from Caldicellulosiruptor sp. strain F32. (c) Heterologous recombination of a full-length XynF in E. coli by high-resolution mass spectroscopy-guided mutagenesis. (d) Specific activities of AbF51 and XynF on the different substrates.
Two truncated mutants of XynF (XynF-TM1 and XynF-TM2) were also produced (Fig. 2c). XynF-TM1 consisted of a concanavalin A-like lectin/glucanase (CALG) domain and a catalytic module in the carboxyl terminus of XynF. XynF-TM2 contained only the catalytic module (Fig. 2a). After heterologous expression, two prominent bands with similar molecular masses (∼50 kDa) were observed on SDS-PAGE (Fig. 2c). Two faint bands, corresponding to the expected Mr (the theoretical Mr values for XynF-TM1 and XynF-TM2 are 92.38 and 66.84, respectively), were also observed (Fig. 2c). High-resolution mass spectrometry (HRMS) analysis of the two mutant samples (Fig. S2) indicated proteolytic cleavage in E. coli (for XynF-TM1, 54,039.0669; for XynF-TM2, 54,038.6772) between Val869 and Tyr870 with an expected Mr (monoisotopic) of 54,038.30. Following a double mutation from Val869/Tyr870 to Leu/Ala, full-length XynF could be expressed and purified in E. coli C43(DE3) (Fig. 2c). These two mutated residues are located in a long segment linking the catalytic domains and are not likely to be important for XynF function. As such, the full-length XynF mutant was then used in subsequent experiments.
Biochemical characterization of the putative AbFs from Caldicellulosiruptor.
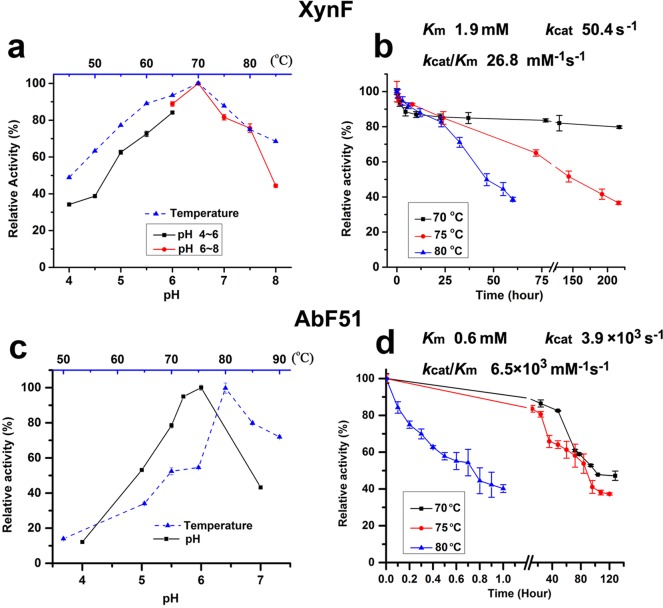
After purification of all putative AbFs, their hydrolysis profiles were determined on relevant substrates (Fig. 2d; see also Table S1 in the supplemental material). Two GH3 and five GH43 hydrolases exhibited very low or undetectable hydrolytic activities on pNP-AraF and on arabinose-containing polysaccharides (Table S1), suggesting that these enzymes were not AbFs. However, AbF51, as well as XynF, had arabinofuranosidase activities on pNP-AraF, sugar beet arabinan (SA), and debranched sugar beet arabinan (DSA) (Fig. 2d). While AbF51 was highly active on pNP-AraF, it exhibited no activity on WAX (Fig. 2d). XynF and AbF51 were most active at pH 6.5 and 6.0, respectively, and at 70°C and 80°C, respectively (Fig. 3a and c). XynF retained 80% activity for more than 250 h at 70°C and had half-lives of 150 h and 50 h at 75°C and 80°C, respectively (Fig. 3b). Similarly, the half-lives of AbF51 at 70°C and 75°C were 100 h and 90 h, respectively (Fig. 3d). For XynF, the Michaelis-Menten constant (Km), turnover number (kcat), and catalytic efficiencies (kcat/Km) of the enzyme were 1.9 mM, 50.4 s−1, and 26.8 s−1 · mM−1, respectively. For AbF51, these parameters were 0.6 mM, 3.9 × 103 s−1, and 6.5 × 103 s−1 · mM−1, respectively (Fig. 3). Neither metal ions nor chemical reagents completely inhibited XynF activity, but Cu2+ inhibited AbF51 activity nearly completely at concentrations of 1 mM and 5 mM (Table 2). Cu2+ and Fe2+ significantly impacted the activities of two AbFs. AbF51 was inactive in the presence of 5 mM SDS, while XynF retained 41% of its initial activity. XynF and ABF51 retained more than 90% of their activity in the presence of 1 mM EDTA. XynF maintained approximately 90% activity in the presence of 5 mM EDTA, whereas AbF51 retained only 24% activity (Table 2). This suggests that XynF is not a metalloprotein.
FIG 3.
Biochemical characterizations of XynF and AbF51. (a) Effect of pH and temperature on the activities of XynF. For optimum pH conditions, citrate buffer was used at pH 4 to 6 (black line) and phosphate buffer at pH 6 to 8 (red line). (b) Thermostability of XynF at different temperatures. (c) Effect of pH and temperature on the activities of AbF51. (d) Thermostability of AbF51 at different temperatures. Wheat arabinoxylan and pNP-AraF were used as substrates for XynF and AbF51, respectively. The kinetic parameters of XynF and AbF51 were examined under optimum pH and temperature conditions.
TABLE 2.
Effects of metal ions and other chemicals on two AbFs
| Metal | Relative activity (%) at indicated metal concn |
|||
|---|---|---|---|---|
| XynF on WAX |
AbF51 on pNP-AraF |
|||
| 1 mM | 5 mM | 1 mM | 5 mM | |
| Control | 100 | 100 | 100 | 100 |
| Ni2+ | 111 ± 2.4 | 95 ± 0.8 | 82 ± 5.4 | 85 ± 4.8 |
| Mn2+ | 105 ± 1.5 | 110 ± 1.7 | 98 ± 2.4 | 117 ± 4.1 |
| Ca2+ | 99 ± 1.9 | 114 ± 2.1 | 96 ± 1.7 | 106 ± 2.6 |
| Cu2+ | 28 ± 1.8 | 18 ± 0.3 | 0.8 ± 0.2 | 2.5 ± 0.3 |
| Mg2+ | 116 ± 1.2 | 104 ± 2.8 | 97 ± 1.1 | 76 ± 2.7 |
| Co2+ | 125 ± 1.5 | 113 ± 1.7 | 78 ± 5.1 | 39 ± 1.6 |
| Fe2+ | 53 ± 1.9 | 30 ± 1.6 | 16 ± 1.0 | 37 ± 1.0 |
| K+ | 96 ± 2.6 | 91 ± 1.8 | 14 ± 0.8 | 17 ± 2.7 |
| SDS | 89 ± 0.8 | 41 ± 1.3 | 23 ± 0.3 | 0.2 ± 0.2 |
| EDTA | 93 ± 0.9 | 91 ± 1.1 | 95 ± 2.1 | 24 ± 1.9 |
Synergistic effects of two AbFs with different xylanases on WAX.
