Abstract
Kinesin processivity, defined as the average number of steps that occur per interaction with a microtubule, is an important biophysical determinant of the motor’s intracellular capabilities. Despite its fundamental importance to the diversity of tasks that kinesins carry out in cells, no existing quantitative model fully explains how structural differences between kinesins alter kinetic rates in the ATPase cycle to produce functional changes in processivity. Here we use high-resolution single-molecule microscopy to directly observe the stepping behavior of kinesin-1 and -2 family motors with different length neck-linker domains. We characterize a one-head-bound posthydrolysis vulnerable state where a kinetic race occurs between attachment of the tethered head to its next binding site and detachment of the bound head from the microtubule. We find that greater processivity is correlated with faster attachment of the tethered head from this vulnerable state. In compliment, we show that slowing detachment from this vulnerable state by strengthening motor-microtubule electrostatic interactions also increases processivity. Furthermore, we provide evidence that attachment of the tethered head is irreversible, suggesting a first passage model for exit from the vulnerable state. Overall, our results provide a kinetic framework for explaining kinesin processivity and for mapping structural differences to functional differences in diverse kinesin isoforms.
Introduction
Kinesin motor proteins drive many active processes in the cell, including vesicle transport (1, 2, 3), DNA and organelle repositioning (4, 5), intraflagellar transport (6, 7), microtubule dynamics control (8, 9, 10), and mitotic spindle organization (11, 12, 13). There are 45 kinesin genes in the human genome, each of which encodes an isoform that is optimized to drive some processes, but is incapable of driving others (14, 15). Understanding the nature of this functional specialization is critical to elucidating the molecular bases of Charcot-Marie-Tooth disease (16), hereditary spastic paraplegia (17, 18), Alzheimer’s disease (19, 20), and the various cancers (21, 22) associated with either kinesin dysfunction or overactivity. Part of kinesin’s functional diversity can be understood by major structural differences between isoforms. For example, kinesin-5 acts as a tetramer instead of the typical dimer (23), and kinesin-14 has its motor domain at its C-terminus rather than its N-terminus (24). However, most kinesins have a similar structure, and their functional specialization thus comes from the tuning of motility parameters such as velocity, processivity, and force sensitivity. Here we focus on processivity, or the number of steps kinesin takes per interaction with a microtubule. Processivity values for different kinesins vary over multiple orders of magnitude from just a few steps (25) to a thousand steps or higher (26, 27), in part enabling them to perform their different tasks in the cell. Despite its fundamental importance, a consensus quantitative model that explains how processivity is tuned between kinesin isoforms is absent from the literature. This is in part due to the inability to measure kinetic intermediates in the ATPase cycle from which processivity can be controlled. Here we apply new high-resolution tracking technology (25, 28, 29, 30, 31) to fill this gap in the literature.
The simplest model for kinesin processivity is the kinetic race model, which posits that kinesin must proceed through a single vulnerable state each turnover of its ATPase cycle (32). In our previous work, we identified a one-head-bound posthydrolysis state as a candidate for the vulnerable state (28, 33). This result suggested that processivity could be controlled by a kinetic race between tethered head attachment and bound head detachment from the one-head-bound intermediate. In this study, we examine these rate constants with high-resolution single-molecule microscopy as we implement four methods to alter processivity: changing the motor domain from kinesin-1 to -2, altering the neck-linker (NL) length, changing the solvent ionic strength, and using slowly dissociating ATP analogs. We find that any method for increasing processivity leads to either the tethered head finding the next microtubule binding site more quickly or the bound head dissociating from the current microtubule binding site more slowly. Thus, we find that the kinetic race model is sufficient for quantitatively explaining processivity under physiological conditions, and that the intracellular capabilities of a given motor in part stem from the tuning of the two rate constants in the race.
Materials and Methods
Constructs and protein preparation
All kinesin proteins used were expressed in BL21(DE3) bacteria (New England Biolabs, Ipswich, MA) with a C-terminal 6× His tag and purified by affinity chromatography followed by buffer exchange as reported in Mickolajczyk et al. (28) and Chen et al. (34). The kinesin-1 construct used was Drosophila KHC truncated at amino acid (aa) 559, and the elongated NL K117 construct was made by adding the 3-aa DAL at position 345 just preceding α7, consistent with previous works (35, 36, 37). The kinesin-2 construct used was human KIF3A motor and NL homodimerized using the kinesin-1 coiled-coil (345–559) (36), and the K214 construct was made by deleting the DAL sequence at the end of the NL and replacing the kinked proline at positon 355 with an alanine (36). The GFP constructs included a C-terminal eGFP directly preceding the His tag (36, 37, 38). The N-terminal avi-tag constructs were described in Mickolajczyk et al. (28). For the kinesin-1 and -2 constructs, the linkers GG and GGAGG, respectively, were added directly downstream of the avi-tag. Biotin was added after induction, typically 0.5–2 h before cell lysis to empirically control the degree of biotinylation of the homodimers. Biotinylation was quantified by comparing the biotin concentration as measured by the colorimetric HABA assay (Sigma-Aldrich, St. Louis, MO) to total protein concentration as indicated by absorbance at 280 nm, and all motors prepared had <0.2 mol of bound biotin per mole of dimer motor. Kinesins used for biochemical assays were truncated at position 406 rather than 560 to enable higher yields (28, 34). Insertions and deletions were made either using Q5 or Gibson Assembly (New England Biolabs).
Single-molecule experiments
Gold nanoparticles were imaged using a custom-built total internal reflection dark field microscope, employing a Sapphire-LP 532 nm laser (5–10 mW at sample; Coherent, Bloomfield, CT). Images were recorded using a Basler Ace acA640-750um CMOS camera (1000 frames per s, 945 μs exposure; Basler, Ahrensburg, Germany) accessed by custom LabVIEW software (National Instruments, Cos Cob, CT). Microtubules were adhered to cleaned coverslips as in Mickolajczyk et al. (28). All assays, unless otherwise noted, were carried out in imaging solution: 0.5 mg/mL casein, 10 μM taxol, 20 mM glucose, 20 μg/mL glucose oxidase, 8 μg/mL catalase, 0.2 mg/mL BSA, 1:200 β-mercaptoethanol, and 2 mM MgATP in BRB80 (80 mM K-PIPES, 1 mM EGTA, 1 mM MgCl2, pH 6.8). Biotinylated motors were incubated with a stoichiometric excess of streptavidin-coated 30-nm gold nanoparticles (BBI Solutions, Cardiff, Wales, UK) on ice for 30 min, then spun down at 20,000 g for 4 min and resuspended at working levels (100–300 pM gold) in imaging solution. Point spread functions were fit using the software FIESTA (39) to obtain (X,Y,t) data.
GFP was imaged by total internal reflection fluorescence microscopy using a model No. TE2000 inverted microscope (Nikon, Melville, NY) as in Mickolajczyk et al. (28) and Shastry and Hancock (36, 37). Frame rates were set to 3 and 20 frames per s for ATP and ADP experiments, respectively. GFP motors were used at 10–300 pM. ATP movies were analyzed using FIESTA (39), and velocity and run length were determined from the returned distance over time trace. Population run lengths were determined by fitting the cumulative density function (after removing runs <5 pixels, 71 nm/pixel) with an exponential with an X offset. Mant run lengths were measured in imaging solution with 0.1 mM mantATP. ADP durations were measured manually from kymographs drawn at positions coincident with Cy5-labeled microtubules in the software ImageJ (National Institutes of Health; http://imagej.nih.gov/ij/) (34). All experiments were performed at 22–23°C.
Stopped-flow spectrofluorometry and steady-state biochemistry
ATP half-site experiments were performed on an Applied Photophysics SX20 spectrofluorometer (Applied Photophysics, Leatherhead, Surrey, UK) (34). Data acquisition and fitting were performed in Pro-Data SX software (Applied Photophysics). One syringe was filled with 2–6 μM microtubules, 10 μM taxol, 500 nM mantADP, and 200–600 nM motor in BRB80 buffer (1:10 molar ratio motor dimer to microtubule). For kinesin-2, 500 nM mantGDP was used instead of mantADP due to the higher affinity of the motor for mantADP (34). The second syringe was filled with 4 mM ATP in BRB80. Excitation was set to 356 nm for the mantADP nucleotide and 450 nm emission was collected using a HQ480SP emission filter (Chroma Technology, Bellows Falls, VT). An integration time of 1 ms was set for the photomultiplier tube. Each experiment contained 1200 μL split into 60 μL shots, and the ensemble average fluorescence transient generated upon mixing was fit with a double exponential in the range of 1–500 ms. The faster rate constant was reported. The experiment was repeated at least nine times for each motor, and data sets were taken on at least two separate days for each motor. Solution exchange of cold versus mantADP to determine relative affinity was done as previously (34). ATPase experiments were carried out as in Chen et al. (34) with 5–10 nM motor. ATP turnover rates were determined from the Michaelis-Menton fit to the microtubule dependence of the observed reaction rate, corrected for initial kinesin concentration as determined by mant-ADP exchange (25). All experiments were performed at 22–23°C.
For details on the Hidden Markov Model, two-dimensional step-finding algorithm, and calculation of rates for the first passage model, see Supporting Material.
Results
High-resolution single-molecule microscopy enables direct measurement of a one-head-bound intermediate in the stepping cycle of kinesins-1 and -2
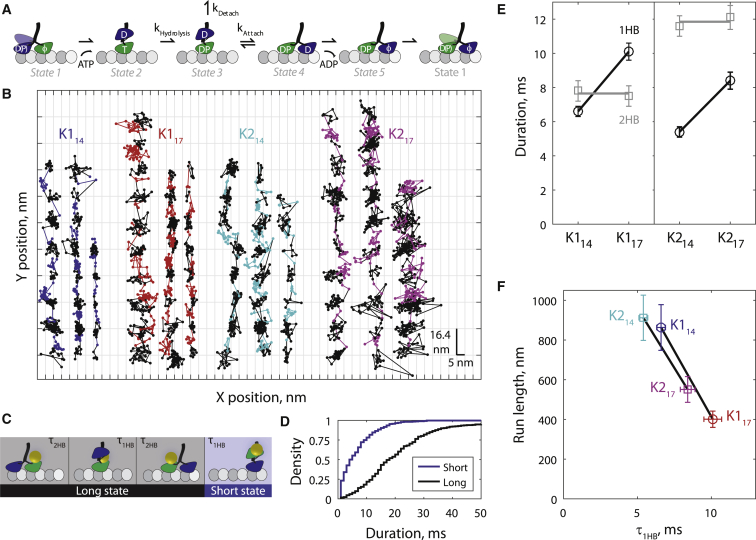
To test the hypothesis that kinesin processivity is set by a kinetic race between attachment of the tethered head and detachment of the bound head when the motor is in the one-head-bound (1HB) posthydrolysis state (Fig. 1 A, state 3), we performed high-resolution single-molecule tracking to measure the 1HB duration for multiple kinesins with different processivities. The motors used were wild-type kinesin-1 (K114), kinesin-1 with the NL extended to 17 aa (K117), wild-type kinesin-2 (K217), and kinesin-2 with the NL shortened to 14 aa (K214). It was previously shown that extending the NL reduces processivity independent of the motor domain (35, 36, 37). For all motors, a 30-nm-diameter gold nanoparticle was bound to the motor domain using biotin-streptavidin (see Materials and Methods), a technique that has been shown both experimentally (28, 30) and theoretically (40) to have little influence on stepping. The motors were observed in an in vitro single-molecule stepping assay under total internal reflection dark field microscopy at 1000 frames per s in 2 mM ATP. Example position versus time traces for the four kinesin constructs, obtained by fitting point spread functions of gold nanoparticles moving along microtubules (39, 41), are shown in Fig. 1 B. Kinesin-1 with a single motor domain tagged has previously been shown to take 16.4 nm hand-over-hand steps (42, 43, 44), and we recently established that at saturating ATP, a one-head-bound intermediate can be measured in which the tethered head is displaced ∼8 nm from its previous binding site (28). Here we clearly observe this intermediate state for all kinesin-1 and -2 constructs tested (Fig. 1 B, colored points). Because only one head is labeled, this short duration intermediate (“short state”; Fig. 1, C and D) represents a single 1HB state. The longer duration intermediate (“long state”), where the labeled motor domain is bound to the microtubule (Fig. 1 B, black points), represents the two-head-bound (2HB) state of the labeled head plus the 1HB and 2HB states of the unlabeled head (Fig. 1 C). To quantify the duration the motors spent in the long and short states, the (X,Y,t) data were fit using a hidden Markov model (see state space diagram in Fig. S1; and see Materials and Methods and Supporting Material for details). From the stepping traces, distributions of at least 300 determinations of each long and short state were built up for each of the four motors studied (K114 distributions shown in Fig. 1 D; see Fig. S2 for all distributions). 1HB and 2HB durations were then calculated from long and short distribution sample means (Fig. 1 E). The key result was that for both kinesin-1 and kinesin-2, extending the NL from 14 to 17 aa increased the 1HB duration, while having little effect on the 2HB duration.