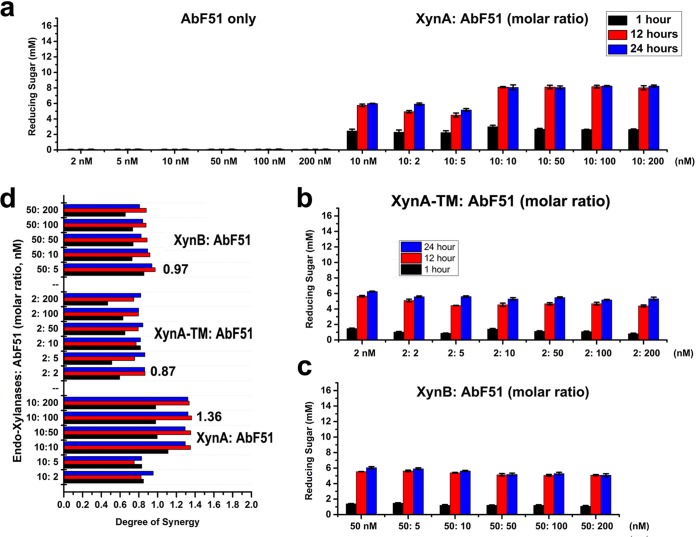
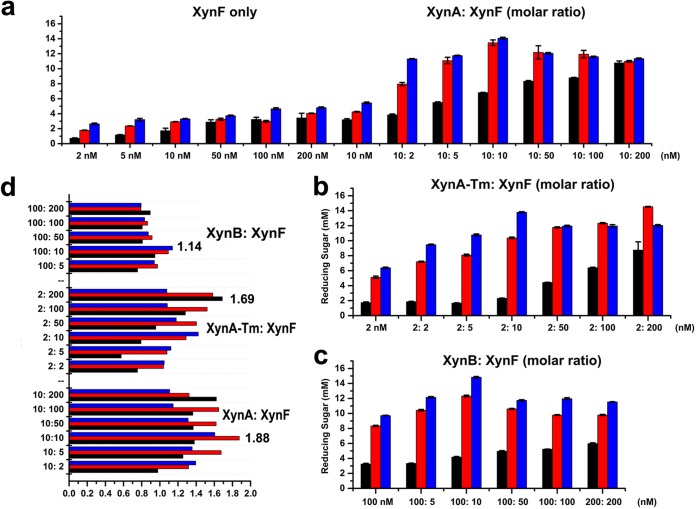
To investigate the synergistic effects of the AbFs, analysis of the reducing saccharides that were released from insoluble WAX by combinations of three different endo-xylanases was performed by the 3,5-dinitrosalicylic acid (DNS) method (22). Different molar ratios were used to observe the synergistic effects of two AbFs with endo-xylanases (Fig. 4 and 5). AbF51 alone did not generate detectable reducing sugars after incubation with WAX (Fig. 4a). When AbF51 was combined with xylanases, the combination of XynA and AbF51 was most effective (Fig. 4a, b, and c). The highest degree of synergy (DOS) (23) (corresponding to a score of 1.36) was obtained with XynA and AbF51 in a molar ratio of 1:10. However, the DOS values determined for AbF51/XynA-TM and AbF51/XynB were lower than 1.0 (Fig. 4d). These results suggested that the XynA xylanase, but not XynA-TM or XynB, was synergistic with AbF51. XynF not only liberated the reducing sugars from WAX (unlike AbF51) but also generated much higher concentrations of reducing sugars than AbF51 when combined with XynA or XynB (Fig. 4 and 5). A 1:1 ratio released the largest amount of reducing sugar for XynA/XynF (Fig. 5a), whereas 1:100 and 1:10 were best for XynA-TM/XynF and XynB/XynF, respectively (Fig. 5b and c). The highest synergy was for XynA/XynF (Fig. 5d).
FIG 4.
Synergistic effects of AbF51 with three endo-xylanases. A series of enzyme combinations (molar ratio) were used. All the reactions were performed at 70°C against wheat arabinoxylan (0.8%) for three time durations (1 h, 12 h, and 24 h). Reducing sugars were measured by the DNS method. (a) Reducing sugars obtained by AbF51 singly or in combination with XynA. (b) Sugar released by the combined activities of AbF51 and XynA-TM. (c) Sugar released by the combined activities of AbF51 and XynB. (d) Degree of synergy obtained by coupling the action of AbF51 with that of the endoxylanases (separately) for three time durations. Degree of synergy is defined as the ratio of the amounts of saccharides released from the simultaneous activities to the sum of that released by the individual enzyme.
FIG 5.
Synergistic effects of XynF with three endo-xylanases (XynA, XynA-TM, and XynB). A series of enzyme combinations (molar ratio) were used. All the reactions were performed as described for Fig. 4. (a) Reducing sugars obtained by XynF singly or in combination with XynA. (b) Sugar released by the combined activities of XynF and XynA-TM. (c) Sugar released by the combined activities of XynF and XynB. (d) Degree of synergy obtained by a coupled action of XynF with the endoxylanases (separately) for three time durations. Degree of synergy is defined as the ratio of the amounts of saccharides released from the simultaneous activities to the sum of that released by the individual enzyme.
Distinct modes of action of AbF51 and XynF on arabino-containing polysaccharides.
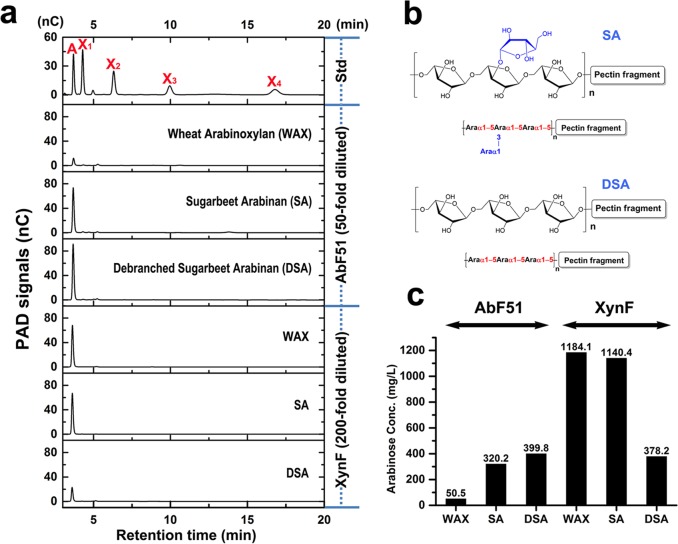
Enzymatic hydrolysis on arabino-based polysaccharides was examined for three natural substrates (WAX, SA, and DSA). WAX, with random α-1,2- and α-1,3-l-arabinofuranosyl residues, was minimally affected by AbF51 (Fig. 6a). However, hydrolysis of the arabinans SA and DSA, both with α-1,5-linked arabinose in the backbone (Fig. 6b), was observed (Fig. 6a). For XynF, WAX and SA, containing the side chains of α-1,2- and/or α-1,3-l-arabinofuranosyl residues, released more arabinose than the linear DSA (Fig. 6a), and XynF had higher arabinofuranosidase activity than AbF51 (Fig. 6c). These results suggest that AbF51 hydrolyzes the α-1,5-l-arabinofuranosyl-linked glycosidic bond whereas XynF was capable of cleaving α-1,2, α-1,3, and α-1,5 bonds of arabino-based polysaccharides.
FIG 6.
Hydrolysis actions of two AbFs on arabinose-based polysaccharides. (a) The natural substrates WAX, SA, and DSA were incubated with AbF51 or XynF for 12 h at 70°C. Hydrolysis products were analyzed by HPAEC-PAD. A, X1, X2, X3, and X4 refer to arabinose, xylose, xylobiose, xylotriose, and xylotetrose, respectively. Std, standard. (b) The chemical structures of SA and DSA. (c) The amounts of arabinose released by AbF51 and XynF on three substrates according to the signal areas of arabinose shown in panel a. AbF51 participant hydrolysis products were diluted 50-fold, and XynF participant products were diluted 200-fold with distilled water. Conc., concentration.
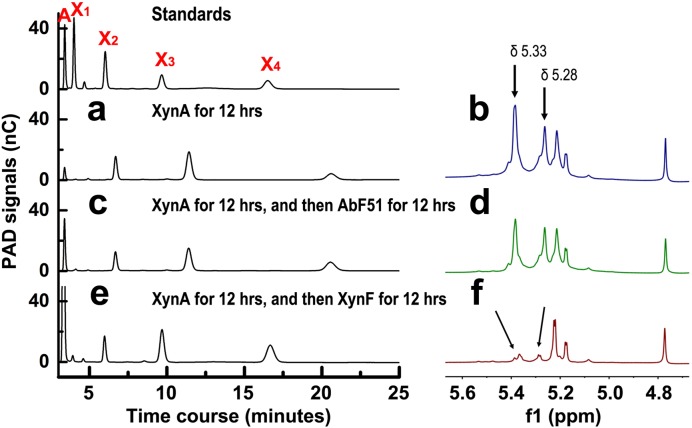
Caldicellulosiruptor XynA is a typical GH11 xylanase, releasing short xylooligosaccharides (XOSs) such as xylobiose, xylotriose, and xylotetraose (22). DNS analysis indicated that XynA was synergistic with AbF51 and XynF (Fig. 4 and 5). After 12 h of digestion by XynA alone, WAX released oligosaccharides that did not match the XOS standards (Fig. 7a). As such, it is possible that the XOSs had arabinose branches. Previous studies provided evidence that the α-1,2- and α-1,3-l-arabinofuranosyl residues in the arabino-branched XOSs could be identified by 1H nuclear magnetic resonance (1H-NMR) spectroscopy (24–27). 1H-NMR analyses also suggested the presence of 2-O-linked (δ 5.28) and 3-O-linked (δ 5.33) arabinoses in the XynA-hydrolyzed WAX products (Fig. 7b and S3a). Following digestion by AbF51, the arabinose signal in high-performance anion-exchange chromatography (HPAEC) was enhanced, whereas those of the others from the arabino-branched XOSs decreased slightly (Fig. 7c). These anomeric signals determined by 1H-NMR (Fig. 7d and S3b) were similar to those shown in Fig. 7b. In case of XynF digestion, the arabinose HPAEC signal was dramatically increased, and the retention times of other peaks were shifted left relative to those of the standard XOSs (Fig. 7e). Also, the specific 1H-NMR signals related to the α-1,2- and α-1,3-bonds disappeared (Fig. 7f and S3c). These results obtained using HPAEC with pulsed amperometric detection (HPAEC-PAD) and 1H-NMR spectroscopy clearly showed that XynF, but not AbF51, was strongly hydrolytic on arabino-branched XOSs.
FIG 7.
Hydrolysis actions of two AbFs on arabinose-branched XOSs. (a and b) WAX (1%) was hydrolyzed by XynA for 12 h at 70°C. The products were examined by HPAEC-PAD (a) and 1H-NMR (b). (c and d) WAX (1%) was hydrolyzed by XynA for 12 h followed by AbF51 for 12 h at 70°C. The products were examined by HPAEC-PAD (c) and 1H-NMR (d). (e and f) WAX (1%) was hydrolyzed by XynA for 12 h followed by XynF for 12 h at 70°C on HPAEC-PAD (e) and 1H-NMR (f). A, X1, X2, X3, and X4 refer to arabinose, xylose, xylobiose, xylotriose, and xylotetrose, respectively. The signals of 2-O-linked (δ 5.28) and 3-O-linked (δ 5.33) arabinoses were assigned.