Figure 1.
Extending the NL reduces processivity by slowing the tethered head attachment rate. (A) Five-state mechanochemical cycle established in Mickolajczyk et al. (28) and Chen et al. (34), in which processivity is set by a kinetic race between attachment of the tethered (blue) head and detachment of the bound (green) head in the 1HB posthydrolysis state (state 3). Here T denotes ATP, D denotes ADP, DP denotes ADP plus phosphate, and φ denotes no nucleotide. (B) Example 1000 frames/s traces of kinesin-1 (K1) and kinesin-2 (K2) motors with 14- and 17-aa NL domains stepping in 2 mM ATP. Horizontal lines denote inferred microtubule binding sites of the nanoparticle-labeled head. Long states on the microtubule (black points) and short states off the microtubule (colored points corresponding to motor type) were determined by fitting to the (X,Y,t) data (details in Supporting Material). Each data point represents 1 ms. (C) Two-state mechanical model in which each 16.4-nm step is separated into 1HB and 2HB states. Long and short states correspond to black and colored points, respectively, in (B). Note: the terms “long” and “short” refer to duration, not molecular configuration. (D) Distributions of N = 370 long states and N = 347 short states for K114; see Fig. S2 for other motor distributions. (E) 1HB and 2HB durations showing that for both kinesin-1 and kinesin-2, lengthening the neck linker from 14 to 17 aa increased the 1HB duration and did not change the 2HB duration. Values were calculated from means of long and short state distributions. Error bars represent propagated error from the SE of long and short state distributions. (F) Run length is negatively correlated with 1HB duration for both kinesin-1 and -2, indicating that motors with long NL have reduced processivity due to a reduced kAttach. Colors denoting motor type correspond to (B). To see this figure in color, go online.
Altering the neck-linker length modulates the processivity of kinesin-1 and -2 by tuning the tethered head attachment rate
We next measured the run lengths (RL) of the four motors using GFP constructs (Fig. 1 F; distributions in Fig. S3). Consistent with previous measurements (35, 36, 37), constructs with the same NL length had similar RLs independent of motor domain. Importantly, a strong negative correlation was observed between the 1HB duration and the run length when comparing the 14 to the 17-aa NL construct of each motor type (Fig. 1 F). From the established mechanochemical cycle (Fig. 1 A), the 1HB state includes both a pre- and posthydrolysis state with detachment occurring from the posthydrolysis state (28, 33). Therefore, the total 1HB duration (Fig. 1 A states 2 and 3) is:
Assuming that the motor rarely returns backward to the 1HB state from the 2HB state (Fig. 1 A state 4→3 transition; discussed in detail below), we can derive the probability of stepping as a simple kinetic race between attachment and detachment:
Because the probability of stepping is proportional to the run length, and the run length is >50 steps for all the motors investigated here, it follows that kAttach ≫kDetach. Moreover, the existing evidence (45) indicates that kHydrolysis is not significantly affected by NL length. Thus, the difference in the τ1HB values for kinesins with the same motor domain but different NL lengths is as follows:
Therefore, the differences in 1HB duration in Fig. 1, E and F are best understood as resulting from differences in kAttach. It follows that for both kinesin-1 and -2, extending the NL from 14 to 17 aa reduces processivity by decreasing kAttach. This result shows that for kinesin-1 and -2, kAttach is primarily determined by the NL domain and not by the motor domain.