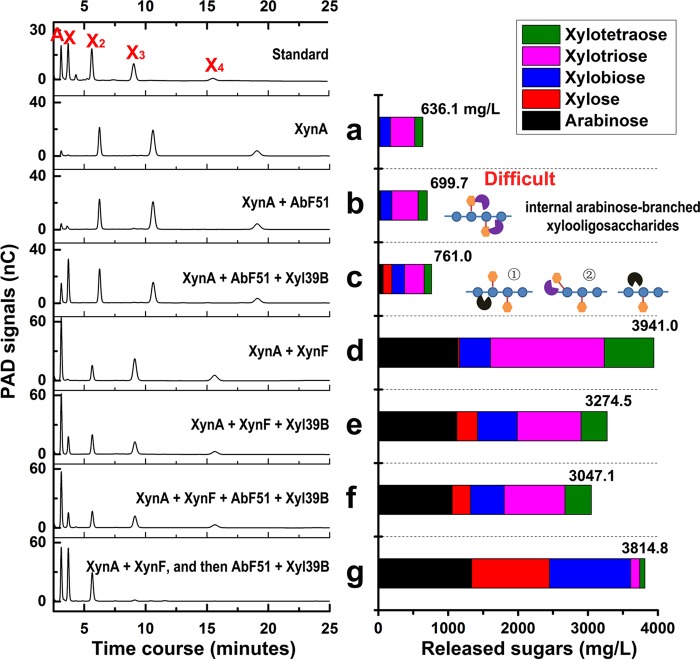
Finally, the effects of combinations of the AbFs with other hemicellulases were examined. After 24 h of incubation of XynA with 1% WAX, the concentration of arabino-branched XOSs reached 636.1 mg/liter (Fig. 8a). Under conditions of incubation with 10 nM AbF51, saccharide levels reached 699.7 mg/liter (Fig. 8b). When a GH39 β-xylosidase from Caldicellulosiruptor sp. strain F32 (Xyl39B) was also added, the arabinose and xylose signals increased, although the main products were still arabino-branched XOSs (Fig. 8c). These results suggest that AbF51 had minimal impact on WAX hydrolysis and that Xyl39B facilitated AbF51 degradation of arabino-branched XOSs. Importantly, the combination of XynF and XynA dramatically increased the level of soluble sugars to 3.9 g/liter (Fig. 8d), a 6-fold increase compared to the level seen with XynA alone. However, addition of Xyl39B or AbF51/Xyl39B to the XynF/XynA mixture decreased the level to 3.3 or 3.0 g/liter, respectively, compared with the XynA/XynF results (Fig. 7d, e, and f). The possibility exists that enzyme competition reduced the hydrolytic efficiency of the hemicellulase mixture (28). In a sequential hydrolysis reaction using XynA/XynF followed by AbF51/Xyl39B for another 12 h, the concentration of released oligosaccharides reached 3.8 g/liter (Fig. 8g), which was close to our previous result (Fig. 8d). The HPAEC signals for xylotriose and xylotetraose disappeared (Fig. 8g).
FIG 8.
Combinational actions of AbF with other hemicellulases on hydrolyzing WAX. WAX (1%) was hydrolyzed by different enzyme combinations, including XynA alone (a), XynA/AbF51 (b), XynA/AbF51/Xyl39B (c), XynA/XynF (d), XynA/XynF/Xyl39B (e), and XynA/XynF/AbF51/Xyl39B (f), for 24 h and then by XynA/XynF for 24 h followed by AbF51/Xyl39B for another 24 h (g). The products were examined by HPAEC-PAD (left panel). The amounts of released sugars were calculated (right panel) according to standard curves of xylo-oligosaccharides. AbF51 participant hydrolysis products were diluted 50-fold, and XynF participant products were diluted 200-fold with distilled water.
DISCUSSION
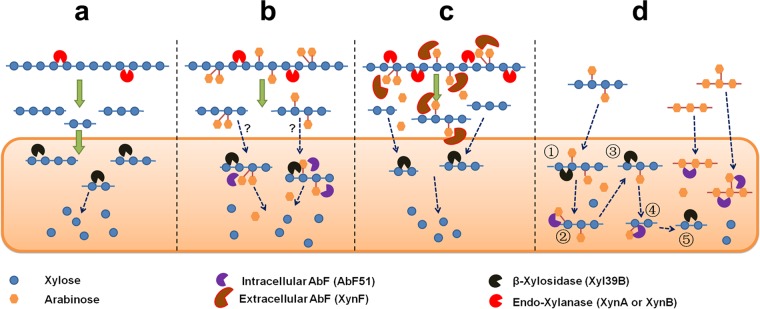
The members of the Caldicellulosiruptor genus constitute a group of extremely thermophilic and strictly anaerobic bacteria that can significantly degrade a wide range of plant biomasses without physical or chemical pretreatment (29). Understanding the novel lignocellulolytic enzymes within this genus is important for achieving higher levels of fermentable saccharides (13). The results seen with a unique multimodular glycoside hydrolase, CelA, which functions as a bifunctional exo- and endo-glucanase, are comparable to or better than those seen with current commercial cellulase formulations (30, 31). However, branched hemicelluloses, such as arabino-xylan, restrict the complete enzymatic hydrolysis of lignocellulosic biomasses. In this study, genus-wide screening of putative AbFs was performed in Caldicellulosiruptor species (Fig. 2a and Table 1), two of which synergistically degraded arabinose-containing polysaccharides (Fig. 9).
FIG 9.
A model in Caldicellulosiruptor species for two AbFs involving arabinose-based polysaccharide degradation. (a) A linear route of xylan degradation by xylanases and xylosidases. (b) An interrogative route of arabinoxylan degradation by intracellular AbF51. (c) A route of arabinoxylan degradation by extracellular XynF in Caldicellulosiruptor species. (d) Intracellular route of degradation involving AbF51 on arabinose-containing oligosaccharides.
GH51 arabinofuranosidases contain the largest number of characterized AbFs, most of which are specifically active on natural substrates (3). Putative AbF51 hydrolases can be identified throughout the members of the Caldicellulosiruptor genus (Table 1). A homolog (Csac_1561; 97% protein identity with AbF51) from C. saccharolyticus was shown to have AbF characteristics and exists as a homo-octamer (32). Moreover, the transcription of AbF51-like enzymes increased significantly when several Caldicellulosiruptor strains were grown on switchgrass or other plant biomass (9, 33, 34), indicating an important role of AbF51 on lignocellulosic degradation.
Most of the GH51 AbFs from plants and fungi are extracellular, whereas most of bacterial versions are cytosolic (3). The AbF51 reported here and its homologs from across the genus Caldicellulosiruptor have no identifiable signal peptide and are presumed to be intracellular enzymes. Many bacterial AbFs show specificity toward arabinan and arabino-oligosaccharides but have minimal activity on AX and arabinose-branched XOSs (3). That observation was confirmed here (Fig. 2d, 6, and 7). Meanwhile, combinations of AbF51 with a GH11 xylanase (XynA) did not significantly release monosaccharides (arabinose and xylose) from the arabinose-branched XOSs (Fig. 8b). However, addition of an intracellular GH39 β-xylosidase (Xyl39B) increased arabinose and xylose release (Fig. 8c). The GH11 xylanase mainly released the internal arabinose-branched XOSs (Fig. 8b, right panel) but not the terminal residues (25, 35). The results indicated that the level of recognition of these internal arabinofuranosyl residues by AbF51 was low (Fig. 8b, right panel). However, β-xylosidase Xyl39B might hydrolyze the nonreducing terminus of these saccharides, exposing the arabinofuranosyl residues at the ends (Fig. 8c, right panel, step 1). Then, AbF51 would be able recognize and digest these exposed arabinofuranosyl residues more readily (step 2). This conjecture points to the possibility that the arabinose-branched XOSs are hydrolyzed by AbF51 and xylosidases in the cytoplasm of Caldicellulosiruptor species.
Biodegradation of linear xylans, such as BWX, to xylose is relatively straightforward. Xylanases randomly cleave the β-1,4 linkages between the xylopyranosyl residues in the xylan backbone, and the released XOSs are transported into cytoplasm and hydrolyzed into xyloses (Fig. 9a). However, the efficiency of transportation of arabinose-branched XOSs into the cytoplasm should be lower than that of the linear XOSs, since branching complicates transport (Fig. 9b). When these branched oligomers are transported into the cytoplasm, their hydrolysis is catalyzed by cytoplasmic AbF51 and xylosidases.
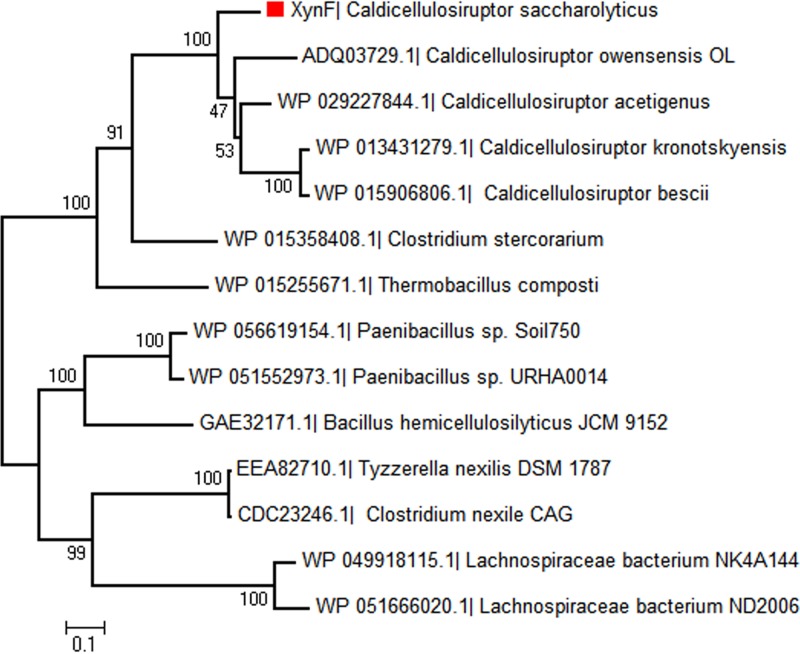
Multimodular XynF-like hydrolases can be identified in genomes of members of the thermophilic genera Caldicellulosiruptor, Thermobacillus, Gracilibacillus, and Paenibacillus and of the mesophilic genera Clostridium, Bacillus, and Lachnospiraceae (family) (Fig. 10). Most of these bacteria are anaerobic and inhabit a variety of environments, revealing the universality of XynF-driven biodegradation of arabino-polysaccharides (36, 37). Only five XynF-like homologs can be identified among 12 genome-sequenced Caldicellulosiruptor species (Table 1). Bergquist et al. (38) suggested that XynF (Csac_2411) was a result of gene fusion. Unsuccessful recombination of full-length XynF in E. coli and the strict α-l-arabinofuranosidase activities of two truncated catalytic segments (10) provided a glimpse into the features of this enzyme. Upregulation of XynF-like homologs at the mRNA and protein levels by Caldicellulosiruptor species growing on unpretreated plant biomass (9, 10, 33, 34) strongly suggests the important role of this enzyme in degrading the arabinose-containing polysaccharides.
FIG 10.
Phylogenetic tree of XynF-like enzymes in bacteria. An alignment of NCBI sequences was performed using clustalx. A maximum likelihood nearest-neighbor interchange tree was then constructed in MEGA 5.1 and tested with 500 bootstrap replicates. Bootstrap values are listed near each branch. RefSeq numbers are listed in parentheses.
According to results presented here, full-length XynF exhibited high arabinofuranosidase activity on complex substrates, such as WAX, SA, and DSA (Fig. 2d, 6, and 8), and worked synergistically with different xylanases in WAX hydrolysis (Fig. 5 and 8). The arabino-branched polysaccharides and XOSs could be completely debranched by XynF (Fig. 6 and 7). Extracellular XynF synergistically acts with xylanases or other hemicellulases on arabino-branched polysaccharides. The debranched XOSs are then released and transported into the cytoplasm and are finally decomposed to xylose by intracellular xylosidases (Fig. 9c). In addition, AbF51, sometimes with xylosidases, acts on arabino-containing oligosaccharides transported into cytoplasm (Fig. 9d).
MATERIALS AND METHODS
Protein production.
The target genes with blunt ends were amplified from the genomic DNA of Caldicellulosiruptor sp. strain F32 and C. saccharolyticus using Kapa HiFi DNA polymerase (Kapa, USA) and cloned into pEASY blunt-E2 vector (TransGen, China), as previously described (22). The primers for cloning the genes are listed in Table 3. E. coli C43 (DE3) cells (Lucigen, USA) harboring a recombinant plasmid containing one of the putative AbF genes were cultured in LB medium at 37°C at 200 rpm until the absorbance at 600 nm reached 0.5. Gene expression in E. coli was then induced at 16°C with the addition of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) overnight. Cell pellets were harvested by centrifugation at 10,000 × g for 20 min at 4°C and were frozen at −80°C.
TABLE 3.