Pre-steady-state kinetics independently support 1HB duration differences
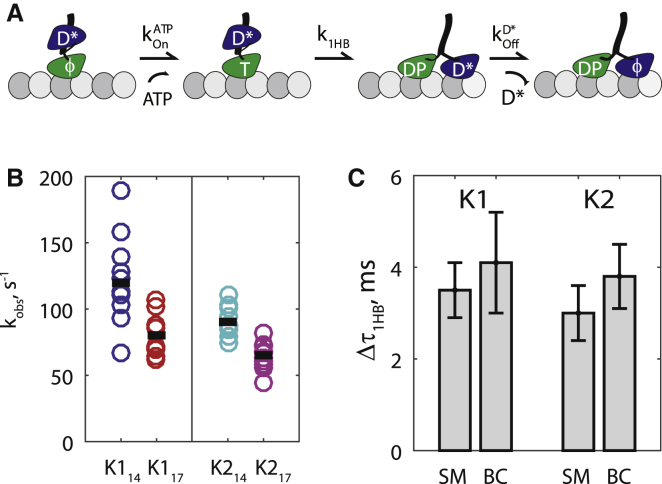
To measure the effect of NL length on 1HB duration in an independent experimental platform, we performed ATP half-site release experiments for all four motor constructs using stopped-flow. Motors were incubated with microtubules and low concentrations of fluorescent 2′(3′)-O-(N-methylanthraniloyl) nucleoside disphosphate (mantNDP) to create a 1HB ATP waiting state with the mantNDP in the tethered head (46, 47, 48). The motor-microtubule complexes were then flushed against ATP, enabling the motors to proceed through their 1HB states, and a drop in fluorescence intensity was detected when they released their mantNDP into solution (Fig. 2 A). In the scheme of Fig. 2 A, if a high ATP concentration (fast ) and a low-affinity mantNDP (fast ) are used, the rate-limiting step in the half-site release process is k1HB. Importantly, extending the NL was recently shown not to affect the mantNDP release rate from the front head in the 2HB state (Fig. 2 A ; see Figs. 5E and 7D in (34)). Thus, by comparing the half-site release rate of 14- to 17-aa NL constructs, differences in 1HB duration (Δτ1HB) can be calculated. Results for half-site release for the four motor constructs are shown in Fig. 2 B (see example transients in Fig. S4). In these experiments mantADP was used for kinesin-1, but due to the high mantADP affinity of kinesin-2 (34), mantGDP was used instead (investigated in detail below). Consistent with previous measurements using different motor constructs (49), elongating the NL was seen to decrease the half-site release rate. The Δτ1HB measured by half-site was in excellent agreement with the Δτ1HB measured by single-molecule (Fig. 2 C), providing a second independent line of evidence that extending the NL reduces processivity by reducing the tethered head attachment rate.
Figure 2.
Extending the neck linker increases the 1HB duration. (A) The sequence of states in the ATP half-site experiment; D∗ denotes a fluorescent nucleotide. (B) Half-site release rates at 2 mM ATP, measured by fitting fluorescence transients (Fig. S4). Open circles represent 9–11 replicates for each motor, with mean value shown as a black bar. (C) Comparison of differences in the 1HB durations (Δτ1HB) between 14- and 17-residue neck-linker constructs for kinesin-1 and kinesin-2 measured by single-molecule (SM) experiments (Fig. 1E) and biochemical (BC) assays (Fig. 2B). Error bars denote propagated error from the SE of short state distributions for single-molecule experiments, and from the SE of half-site values for biochemical assays. To see this figure in color, go online.
Altering ionic strength modulates the processivity of kinesin-1 and -2 by tuning the bound head detachment rate
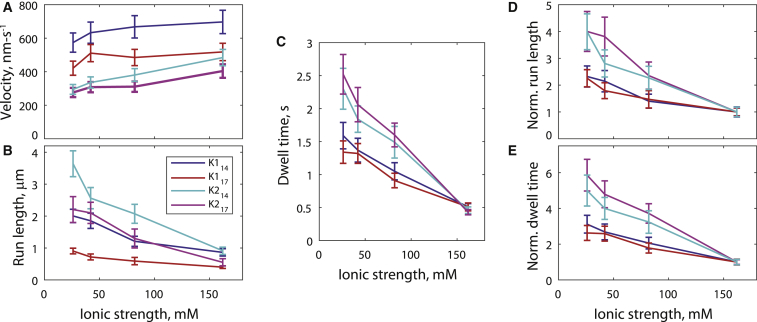
We next investigated the effect of reducing the ionic strength on the velocity and run length of all four motor constructs. Each motor was measured in BRB80 (as above in Figs. 1 and 2), and in buffers containing 40, 20, and 12 mM PIPES. Consistent with previous kinesin-1 measurements (36), reducing the ionic strength was seen to slightly decrease the velocity of all motors (Fig. 3 A) and to increase the run lengths of all motors to different extents (Fig. 3 B; distributions in Fig. S3). Under the model that processivity is controlled by a kinetic race between tethered head attachment and bound head detachment in the 1HB state, an increase in processivity due to an increase in kAttach must lead to a faster, not a slower velocity. Thus, the measured increase in processivity at low ionic strength more likely stems from a decrease in kDetach. To test this claim, we measured the microtubule dwell time of all four motors at saturating ADP as a function of ionic strength (Fig. 3 C; see distributions and example kymographs in Fig. S5). Consistent with the prediction that low ionic strength reduces detachment rates, the dwell time increased as the ionic strength was reduced for all four motors. Strikingly, as highlighted when the run lengths (Fig. 3 D) and ADP dwell times (Fig. 3 E) were normalized to their values in BRB80, increases in both parameters were shared by each motor type independent of NL length: K114 and K117 both had an ∼2.5-fold increase in RL and ADP dwell time, whereas K214 and K217 both had an ∼4–6-fold increase in RL and ADP dwell time. Thus, kDetach can be adjusted by altering the electrostatic interactions between the motor domain and the microtubule, and tuning kDetach is a means of controlling RL in an approximately proportional fashion. The fact that elongating the NL did not affect the ADP dwell time bolsters the claim that changes in RL due to differences in NL length stem from changes in kAttach.
Figure 3.
Ionic strength alters processivity by tuning the detachment rate from the 1HB vulnerable state. (A) Velocities in 2 mM ATP, (B) run lengths in 2 mM ATP, and (C) dwell times in 2 mM ADP for GFP motors in BRB12, BRB20, BRB40, and BRB80 buffers. Velocities are shown as mean ± SE with a 10% error added for ∼1°C variability in temperature (61). Run lengths and dwell times were determined by distribution fitting with error bars determined by bootstrapping (52). (D) Run lengths and (E) ADP dwell times normalized to BRB80 values (with propagated error) to emphasize the pairing by motor domain rather than by neck-linker length. All panels are color-coded by legend in (B), consistent with Figs. 1 and 2. To see this figure in color, go online.