Primers used in this study
| Clone | Primer orientation | Primer sequence |
|---|---|---|
| GH3-1 | Forward | ATGTCAGATAAAATAAAAGAGTTGATATC |
| Reverse | CTGCTTATTTAACTCCCTGAGCC | |
| GH3-2 | Forward | ATGTCAATTGAAAAGAAAGTGAATGATC |
| Reverse | TTCGCACCAAACTTTGCTAAAGTAG | |
| GH43-1 | Forward | ATGAAGAAGGTCTTTGCAAAAAAGG |
| Reverse | TTTTACCTGCAAATCCCCTTC | |
| GH43-2 | Forward | ATGTTCAAACTCCAAAGGGTG |
| Reverse | ATCGAATTTTATCTTTTCTTTATTGAAC | |
| GH43-3 | Forward | ATGATTTCAACTTTATGGGACCCAAA |
| Reverse | ACACTCATCTTCCTTTCTAAAGTTTTC | |
| GH43-4 | Forward | ATGATAAATGTGCCAAAGCCAC |
| Reverse | AAACAAATTAGGAAGACAAAAGTC | |
| GH43-5 | Forward | ATGGATATTAAAATTATAGGGCAATC |
| Reverse | TTCCTGACTTTCTCTCATGAG | |
| GH51 | Forward | ATGAAAAAAGCAAAAGTCATCTAGG |
| Reverse | TTCTCTTTTCTTTAATCTAATTACATTCC | |
| XynF | Forward | TATAGTTCATCTAACCAACAGGGAG |
| Reverse | CTTTACAAATTTCCAGCAATCCATATC | |
| XynF-protease inhibition | Forward | CAACTATAGGAGTGAAAGTGGACTGACAAAGGTAAAATTTGTAGC |
| Reverse | GCTACAAATTTTACCTTTGTCAGTCCACTTTCACTCCTATAGTTG |
For protein purification, the resuspended cells were disrupted by ultrasonication on ice, and cell debris was removed by centrifugation (13,000 × g for 30 min at 4°C). Since the proteins from Caldicellulosiruptor species were expected to be thermostable, the extractions were heated at 65°C for 30 min in a water bath, cooled on ice, and again centrifuged at 4°C (12,000 × g for 30 min). Each protein with a His6 tag was purified using nickel-nitrilotriacetic acid (Ni-NTA)-sefinose resin (Sangon, China), according to the manufacturer's instructions. Briefly, the resin was incubated with supernatant for 30 min and then washed with 100 ml binding buffer (50 mM Tris-HCl, 300 mM NaCl, 30 mM imidazole, pH 8.0), and the protein was eluted with binding buffer supplemented with imidazole at 250 mM. The eluted proteins were concentrated using Amicon Ultra filters (Millipore, Ireland). Protein concentrations were measured using the Bradford method with bovine serum albumin as a standard.
Substrate specificity.
The substrate specificities of AbF51 and XynF were screened with different substrates, including synthetic substrates pNP-AraF, pNP-XylP, and pNP-GluP (Sigma-Aldrich, USA) and natural substrates BWX, WAX, SA, and DSA (Megazyme, Ireland). For synthetic substrates, the reactions were performed as follows: 100 μl of enzyme at an appropriate concentration and 100 μl of 2 mM substrate in phosphate buffer were mixed and incubated at 70°C for 10 min. A 1-ml volume of 0.2 mM Na2CO3 was added to quench the reaction. The absorbance of released p-nitrophenol was measured at 415 nm. For natural polysaccharides, a 3,5-dinitrosalicylic acid (DNS) assay was performed as follows. A 25-μl volume of enzyme at an appropriate concentration was incubated with 50 μl of 1% substrate at 70°C for 1 h. The mixture was combined with 100 μl DNS, and the resultant mixture was then boiled for 5 min. Distilled water (325 μl) was added, and the absorbance of released arabinose was measured at 540 nm. One unit of enzyme activity was defined as the amount of enzyme that released 1 μmol of p-nitrophenol (or of the reducing ends of sugar in the DNS assay) per min under the optimum conditions. All of the experiments were performed in triplicate.
Biochemical characterization.
The optimum pH levels of two AbFs were determined by incubation at 70°C for 10 min at different pHs ranging from 4 to 8 in sodium citrate buffer (approximate pH, 4.0 to 6.0) and phosphate buffer (approximate pH, 6.0 to 8.9). For AbF51, phosphate buffer (approximate pH, 4.0 to 7.0) was used. The effect of temperature on enzyme activity was determined in phosphate buffer at the respective pH levels by using different temperatures ranging from 40°C to 90°C. To determine the kinetic parameters, various concentrations (0.25 mM to 1.75 mM for AbF51 and 0.5 mM to 3.5 mM for XynF) of pNP-AraF were used. The AbF51 and XynF enzymes were used at final concentrations of 0.1 μg and 3.5 μg, respectively. Reactions were performed under optimum pH and temperature conditions. Released para-nitrophenols were measured using the Beer-Lambert law. Km (measured in millimoles) and kcat (measured per second) were calculated from the Michaelis-Menten equation with GraphPad Prism 5.01 software (San Diego, USA).
For analysis of effects of metal ions and chemical reagents on the AbFs, a series of chemicals, such as NiCl2, CaCl2, CuSO4, MgSO4, CoCl2, FeSO4, MnCl2, KCl, SDS, and EDTA, with 1 mM and 5 mM concentrations, were analyzed using the enzyme assay protocols described above. Half-lives of two AbFs were monitored by a time course of thermal inactivation at different temperatures (70, 75, and 80°C). At different time points, aliquots of samples were withdrawn and the enzyme activity was measured immediately. Three replicate analyses were performed for each assay, and all the results were expressed as relative (percent) activity levels.
Synergistic effects of multiple GHs.
To investigate synergistic effects of two arabinofuranosidases with other hemicellulases, xylanases XynA, XynA-TM, and XynB and xylosidase Xyl39B from Caldicellulosiruptor sp. strain F32 were produced as previously described (22). To determine the extent of synergy, different molar ratios (expressed in nanomoles) were used to obtain higher reducing ends from WAX. All the reactions were performed at pH 6 and 70°C in citrate buffer, and then the reactions were quenched by boiling for 10 min. Hydrolysis products were analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) using a Dionex ICS-3000 system (Sunnyvale, CA, USA). Samples were separated with a CarbonPac PA10 analytical column (Thermo Scientific) (4 by 250 mm). The samples were eluted at the speed of 1 ml/min via the use of an eluent consisting of 200 mM NaOH. In order to assess the synergy of each mixture, the degree of synergy (DOS) (23, 39) was calculated using the following formula: DOS = (total sugars released from enzyme mixture)/(total sugars released from incubation with individual enzymes).
1H-NMR analysis of products.
A reaction mixture containing 12 ml of 3% insoluble WAX in reaction buffer (20 mM Na-acetate, 50 mM NaCl, pH 6.5) was incubated with XynA xylanase at 70°C for 12 h. The mixture was centrifuged and boiled for 10 min to inactivate XynA. The supernatant was divided into three portions (4 ml each). Fresh AbF51 or XynF (10 nM) was added to the mixture for another 12 h. The reaction mixture was boiled for 10 min and centrifuged again. Hydrolysis products were lyophilized and dissolved in D2O to replace the H in the samples with D. 1H-NMR spectra were recorded on Bruker Avance III 600 spectrometers using D2O at 30°C.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cheng Li for HPAEC-PAD, Chao Chen for 1H-NMR, and Fa-Li Bai for HRMS.
This work was supported by grants from the National Natural Science Foundation of China (no. 31400060) to M.L. This work was also supported by grants from the Shandong Province Natural Science Funds for Distinguished Young Scholar (no. JQ201507) and from the Key Scientific and Technological Project of Shandong Province (no. 2015ZDXX0403A01) to F.-L.L. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00574-17.
REFERENCES
- 1.Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annu Rev Plant Biol 61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 2.Rahman AK, Kato K, Kawai S, Takamizawa K. 2003. Substrate specificity of the α-l-arabinofuranosidase from Rhizomucor pusillus HHT-1. Carbohydr Res 338:1469–1476. doi: 10.1016/S0008-6215(03)00203-9. [DOI] [PubMed] [Google Scholar]
- 3.Lagaert S, Pollet A, Courtin CM, Volckaert G. 2014. β-Xylosidases and α-l-arabinofuranosidases: accessory enzymes for arabinoxylan degradation. Biotechnol Adv 32:316–332. doi: 10.1016/j.biotechadv.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Shallom D, Belakhov V, Solomon D, Gilead-Gropper S, Baasov T, Shoham G, Shoham Y. 2002. The identification of the acid-base catalyst of α-arabinofuranosidase from Geobacillus stearothermophilus T-6, a family 51 glycoside hydrolase. FEBS Lett 514:163–167. doi: 10.1016/S0014-5793(02)02343-8. [DOI] [PubMed] [Google Scholar]
- 5.Saha BC. 2000. α-l-Arabinofuranosidases: biochemistry, molecular biology and application in biotechnology. Biotechnol Adv 18:403–423. doi: 10.1016/S0734-9750(00)00044-6. [DOI] [PubMed] [Google Scholar]
- 6.Osaki S, Kimura T, Sugimoto T, Hizukuri S, Iritani N. 2001. l-Arabinose feeding prevents increases due to dietary sucrose in lipogenic enzymes and triacylglycerol levels in rats. J Nutr 131:796–799. [DOI] [PubMed] [Google Scholar]
- 7.Seri K, Sanai K, Matsuo N, Kawakubo K, Xue C, Inoue S. 1996. l-Arabinose selectively inhibits intestinal sucrase in an uncompetitive manner and suppresses glycemic response after sucrose ingestion in animals. Metabolism 45:1368–1374. doi: 10.1016/S0026-0495(96)90117-1. [DOI] [PubMed] [Google Scholar]
- 8.Krog-Mikkelsen I, Hels O, Tetens I, Holst JJ, Andersen JR, Bukhave K. 2011. The effects of l-arabinose on intestinal sucrase activity: dose-response studies in vitro and in humans. Am J Clin Nutr 94:472–478. doi: 10.3945/ajcn.111.014225. [DOI] [PubMed] [Google Scholar]
- 9.Dam P, Kataeva I, Yang SJ, Zhou F, Yin Y, Chou W, Poole FL II, Westpheling J, Hettich R, Giannone R, Lewis DL, Kelly R, Gilbert HJ, Henrissat B, Xu Y, Adams MW. 2011. Insights into plant biomass conversion from the genome of the anaerobic thermophilic bacterium Caldicellulosiruptor bescii DSM 6725. Nucleic Acids Res 39:3240–3254. doi: 10.1093/nar/gkq1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.VanFossen AL, Ozdemir I, Zelin SL, Kelly RM. 2011. Glycoside hydrolase inventory drives plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Biotechnol Bioeng 108:1559–1569. doi: 10.1002/bit.23093. [DOI] [PubMed] [Google Scholar]
- 11.Blumer-Schuette SE, Giannone RJ, Zurawski JV, Ozdemir I, Ma Q, Yin Y, Xu Y, Kataeva I, Poole FL II, Adams MW, Hamilton-Brehm SD, Elkins JG, Larimer FW, Land ML, Hauser LJ, Cottingham RW, Hettich RL, Kelly RM. 2012. Caldicellulosiruptor core and pangenomes reveal determinants for noncellulosomal thermophilic deconstruction of plant biomass. J Bacteriol 194:4015–4028. doi: 10.1128/JB.00266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng DD, Ying Y, Zhang KD, Lu M, Li FL. 2015. Depiction of carbohydrate-active enzyme diversity in Caldicellulosiruptor sp. F32 at the genome level reveals insights into distinct polysaccharide degradation features. Mol Biosyst 11:3164–3173. [DOI] [PubMed] [Google Scholar]
- 13.Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. 2008. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr Opin Biotechnol 19:210–217. doi: 10.1016/j.copbio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkens C, Andersen S, Petersen BO, Li A, Busse-Wicher M, Birch J, Cockburn D, Nakai H, Christensen HE, Kragelund BB, Dupree P, McCleary B, Hindsgaul O, Hachem MA, Svensson B. 2016. An efficient arabinoxylan-debranching α-l-arabinofuranosidase of family GH62 from Aspergillus nidulans contains a secondary carbohydrate binding site. Appl Microbiol Biotechnol 100:6265–6277. doi: 10.1007/s00253-016-7417-8. [DOI] [PubMed] [Google Scholar]
- 16.Bouraoui H, Desrousseaux ML, Ioannou E, Alvira P, Manai M, Remond C, Dumon C, Fernandez-Fuentes N, O'Donohue MJ. 2016. The GH51 α-l-arabinofuranosidase from Paenibacillus sp. THS1 is multifunctional, hydrolyzing main-chain and side-chain glycosidic bonds in heteroxylans. Biotechnol Biofuels 9:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Shi P, Ma R, Luo H, Huang H, Yang P, Yao B. 2015. A new GH43 α-arabinofuranosidase from Humicola insolens Y1: biochemical characterization and synergistic action with a xylanase on xylan degradation. Appl Biochem Biotechnol 175:1960–1970. doi: 10.1007/s12010-014-1416-y. [DOI] [PubMed] [Google Scholar]
- 18.Maehara T, Fujimoto Z, Ichinose H, Michikawa M, Harazono K, Kaneko S. 2014. Crystal structure and characterization of the glycoside hydrolase family 62 α-l-arabinofuranosidase from Streptomyces coelicolor. J Biol Chem 289:7962–7972. doi: 10.1074/jbc.M113.540542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tateishi A, Mori H, Watari J, Nagashima K, Yamaki S, Inoue H. 2005. Isolation, characterization, and cloning of α-l-arabinofuranosidase expressed during fruit ripening of Japanese pear. Plant Physiol 138:1653–1664. doi: 10.1104/pp.104.056655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichinose H, Yoshida M, Fujimoto Z, Kaneko S. 2008. Characterization of a modular enzyme of exo-1,5-α-l-arabinofuranosidase and arabinan binding module from Streptomyces avermitilis NBRC14893. Appl Microbiol Biotechnol 80:399–408. doi: 10.1007/s00253-008-1551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guais O, Tourrasse O, Dourdoigne M, Parrou JL, Francois JM. 2010. Characterization of the family GH54 α-l-arabinofuranosidases in Penicillium funiculosum, including a novel protein bearing a cellulose-binding domain. Appl Microbiol Biotechnol 87:1007–1021. doi: 10.1007/s00253-010-2532-4. [DOI] [PubMed] [Google Scholar]