Slowing the ADP off-rate modulates processivity by shunting the motor backward into the 1HB state
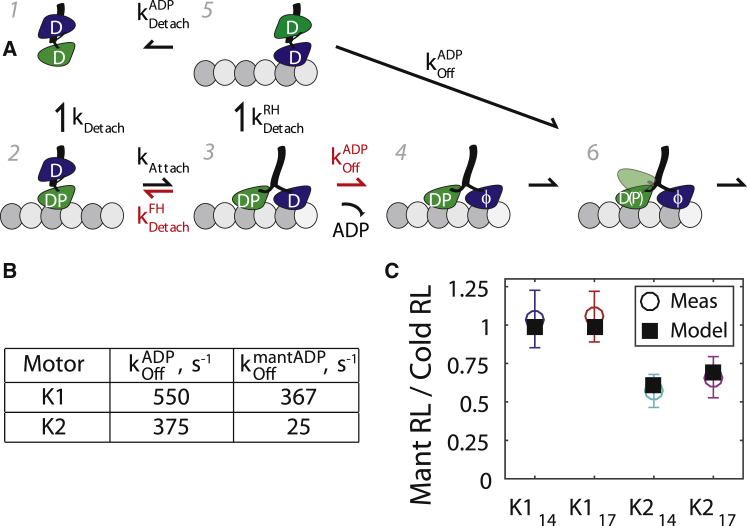
The data in Figure 1, Figure 2, Figure 3 establish that kinesin-1 and-2 processivity can be understood as a kinetic race between kAttach and kDetach when the motor is in the vulnerable 1HB state (Fig. 4 A, state 2). However, completion of a forward step also involves ADP release to lock in the gains made by the attachment step. Hence, there is a second kinetic race after tethered head attachment—either ADP is released to complete the step, or the attachment step is reversed and the motor returns to the vulnerable 1HB state (Fig. 4 A, state 3 → 4 versus 3 → 2 transition). Depending on the relative kinetic parameters, this process can be described in two ways. In a “first passage model”, kAttach is relatively slow and ADP release follows rapidly such that the motor rarely reverts to the 1HB state. In this sense, tethered head attachment is the irreversible ratcheting step. Alternatively, in a “rapid equilibrium model”, both kAttach and the reverse rate are very fast, and the equilibrium favors the 1HB state sufficiently that it is detected in single-molecule tracking (28). Here, the rare outpacing of by locks the motor into the 2HB state (Fig. 4 A, equilibrium between states 2 and 3 until the 3 → 4 transition occurs), and ADP release is thus the irreversible ratcheting step. To test between these models, we measured the run lengths of our four GFP constructs in mantATP. Although the off-rates for mant- and cold ADP are similar for kinesin-1 (Fig. 4 B; Fig. S6 A) (28), kinesin-2 has a 15-fold lower off-rate for mantADP than cold ADP (Fig. 4 B) (34). Thus, under the first passage model the kinesin-2 RL should drop only slightly in mantADP, whereas under the rapid equilibrium model the kinesin-2 RL should drop ∼15-fold in mantATP. We found that in mantATP the RL of K214 and K217 only dropped roughly twofold, thus ruling out the rapid equilibrium model (Fig. 4 C, distributions in Fig. S6 B). As expected from the similarity in their ADP and mantADP off-rates, the K114 and K117 RLs were not measurably different in mantATP versus cold ATP.
Figure 4.
Tethered head attachment follows a first passage model. (A) Mechanochemical model based on Fig. 1A, showing allowed transitions. When ADP release (state 3 → 4) is significantly slowed, front-head (FH) or rear-head (RH) detachment from state 3 may occur, forcing kinetic flux into vulnerable states (2, 5). In the first passage model, is small relative to , whereas in the rapid equilibrium model, is very fast relative to (red arrows). (B) Previous determinations of by front-head nucleotide exchange experiments for kinesin-1 (28) and kinesin-2 (34). These rates are independent of neck-linker length (34). (C) Run lengths in mantATP relative to run lengths in unlabeled (cold) ATP for each motor. Consistency of experiment and model (black squares) supports first passage model for kAttach. To see this figure in color, go online.
To ensure quantitative consistency of the first passage model, we modeled the six-state process shown in Fig. 4 A as a Markov chain. We solved for kAttach and kDetach using our cold ATP RL and Δτ1HB values (Figs. 1 A and 2 C), used literature values for or (28, 34), and used measured values for (Fig. 3 C; and see Table S1 and Supporting Material for details). Using transition probabilities calculated from the rate constants, we determined the run length by calculating the average number of times the motor entered the ATP waiting state before detaching (Fig. 4 A state 6). Comparing the modeled mantATP versus cold ATP RL (changing only to as in Fig. 4 B), we were able to nearly perfectly replicate the experimental data (Fig. 4 C), validating the first passage model. Hence in physiological ATP, tethered head attachment is the ratcheting step of the cycle and locking-in steps via ADP release occurs with nearly 100% probability.
Discussion
By changing the identity of the motor domain, the length of the NL domain, the ionic strength of the solvent, and the nucleotide off-rate, we demonstrate here that kinesin processivity can be quantitatively described as a kinetic race between tethered head attachment and bound head detachment. This model should generalize to N-terminal kinesins beyond kinesin-1 and -2, and it provides a quantitative framework for understanding how small structural differences between isoforms can tune rates in the ATPase cycle to yield functional differences in motor behavior.
Elongating the neck-linker tunes processivity by changing the tethered head attachment rate
Elongating the NL has been previously shown to reduce processivity (36, 37, 43, 49), but the underlying mechanism has remained mysterious. Using high-resolution tracking and stopped-flow, we show here that NL mutants alter the vulnerable state duration, and that independent of motor domain there is a strong negative correlation between vulnerable state duration and processivity (Fig. 1 F). Recent gold-nanoparticle tracking results from Isojima et al. (30) showed that human kinesin-1, with cys-lite modifications in 12 mM PIPES buffer containing saturating ATP, had an increased 1HB duration when the NL was shortened to 13 aa, and slightly decreased duration when the NL was increased to 20 aa (six-glycine insert). However, kinesins with a 13-aa NL have a reduced run length (37) and Cys-lite mutants with various 20-aa NLs have a surprisingly increased run length (50). Hence, their results follow the same negative correlation between vulnerable state duration and processivity. Additionally, our 17-aa NL constructs did not show the unbinding events, sideways steps, or futile hydrolysis (Fig. 1; Fig. S7) that 20-aa NL cys-lite did (30, 43), indicating that not all elongated NL constructs should be lumped together for comparison.
One possible explanation for why elongating the NL increases the 1HB duration is that the longer NL increases the conformational search volume of the tethered head. However, an argument against this model is that Drosophila kinesin-1 constructs with NLs from 15–20 residues all have roughly the same RL (27, 36). A second possibility is that elongating the NL alters the interaction of the tethered head with the bound head while in the 1HB state. Alonso et al. (51) used biochemical and structural data to argue that in the ATP waiting state, the tethered head is nestled next to the bound head, and chemical events in the bound head release the tethered head to step to the next binding site. If extending the NL disrupted the ability of the bound head to free the tethered head, this would increase the measured 1HB duration and time spent in the vulnerable state. A third possibility is that a NL of 14 aa provides the ideal geometry for the tethered head to find its next binding site, perhaps because the docked NL is rigid and any lengthening or shortening of this domain causes misregistration between the tethered head and the next binding site, thereby reducing the attachment rate.
Electrostatic interactions stabilize the kinesin-microtubule complex in the vulnerable state
We found that adding or subtracting the 3-aa DAL to the NL affected the tethered head attachment rate but not the bound head detachment rate. Thus, kinesins were paired by NL length when observing attachment (Fig. 1) and paired by motor domain when observing detachment (Fig. 3). The bound-head detachment rate was evaluated by proxy using the dwell time in saturating ADP, an approach justified by the close agreement of the measured values (Fig. 3 C) with the kDetach values calculated from measured RLs and Δτ1HB (Table S1). The effect most likely comes from electrostatic interactions between positively charged residues in kinesin and negatively charged residues in the microtubule, particularly in the tubulin C-terminal tail (52). Consistent with this, cleaving off the C-terminal tail with subtilisin has been shown to decrease kinesin processivity (52, 53, 54). It is notable that kDetach can also be affected by adding positive residues outside the motor domain. Adding lysines to the neck coil has been shown to increase processivity (52). Adding lysines when elongating the kinesin-1 NL has been shown to decrease the run length in BRB80 but not in BRB12 (36, 43), a result best understood by offsetting changes in kAttach (longer NL) and kDetach (ionic strength dependence). We verified this by showing that K117 with a lysine (KAL inserted) had a stronger ionic strength dependence of run length and dwell time in ADP than K117 without a lysine (DAL inserted; Fig. S8).
Slowing ADP release reduces processivity by reversing tethered head attachment
When mantATP is used to power kinesin-2 stepping, the slow mantADP release rate increases the probability that the newly bound front head fails to complete its step by releasing its ADP, and instead reverts to the 1HB vulnerable state. This effect only decreases run length approximately twofold (Fig. 4 C), which is surprisingly mild and suggests a first passage model: attachment of the tethered head is the irreversible ratcheting step, and rapid ADP release subsequently locks-in the 1HB to 2HB transition. Our results and model predictions show that kinesin-1 and -2 both rectify steps with nearly 100% efficiency in cold ATP, meaning that the vulnerable state is only entered once per cycle the majority of the time. In Fig. 4, the efficiency of capturing the forward step was compromised chemically by using a high-affinity nucleotide analog. It can also be compromised mechanically by increasing intramolecular tension: Isojima et al. (30) showed that kinesin-1 with a 13-residue NL occasionally returned to the 1HB state after tapping down on the microtubule-binding site, indicating an increased . Multiple isoforms of kinesin have been shown to undergo one-dimensional diffusion when forced into the ADP state (26, 55, 56, 57, 58), and some low-processivity kinesins such as MCAK utilize diffusion in the ADP or ADP-Pi state in the cell to perform their role in mitosis (58). It follows that these kinesins have a greatly slowed ADP off-rate (59), such that the efficiency of locking-in steps is reduced and kinetic flux is forced into the 1HB ADP-Pi and ADP states.
Assisting loads decrease run length by enhancing detachment
This work also provides insights into how external loads alter kinesin processivity. Optical trapping experiments on wild-type kinesin-1 have shown that hindering loads reduce motor velocity and run length, whereas assisting loads have no effect on velocity but strongly reduce run length (27, 33, 60). Our results show that either kAttach or kDetach can affect run length, but of these two only kAttach affects velocity. Thus, the decrease in run length under hindering load may result from changes to kAttach and kDetach, but the decrease in run length under assisting load must result solely from changes to kDetach. This makes intuitive sense, as it not clear how an assisting load would decrease kAttach, and assisting loads have been shown to rescue the irregular stepping of mutants (43). It also means that assisting loads must speed kDetach to a greater degree than do hindering loads. External loads may also increase the front head or rear head detachment rates from the 2HB state (Fig. 4 A, states 3 → 2 and 3 → 5 transitions, respectively), and thus increase the flux into vulnerable states in a fashion similar to mantATP (Fig. 4).
Here we show that processivity, an important biophysical determinant of intracellular function, is set by a kinetic race between attachment of the tethered head and detachment of the bound head when kinesin is in a vulnerable one-head-bound state. This simple and general model provides a link between kinesin structure and function that should apply across the kinesin superfamily in both healthy and disease states.
Author Contributions
K.J.M. built the total internal reflection dark field microscope, designed experiments, performed experiments, analyzed data, and wrote the article. W.O.H. designed experiments and wrote the article.
Acknowledgments
We are grateful to David Arginteanu for his help with protein preparation, the labs of Philipp Kukura, Zev Bryant, and Aaron Hoskins for microscopy advice, Greg Hoeprich for advice on run length measurements, and Scott Chen for his help on solution mantADP-ADP exchange data.