- 22.Meng DD, Ying Y, Chen XH, Lu M, Ning K, Wang LS, Li FL. 2015. Distinct roles for carbohydrate-binding modules of glycoside hydrolase 10 (GH10) and GH11 xylanases from Caldicellulosiruptor sp. strain F32 in thermostability and catalytic efficiency. Appl Environ Microbiol 81:2006–2014. doi: 10.1128/AEM.03677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang M, Zhang KD, Zhang PY, Zhou X, Ma XQ, Li FL. 2016. Synergistic cellulose hydrolysis dominated by a multi-modular processive endoglucanase from Clostridium cellulosi. Front Microbiol 7:932. doi: 10.3389/fmicb.2016.00932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann RA, Leeflang BR, de Barse MM, Kamerling JP, Vliegenthart JF. 1991. Characterisation by 1H-n.m.r. spectroscopy of oligosaccharides, derived from arabinoxylans of white endosperm of wheat, that contain the elements →4)[α-l-Araf-(1-ar3)]-β-d-Xylp-(1→or →4)[α-l-Araf-(1→2)][α-l-Araf-(1→3)]-β-d-Xylp-(1→. Carbohydr Res 221:63–81. doi: 10.1016/0008-6215(91)80049-S. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann RA, Geijtenbeek T, Kamerling JP, Vliegenthart JF. 1992. 1H-N.m.r. study of enzymically generated wheat-endosperm arabinoxylan oligosaccharides: structures of hepta- to tetradeca-saccharides containing two or three branched xylose residues. Carbohydr Res 223:19–44. doi: 10.1016/0008-6215(92)80003-J. [DOI] [PubMed] [Google Scholar]
- 26.Borsenberger V, Dornez E, Desrousseaux ML, Massou S, Tenkanen M, Courtin CM, Dumon C, O'Donohue MJ, Faure R. 2014. A 1H NMR study of the specificity of α-l-arabinofuranosidases on natural and unnatural substrates. Biochim Biophys Acta 1840:3106–3114. doi: 10.1016/j.bbagen.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Ferré H, Broberg A, Duus JO, Thomsen KK. 2000. A novel type of arabinoxylan arabinofuranohydrolase isolated from germinated barley - analysis of substrate preference and specificity by nano-probe NMR. Eur J Biochem 267:6633–6641. doi: 10.1046/j.1432-1327.2000.01758.x. [DOI] [PubMed] [Google Scholar]
- 28.Igarashi K, Uchihashi T, Koivula A, Wada M, Kimura S, Okamoto T, Penttila M, Ando T, Samejima M. 2011. Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science 333:1279–1282. doi: 10.1126/science.1208386. [DOI] [PubMed] [Google Scholar]
- 29.Yang SJ, Kataeva I, Hamilton-Brehm SD, Engle NL, Tschaplinski TJ, Doeppke C, Davis M, Westpheling J, Adams MWW. 2009. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl Environ Microbiol 75:4762–4769. doi: 10.1128/AEM.00236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunecky R, Alahuhta M, Xu Q, Donohoe BS, Crowley MF, Kataeva IA, Yang SJ, Resch MG, Adams MWW, Lunin VV, Himmel ME, Bomble YJ. 2013. Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science 342:1513–1516. doi: 10.1126/science.1244273. [DOI] [PubMed] [Google Scholar]
- 31.Yi ZL, Su XY, Revindran V, Mackie RI, Cann I. 2013. Molecular and biochemical analyses of CbCel9A/Cel48A, a highly secreted multi-modular cellulase by Caldicellulosiruptor bescii during growth on crystalline cellulose. PLoS One 8:e84172. doi: 10.1371/journal.pone.0084172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim YR, Yoon RY, Seo ES, Kim YS, Park CS, Oh DK. 2010. Hydrolytic properties of a thermostable α-l-arabinofuranosidase from Caldicellulosiruptor saccharolyticus. J Appl Microbiol 109:1188–1197. doi: 10.1111/j.1365-2672.2010.04744.x. [DOI] [PubMed] [Google Scholar]
- 33.Lochner A, Giannone RJ, Keller M, Antranikian G, Graham DE, Hettich RL. 2011. Label-free quantitative proteomics for the extremely thermophilic bacterium Caldicellulosiruptor obsidiansis reveal distinct abundance patterns upon growth on cellobiose, crystalline cellulose, and switchgrass. J Proteome Res 10:5302–5314. doi: 10.1021/pr200536j. [DOI] [PubMed] [Google Scholar]
- 34.Lochner A, Giannone RJ, Rodriguez M, Shah MB, Mielenz JR, Keller M, Antranikian G, Graham DE, Hettich RL. 2011. Use of label-free quantitative proteomics to distinguish the secreted cellulolytic systems of Caldicellulosiruptor bescii and Caldicellulosiruptor obsidiansis. Appl Environ Microbiol 77:4042–4054. doi: 10.1128/AEM.02811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paës G, Berrin JG, Beaugrand J. 2012. GH11 xylanases: structure/function/properties relationships and applications. Biotechnol Adv 30:564–592. doi: 10.1016/j.biotechadv.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Gao M, Liu ZZ, Zhou YG, Liu HC, Ma YC, Wang L, Chen SF, Ji XC. 2012. Gracilibacillus kekensis sp nov., a moderate halophile isolated from Keke Salt Lake. Int J Syst Evol Microbiol 62:1032–1036. doi: 10.1099/ijs.0.030858-0. [DOI] [PubMed] [Google Scholar]
- 37.Ying Y, Meng DD, Chen XH, Li FL. 2013. An extremely thermophilic anaerobic bacterium Caldicellulosiruptor sp. F32 exhibits distinctive properties in growth and xylanases during xylan hydrolysis. Enzyme Microb Technol 53:194–199. doi: 10.1016/j.enzmictec.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Bergquist PL, Gibbs MD, Morris DD, Te'o VS, Saul DJ, Moran HW. 1999. Molecular diversity of thermophilic cellulolytic and hemicellulolytic bacteria. FEMS Microbiol Ecol 28:99–110. doi: 10.1016/S0168-6496(98)00078-6. [DOI] [Google Scholar]
- 39.Zhang T, Meng X, Zhang T. 2015. Global dynamics of a virus dynamical model with cell-to-cell transmission and cure rate. Comput Math Methods Med 2015:758362. doi: 10.1155/2015/758362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.