This work was funded by National Institutes of Health (NIH) grant R01 GM076476 to W.O.H.
Editor: Steven Rosenfeld.
Footnotes
Supporting Materials and Methods, eight figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30511-8.
Supporting Citations
References (62, 63) appear in the Supporting Material.
Supporting Material
References
- 1.Vale R.D., Reese T.S., Sheetz M.P. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ray K., Perez S.E., Goldstein L.S. Kinesin-II is required for axonal transport of choline acetyltransferase in Drosophila. J. Cell Biol. 1999;147:507–518. doi: 10.1083/jcb.147.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall D.H., Hedgecock E.M. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- 4.Granger E., McNee G., Woodman P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin. Cell Dev. Biol. 2014;31:20–29. doi: 10.1016/j.semcdb.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farina F., Pierobon P., Cappello G. Kinesin KIFC1 actively transports bare double-stranded DNA. Nucleic Acids Res. 2013;41:4926–4937. doi: 10.1093/nar/gkt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole D.G., Diener D.R., Rosenbaum J.L. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou G., Blacque O.E., Scholey J.M. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y., Hancock W.O. Kinesin-5 is a microtubule polymerase. Nat. Commun. 2015;6:8160. doi: 10.1038/ncomms9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner M.K., Zanic M., Howard J. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 2011;147:1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 10.Gumy L.F., Chew D.J., Fawcett J.W. The kinesin-2 family member KIF3C regulates microtubule dynamics and is required for axon growth and regeneration. J. Neurosci. 2013;33:11329–11345. doi: 10.1523/JNEUROSCI.5221-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawin K.E., LeGuellec K., Mitchison T.J. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- 12.Waitzman J.S., Rice S.E. Mechanism and regulation of kinesin-5, an essential motor for the mitotic spindle. Biol. Cell. 2014;106:1–12. doi: 10.1111/boc.201300054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross R.A., McAinsh A. Prime movers: the mechanochemistry of mitotic kinesins. Nat. Rev. Mol. Cell Biol. 2014;15:257–271. doi: 10.1038/nrm3768. [DOI] [PubMed] [Google Scholar]
- 14.Hirokawa N., Takemura R. Kinesin superfamily proteins and their various functions and dynamics. Exp. Cell Res. 2004;301:50–59. doi: 10.1016/j.yexcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Miki H., Okada Y., Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka Y., Hirokawa N. Mouse models of Charcot-Marie-Tooth disease. Trends Genet. 2002;18:S39–S44. doi: 10.1016/s0168-9525(02)02839-1. [DOI] [PubMed] [Google Scholar]
- 17.Reid E., Kloos M., Marchuk D.A. A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10) Am. J. Hum. Genet. 2002;71:1189–1194. doi: 10.1086/344210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klebe S., Lossos A., Stevanin G. KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: distinct phenotypes according to the nature of the mutations. Eur. J. Hum. Genet. 2012;20:645–649. doi: 10.1038/ejhg.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stokin G.B., Goldstein L.S.B. Linking molecular motors to Alzheimer’s disease. J. Physiol. Paris. 2006;99:193–200. doi: 10.1016/j.jphysparis.2005.12.085. [DOI] [PubMed] [Google Scholar]
- 20.Hirokawa N., Takemura R. Molecular motors in neuronal development, intracellular transport and diseases. Curr. Opin. Neurobiol. 2004;14:564–573. doi: 10.1016/j.conb.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Rath O., Kozielski F. Kinesins and cancer. Nat. Rev. Cancer. 2012;12:527–539. doi: 10.1038/nrc3310. [DOI] [PubMed] [Google Scholar]
- 22.Sakowicz R., Finer J.T., Wood K.W. Antitumor activity of a kinesin inhibitor. Cancer Res. 2004;64:3276–3280. doi: 10.1158/0008-5472.can-03-3839. [DOI] [PubMed] [Google Scholar]
- 23.Kashina A.S., Baskin R.J., Scholey J.M. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Endow S.A., Henikoff S., Soler-Niedziela L. Mediation of meiotic and early mitotic chromosome segregation in Drosophila by a protein related to kinesin. Nature. 1990;345:81–83. doi: 10.1038/345081a0. [DOI] [PubMed] [Google Scholar]
- 25.Chen G.Y., Mickolajczyk K.J., Hancock W.O. The kinesin-5 chemomechanical cycle is dominated by a two-heads-bound state. J. Biol. Chem. 2016;291:20283–20294. doi: 10.1074/jbc.M116.730697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soppina V., Norris S.R., Verhey K.J. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc. Natl. Acad. Sci. USA. 2014;111:5562–5567. doi: 10.1073/pnas.1400759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreasson J.O., Milic B., Block S.M. Examining kinesin processivity within a general gating framework. eLife. 2015;4:e07403. doi: 10.7554/eLife.07403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mickolajczyk K.J., Deffenbaugh N.C., Hancock W.O. Kinetics of nucleotide-dependent structural transitions in the kinesin-1 hydrolysis cycle. Proc. Natl. Acad. Sci. USA. 2015;112:E7186–E7193. doi: 10.1073/pnas.1517638112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortega-Arroyo J., Kukura P. Interferometric scattering microscopy (iSCAT): new frontiers in ultrafast and ultrasensitive optical microscopy. Phys. Chem. Chem. Phys. 2012;14:15625–15636. doi: 10.1039/c2cp41013c. [DOI] [PubMed] [Google Scholar]
- 30.Isojima H., Iino R., Tomishige M. Direct observation of intermediate states during the stepping motion of kinesin-1. Nat. Chem. Biol. 2016;12:290–297. doi: 10.1038/nchembio.2028. [DOI] [PubMed] [Google Scholar]
- 31.Andrecka J., Takagi Y., Kukura P. 1st Ed. Elsevier; Amsterdam, the Netherlands: 2016. Interferometric Scattering Microscopy for the Study of Molecular Motors, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hancock W.O., Howard J. Kinesin’s processivity results from mechanical and chemical coordination between the ATP hydrolysis cycles of the two motor domains. Proc. Natl. Acad. Sci. USA. 1999;96:13147–13152. doi: 10.1073/pnas.96.23.13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milic B., Andreasson J.O.L., Block S.M. Kinesin processivity is gated by phosphate release. Proc. Natl. Acad. Sci. USA. 2014;111:14136–14140. doi: 10.1073/pnas.1410943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen G.-Y., Arginteanu D.F.J., Hancock W.O. Processivity of the kinesin-2 KIF3A results from rear head gating and not front head gating. J. Biol. Chem. 2015;290:10274–10294. doi: 10.1074/jbc.M114.628032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoeprich G.J., Thompson A.R., Berger C.L. Kinesin’s neck-linker determines its ability to navigate obstacles on the microtubule surface. Biophys. J. 2014;106:1691–1700. doi: 10.1016/j.bpj.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shastry S., Hancock W.O. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr. Biol. 2010;20:939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shastry S., Hancock W.O. Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. USA. 2011;108:16253–16258. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muthukrishnan G., Zhang Y., Hancock W.O. The processivity of kinesin-2 motors suggests diminished front-head gating. Curr. Biol. 2009;19:442–447. doi: 10.1016/j.cub.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruhnow F., Zwicker D., Diez S. Tracking single particles and elongated filaments with nanometer precision. Biophys. J. 2011;100:2820–2828. doi: 10.1016/j.bpj.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo S.-K., Wang P.-Y., Xie P. A model of processive movement of dimeric kinesin. J. Theor. Biol. 2017;414:62–75. doi: 10.1016/j.jtbi.2016.11.023. [DOI] [PubMed] [Google Scholar]
- 41.Yildiz A., Forkey J.N., Selvin P.R. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 42.Yildiz A., Tomishige M., Selvin P.R. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 43.Yildiz A., Tomishige M., Vale R.D. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toprak E., Yildiz A., Selvin P.R. Why kinesin is so processive. Proc. Natl. Acad. Sci. USA. 2009;106:12717–12722. doi: 10.1073/pnas.0808396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dogan M.Y., Can S., Yildiz A. Kinesin’s front head is gated by the backward orientation of its neck linker. Cell Reports. 2015;10:1967–1973. doi: 10.1016/j.celrep.2015.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma Y.Z., Taylor E.W. Interacting head mechanism of microtubule-kinesin ATPase. J. Biol. Chem. 1997;272:724–730. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- 47.Brendza K.M., Sontag C.A., Gilbert S.P. A kinesin mutation that uncouples motor domains and desensitizes the γ-phosphate sensor. J. Biol. Chem. 2000;275:22187–22195. doi: 10.1074/jbc.M001124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hackney D.D. Pathway of ADP-stimulated ADP release and dissociation of tethered kinesin from microtubules. Implications for the extent of processivity. Biochemistry. 2002;41:4437–4446. doi: 10.1021/bi0159229. [DOI] [PubMed] [Google Scholar]
- 49.Hackney D.D., Stock M.F., Patterson R.A. Modulation of kinesin half-site ADP release and kinetic processivity by a spacer between the head groups. Biochemistry. 2003;42:12011–12018. doi: 10.1021/bi0349118. [DOI] [PubMed] [Google Scholar]
- 50.Clancy B.E., Behnke-Parks W.M., Block S.M. A universal pathway for kinesin stepping. Nat. Struct. Mol. Biol. 2011;18:1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alonso M.C., Drummond D.R., Cross R.A. An ATP gate controls tubulin binding by the tethered head of kinesin-1. Science. 2007;316:120–123. doi: 10.1126/science.1136985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorn K.S., Ubersax J.A., Vale R.D. Engineering the processive run length of the kinesin motor. J. Cell Biol. 2000;151:1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z., Sheetz M.P. The C-terminus of tubulin increases cytoplasmic dynein and kinesin processivity. Biophys. J. 2000;78:1955–1964. doi: 10.1016/S0006-3495(00)76743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lakämper S., Meyhöfer E. The E-hook of tubulin interacts with kinesin’s head to increase processivity and speed. Biophys. J. 2005;89:3223–3234. doi: 10.1529/biophysj.104.057505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okada Y., Hirokawa N. A processive single-headed motor: kinesin superfamily protein KIF1A. Science. 1999;283:1152–1157. doi: 10.1126/science.283.5405.1152. [DOI] [PubMed] [Google Scholar]
- 56.Chandra R., Endow S.A., Salmon E.D. An N-terminal truncation of the NCD motor protein supports diffusional movement of microtubules in motility assays. J. Cell Sci. 1993;104(Pt 3):899–906. doi: 10.1242/jcs.104.3.899. [DOI] [PubMed] [Google Scholar]
- 57.Hunter A.W., Caplow M., Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helenius J., Brouhard G., Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 59.Friel C.T., Howard J. The kinesin-13 MCAK has an unconventional ATPase cycle adapted for microtubule depolymerization. EMBO J. 2011;30:3928–3939. doi: 10.1038/emboj.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andreasson J.O.L., Shastry S., Block S.M. The mechanochemical cycle of mammalian kinesin-2 KIF3A/B under load. Curr. Biol. 2015;25:1166–1175. doi: 10.1016/j.cub.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nara I., Ishiwata S. Processivity of kinesin motility is enhanced on increasing temperature. Biophysics (Nagoya-shi) 2006;2:13–21. doi: 10.2142/biophysics.2.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y., Deffenbaugh N.C., Hancock W.O. Molecular counting by photobleaching in protein complexes with many subunits: best practices and application to the cellulose synthesis complex. Mol. Biol. Cell. 2014;25:3630–3642. doi: 10.1091/mbc.E14-06-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viterbi A. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans. Inf. Theory. 1967;13:260–269. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.